Two forms of NADH-cytochrome b5 reductase (b5R), an erythrocyte-restricted soluble form, active in methemoglobin reduction, and a ubiquitous membrane-associated form involved in lipid metabolism, are produced from one gene. In the rat, the two forms are generated from alternative transcripts differing in the first exon, however, biogenesis of human b5R was less understood. Recently, two different transcripts (M and S), differing in the first exon were also described in humans. Here, we have investigated the tissue-specificity and the role of the S-transcript in the generation of soluble b5R. By RNase protection assays designed to simultaneously detect alternative b5R transcripts in the same sample, the S transcript was undetectable in nonerythroid and in erythroleukemic K562 cells induced to differentiate, but was present in terminal erythroblast cultures, and represented a major b5R transcript in reticulocytes. Analysis of the translation products of the M- and S-transcripts in HeLa cells transfected with the corresponding cDNAs demonstrated that the S-transcript generates soluble b5R, presumably from an internal initiation codon. Our results indicate that the S-transcript is expressed at late stages of erythroid maturation to generate soluble b5R.
DEFICIENCY OF THE enzyme NADH-cytochrome b5 reductase (b5R) causes two types of hereditary methemoglobinemia: type I, in which the enzyme deficit is restricted to red blood cells and the symptoms of the disease are limited to mild cyanosis and type II, in which the deficit is generalized, with severe consequences for several organs, notably the central nervous system, and reduction of the life span of affected individuals who rarely reach adulthood.1 Not only does the enzyme deficit in the two types of methemoglobinemia have a different tissue distribution, but it also involves two, biochemically distinct forms of b5R: erythrocytes contain a soluble reductase, which together with a soluble form of cytochrome b5, constitutes the principal system responsible for methemoglobin (MetHb) reduction; in the other cells of the organism, b5R is bound to membranes via a hydrophobic, myristoylated N-terminus and is involved in many aspects of lipid metabolism, as well as in ascorbate regeneration.2-4 The catalytic domain of membrane-bound b5R is located in the cytosol and is identical to the soluble enzyme.5 Identification of point mutations responsible for type II, generalized, hereditary methemoglobinemia (in which both forms of the enzyme are lacking) has clearly demonstrated that soluble and membrane-bound b5R are products of the same gene.2 6-9
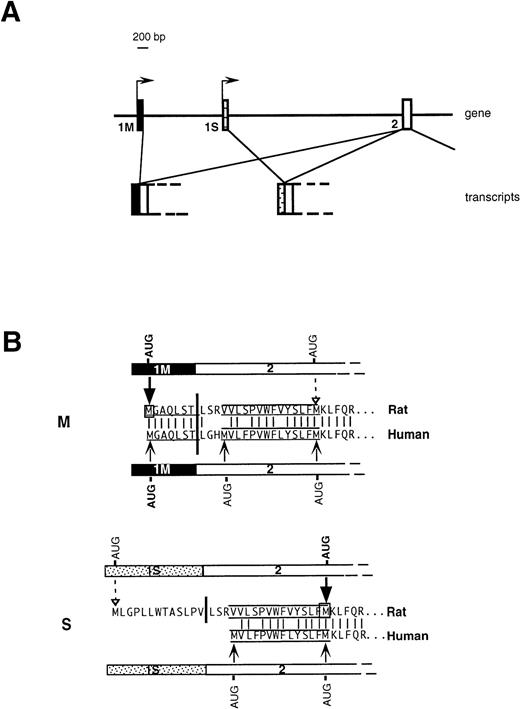
An important issue, relevant to the understanding of the molecular basis of hereditary methemoglobinemia, is the biogenetic relationship between soluble and membrane-bound b5R. We have previously addressed this question in the rat10 and demonstrated that the two enzyme forms are generated from two different mRNAs transcribed from alternative promoters (Fig 1A). A “housekeeping” promoter precedes the ubiquitously expressed first exon (1M). Transcription from this promoter generates the mRNA coding for the membrane-bound form. 1M contains the 5′ untranslated region followed by 21 nucleotides, which code for the N-terminal consensus sequence determining myristoylation (underlined sequence in Fig 1B). The membrane-anchoring domain of b5R contains an additional 14 uncharged residues, but these are encoded in the second exon (labeled 2 in Fig 1A), which is common in the two transcripts and which lies ≈7,000 bp downstream in the gene. Approximately 2,000 bp downstream to 1M is an exon (1S), which is preceded by a 5′ flanking region with characteristics of an erythroid promoter, and which is expressed specifically in erythroid precursors. Translation of the 1S-containing transcript generates soluble b5R by initiating from a downstream AUG located at the end of the sequence specifying the 14 uncharged residues of the membrane anchor (Fig 1B). Although this AUG lies within the common exon 2, it is not used in the 1M-containing mRNA. This is because the initiation codon of 1M, which lies in an optimal context11 arrests the scanning process, thus preventing the use of the downstream AUG. 1S itself contains an in frame AUG, however, this is in an unfavorable context for initiation and is used very inefficiently. We will refer to the 1M- and 1S-containing transcripts as M- and S-mRNA, respectively (rather than L- and R-mRNA, as in our previous work).10
Organization of 5′ region of b5R gene (A) and predicted translation products of rat and human transcripts (B). (A) In rats and humans alternative promoters generate two transcripts differing in the first exon (1M or 1S). (B) M and S show the 5′ portions of b5R M- and S-transcripts in humans and rat and the corresponding deduced amino acid sequences. The myristoylation consensus is underlined and the 14 uncharged residues, which complete the membrane anchor, are overlined and underlined. The heavy vertical bars indicate the junction between first and second exon. The light vertical lines show positions of amino acid identity between man and rat. Translation of the rat M transcript initiates exclusively from the first AUG contained in the 1M exon (heavy arrow), so that the internal AUG of exon 2 is not used (dashed arrow).10 Also the human 1M exon contains an AUG in an optimal context for initiation, however the possible use of the two downstream AUG codons has not been directly tested. The rat 1S exon contains an in-frame AUG in a poor context for initiation, which is used inefficiently (dashed arrow), so that the main translation product is from the downstream AUG (heavy arrow), with generation of soluble b5R.10 The human 1S exon is devoid of AUG codons, however, exon 2 contains an additional AUG at the beginning of the sequence coding for the hydrophobic stretch, which could in principle be used as initiation codon.
Organization of 5′ region of b5R gene (A) and predicted translation products of rat and human transcripts (B). (A) In rats and humans alternative promoters generate two transcripts differing in the first exon (1M or 1S). (B) M and S show the 5′ portions of b5R M- and S-transcripts in humans and rat and the corresponding deduced amino acid sequences. The myristoylation consensus is underlined and the 14 uncharged residues, which complete the membrane anchor, are overlined and underlined. The heavy vertical bars indicate the junction between first and second exon. The light vertical lines show positions of amino acid identity between man and rat. Translation of the rat M transcript initiates exclusively from the first AUG contained in the 1M exon (heavy arrow), so that the internal AUG of exon 2 is not used (dashed arrow).10 Also the human 1M exon contains an AUG in an optimal context for initiation, however the possible use of the two downstream AUG codons has not been directly tested. The rat 1S exon contains an in-frame AUG in a poor context for initiation, which is used inefficiently (dashed arrow), so that the main translation product is from the downstream AUG (heavy arrow), with generation of soluble b5R.10 The human 1S exon is devoid of AUG codons, however, exon 2 contains an additional AUG at the beginning of the sequence coding for the hydrophobic stretch, which could in principle be used as initiation codon.
Less information on the biogenesis of human soluble b5R is available. Although the entire gene, located on chromosome 22,12,13had been cloned and sequenced already some years ago, it was thought that the soluble form of the enzyme was generated from the membrane-bound form by posttranslational proteolysis occurring in erythrocyte precursors.14 Recently, however, Du et al15 identified in RNA from various human cell types a transcript containing an exon homologous to the rat 1S exon, which could be responsible for the generation of soluble b5R. Whereas the general organization of the 5′ portion of the b5R gene thus appears to be similar in humans and rats (Fig 1A), the human S transcript does present some interesting differences with respect to its rat counterpart: human 1S does not contain an in frame AUG, while human exon 2 contains, in addition to the AUG that serves as initiation codon for soluble b5R in the rat, an upstream AUG, which specifies a met residue at the beginning of the stretch of 14 uncharged residues (Fig 1B). This AUG, which is in a semifavorable context for initiation (G in position +4-),11 could arrest scanning, preventing use of the downstream initiation codon. In this case, a b5R with a short hydrophobic, nonmyristoylated, N-terminal extension would be generated (Fig 1B). Another difference is in the 5′ flanking sequences of 1S: in humans, this region appeared to lack typical features of erythroid promoters.16 Moreover, because Du et al15 detected the S-transcript by reverse transcription (RT) followed by polymerase chain reaction (PCR) in several human tissues, they concluded that its expression is not tissue-specific. Thus, the role of the recently identified human 1S exon in the genesis of erythrocyte soluble b5R remains to be clarified.
In the present study, we have investigated the tissue specificity of 1S expression in human cell lines and erythroid precursors by RNase protection assays and characterized its translation product(s) after transfection of the cDNA into a human cell line. The results of our experiments indicate that human S-RNA is expressed in a tissue-specific fashion, and that, like its rat counterpart, it generates the soluble form of b5R.
MATERIALS AND METHODS
Cell culture.
The human erythroleukemia cell line K562 and the human rhabdomyosarcoma cell line TE671 were grown in RPMI 1640 medium, supplemented with heat-inactivated 10% fetal calf serum (FCS) in a 5% CO2incubator. K562 cells were induced to differentiate by incubation in medium supplemented with 20 μmol/L hemin (Sigma Chemical Co, St Louis, MO) for 4 days. Induction of globin synthesis was assessed with the benzidine reaction. Benzidine positive cells were generally ≈70% of the total cell population, in comparison to 5% to 10% in untreated samples.
HeLa cells were grown in a 10% CO2 incubator in Dulbecco's modified Eagle's medium (DMEM), supplemented with 1 mmol/L sodium pyruvate and with 10% heat inactivated FCS.
Erythroid cell cultures derived from normal human peripheral blood were set up as described by Fibach et al,17 but 10 ng/mL stem cell factor (Genzyme, Cambridge, MA) were included in the erythropoietin-dependent second phase of the culture. Human recombinant erythropoietin was from Cylag (Sulzbach, Germany). After 19 to 20 days of culture, cells were harvested and counted. Cell viability was determined by Trypan blue exclusion; cellular morphology and Hb-containing cells were assessed on cytocentrifuge slides stained with May Grunwald's Giemsa and benzidine-HCl.18 The erythroblast RNA used for the experiments in Figs 2 and 3 of this report were from a terminal culture containing 72% of erythroid cells (of which ≈70% were orthochromatic erythroblasts and ≈30% were polychromatophilic cells), with the nonerythroid component due to leukocytes or undifferentiated cells.
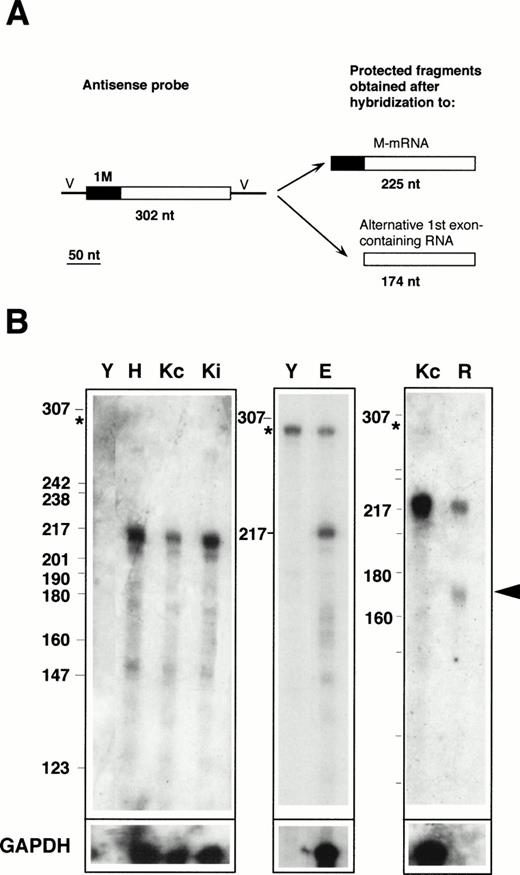
RNase protection with the 1M-containing probe. (A) Design of the experiment. The antisense probe contained the 1M exon (filled rectangle), the second, and part of the third exon common to all b5R transcripts (open rectangle), and portions of the vector sequence (single lines marked V). After hybridization to b5R mRNA, which contains 1M, the entire probe (except for the vector sequence) is expected to be protected from RNase digestion. In contrast, transcripts with an alternative first exon will only partially protect the probe, with generation of a smaller protected species. (B) Electrophoretic analysis of protected fragments. A total of 2 μg of poly A(+) RNA from HeLa cells (lane H) or from K562 cells induced (lane Ki) or not induced (lanes Kc) to differentiate, 17 μg of total RNA from erythroblasts (lane E), or 10 μg of total RNA from reticulocytes (lane R) or from torulla yeast (lane Y) were hybridized overnight with 40,000 dpm of 1M antisense probe at 43°C. One half of the same RNA samples (or one fifth in the case of erythroblast RNA) was hybridized to 40,000 dpm of GAPDH antisense probe (bottom row). Digestion was with 2.50 U/mL of RNase A and 100 U/mL of RNase T1 at 37°C for 1 hours. Numbers on the left of the panels indicate the size (in nucleotides) and position of Msp 1-digested pBR322 DNA marker fragments. The asterisk at the left of the panels indicates the position of the undigested probe. Some probe remained undigested in the samples shown in the middle panel. RNA from reticulocytes shows a clearly detectable partially protected probe fragment (arrowhead), predicted to be generated after hybridization to a b5R transcript with an alternative first exon.
RNase protection with the 1M-containing probe. (A) Design of the experiment. The antisense probe contained the 1M exon (filled rectangle), the second, and part of the third exon common to all b5R transcripts (open rectangle), and portions of the vector sequence (single lines marked V). After hybridization to b5R mRNA, which contains 1M, the entire probe (except for the vector sequence) is expected to be protected from RNase digestion. In contrast, transcripts with an alternative first exon will only partially protect the probe, with generation of a smaller protected species. (B) Electrophoretic analysis of protected fragments. A total of 2 μg of poly A(+) RNA from HeLa cells (lane H) or from K562 cells induced (lane Ki) or not induced (lanes Kc) to differentiate, 17 μg of total RNA from erythroblasts (lane E), or 10 μg of total RNA from reticulocytes (lane R) or from torulla yeast (lane Y) were hybridized overnight with 40,000 dpm of 1M antisense probe at 43°C. One half of the same RNA samples (or one fifth in the case of erythroblast RNA) was hybridized to 40,000 dpm of GAPDH antisense probe (bottom row). Digestion was with 2.50 U/mL of RNase A and 100 U/mL of RNase T1 at 37°C for 1 hours. Numbers on the left of the panels indicate the size (in nucleotides) and position of Msp 1-digested pBR322 DNA marker fragments. The asterisk at the left of the panels indicates the position of the undigested probe. Some probe remained undigested in the samples shown in the middle panel. RNA from reticulocytes shows a clearly detectable partially protected probe fragment (arrowhead), predicted to be generated after hybridization to a b5R transcript with an alternative first exon.
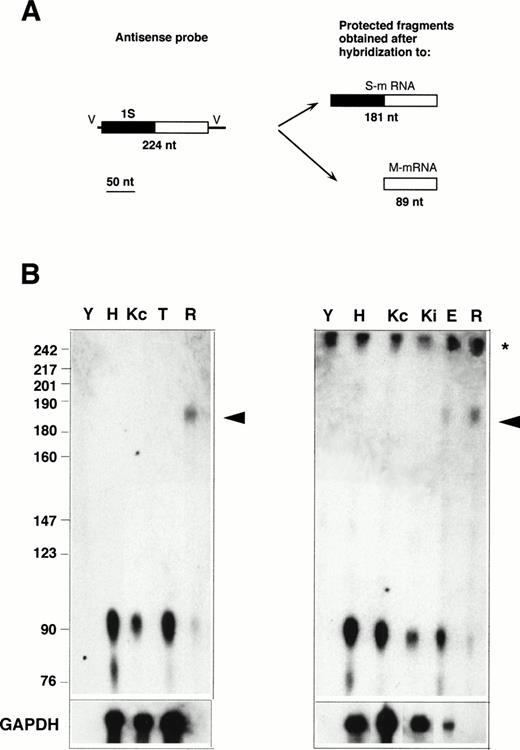
RNase protection with the 1S-containing probe. (A) Design of the experiment. The antisense probe contained the 1S exon (filled rectangle), part of the second exon common to all b5R transcripts (open rectangle), and portions of the vector sequence (single lines marked V). After hybridization to a b5R mRNA, which contains 1S, the entire probe (except for the vector sequence) is expected to be protected from RNase digestion. In contrast, transcripts with the 1M exon will only partially protect the probe, with generation of a smaller protected species. (B) Electrophoretic analysis of protected fragments. A total of 10 μg of total RNA from HeLa (lane H), TE672 (lane T), induced or not induced K562 cells (lanes Kc and Ki on right panel), reticulocytes (lane R) or torulla yeast (lane Y), 2 μg of polyA+ RNA from noninduced K562 cells (lane Kc on left panel), 26 μg of total RNA from primary erythroblast cultures (lane E) were hybridized overnight with 40,000 dpm of 1S antisense probe. One half (left panel) or 1 μg (right panel) of the same RNA samples was hybridized to GAPDH antisense probe (bottom row). Hybridization and digestion conditions were as in the legend to Fig 2. Numbers on the left indicate molecular weight markers as in Fig 2. The asterisk at the right of the panels indicates the position of the undigested probe. Some probe remained undigested in the samples of the right panel. The arrowhead indicates the position of the fully protected probe, due to the presence of S-transcript in reticulocyte and erythroblast RNA. The partially protected probe of 89 nt is visible in all samples.
RNase protection with the 1S-containing probe. (A) Design of the experiment. The antisense probe contained the 1S exon (filled rectangle), part of the second exon common to all b5R transcripts (open rectangle), and portions of the vector sequence (single lines marked V). After hybridization to a b5R mRNA, which contains 1S, the entire probe (except for the vector sequence) is expected to be protected from RNase digestion. In contrast, transcripts with the 1M exon will only partially protect the probe, with generation of a smaller protected species. (B) Electrophoretic analysis of protected fragments. A total of 10 μg of total RNA from HeLa (lane H), TE672 (lane T), induced or not induced K562 cells (lanes Kc and Ki on right panel), reticulocytes (lane R) or torulla yeast (lane Y), 2 μg of polyA+ RNA from noninduced K562 cells (lane Kc on left panel), 26 μg of total RNA from primary erythroblast cultures (lane E) were hybridized overnight with 40,000 dpm of 1S antisense probe. One half (left panel) or 1 μg (right panel) of the same RNA samples was hybridized to GAPDH antisense probe (bottom row). Hybridization and digestion conditions were as in the legend to Fig 2. Numbers on the left indicate molecular weight markers as in Fig 2. The asterisk at the right of the panels indicates the position of the undigested probe. Some probe remained undigested in the samples of the right panel. The arrowhead indicates the position of the fully protected probe, due to the presence of S-transcript in reticulocyte and erythroblast RNA. The partially protected probe of 89 nt is visible in all samples.
Reticulocytes.
Heparinized blood, obtained from anemic patients, was passed through a microcrystalline cellulose-α–cellulose (Sigma) column to remove leukocytes.19 The percentage of reticulocytes in the final preparation determined by staining with brilliant cresyl blue was ≈15%. Leukocytes were undetectable.
RNA extraction.
Total RNA was isolated from cultured cells using the Total RNA Fast ΙΙ Isolation Kit (Molecular Systems, San Diego, CA). Poly A+ RNA was obtained from the total RNA using the Oligotex mRNA Kit from Qiagen GmbH (Hilden, Germany). To extract total RNA from reticulocytes, the reticulocyte-enriched red blood cell preparation described above was homogenized with a solution containing 4 mol/L guanidinium thiocyanate, 25 mmol/L sodium citrate pH 7, 0.5% sarcosyl, 0.1 mol/L 2-mercaptoethanol (2 mL of solution per 109cells). The Chomczynski-Sacchi protocol20was followed until the first isopropanol precipitation. The resulting pellet was resuspended with the homogenization buffer of the Total RNA Fast ΙΙ Isolation Kit and processed further according to the instructions of the manufacturer. Approximately 50 μg total RNA/14 × 109 erythrocytes were obtained.
Cloning of the 5′ extremities of b5R encoding transcripts.
To obtain the 5′ extremities of M and S cDNAs, 250 ng of poly A+ RNA obtained from TE671 cells was subjected to RT-PCR using the First Strand Synthesis Kit from Stratagene (La Jolla, CA) and recombinant Moloney murine leukemia virus Reverse Transcriptase supplied by the kit. All amplification reactions were performed in a DNA thermal cycler from Perkin Elmer-Cetus (Norwalk, CT).
For the 5′ extremity of M cDNA, the following oligonucleotides were used (underlined nucleotides correspond to extra sequences containing restriction sites): upper primer 5′-GGGAATTCCGGCGACAGAGCGAGCG-3′ (nts -32 to -16 in the 5′ UT of 1M of the b5R gene sequence in Tomatsu et al14; lower primer: 5′-CCCTCTA GATGGCGGCAGGGCAAAGC-3′ contained in exon 3 (nts 17671 to 17655 in Tomatsu et al14). After 30 cycles (1 minute at 94°C, 1 minute at 60°C, and 2 minutes at 72°C), a fragment of 230 nucleotides was obtained as expected.
To obtain the 5′ extremity of S cDNA, two consecutive amplifications were required. For the first amplification, the upper primer was (underlined nucleotides as above): 5′-CCCAAGCT TAAAACATTGGAGCTTGG-3′, corresponding to nts 2189-2210 (or 2761-2782 by the numbering of Du et al15) in the 1S exon; the lower primer was the same one used to obtain M cDNA. After 30 cycles of amplification (1 minute at 94°C, 1 minute at 54°C, and 2 minutes at 72°C) and electrophoretic analysis of the samples, no fragment of the expected size (270 nts) could be detected by ethidium bromide staining. The entire PCR sample was loaded on a preparative agarose gel, the region corresponding to 200-300 nts was excised, and the DNA was eluted. All of the eluted material was subjected to a second round of amplification, in which the upper primer of the first extension was maintained, but a nested lower primer was used: 5′-CTTGATGTCGGGGTTCTCGAG-3′ corresponding to nts 12364-12344 in exon 214 and containing an internalXho 1 restriction site. At the end of this second amplification (performed under the same conditions as the first one), a 205-nt fragment was obtained as expected.
The PCR-generated M and S fragments were digested and subcloned into appropriate vectors, either to produce labeled antisense RNA probes or to reconstruct the entire M and S cDNAs from a b5R clone lacking the 5′ extremity (see below).
Antisense RNA probes and RNase protection experiments.
The 230-bp M fragment and the 205-bp S fragment were inserted, respectively, into the EcoR1-Xba 1 sites of pBluescript II KS (+) and into the Hind3-Sal 1 restriction sites of pGEM3. The plasmids, truncated at the Hind2 and Hind3 sites for M and S, respectively, were used to generate32P-cytidine triphosphate (CTP) antisense-labeled transcripts with T7 RNA polymerase. A 316-bp fragment of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cloned into the pTRI plasmid (Ambion Inc, Austin, TX) was used as control. The plasmid was linearized with Hind3 and generated a 404-bp fragment when transcribed with T7 RNA polymerase. Production of32P-CTP–labeled antisense transcripts, hybridization of the probes to the total or poly A+ RNA from human cells, and digestion with RNase A+T1 cocktail were performed with reagents of the Maxi Script and RPA 2 kits from Ambion Inc. Amounts of probe and RNA, as well as temperatures of hybridization, are specified in the figure legends. Protected probe fragments were analyzed on 6% sequencing gels. To provide DNA molecular size markers, Msp1-digested pBR322 DNA (New England Biolabs, Beverly, MA) was labeled with 32P-dCTP using the Klenow fragment of DNA polymerase I. Band intensities of the autoradiograms were determined with the NIH-Image program (version 1.55).
Transient transfection of HeLa cells.
The entire coding sequences of the M- and S-transcripts were reconstructed by splicing the fragments obtained by RT-PCR to an incomplete human reductase cDNA lacking the 5′ extremity and inserted into pUC13 (b5R-pUC13) kindly provided by Dr T. Yubisui (Kochi Medical School, Japan).5 The RT-PCR–generated M fragment (see above) was excised from Bluescript with Sal 1 and Xho 1 and subcloned into b5R-pUC13. The RT-PCR–generated S fragment was subcloned into the Hind3 andXho 1 sites of b5R-pUC13. The resulting M and S constructs were subcloned into the modified mammalian expression vector pCB6.21 22
HeLa cells were plated at ≈20% of confluence, in 100-mm Petri dishes or on 1.7 × 1.7 cm coverslips, and transfected 12 to 24 hours later by the calcium phosphate method,23 with pCB6 containing M- or S-cDNA (1 μg DNA/cm2 of culture dish). Thirty-six hours after transfection cells were either fixed or collected for biochemical analyses.
Immunofluorescence.
Transfected HeLa cells, grown on glass coverslips, were fixed in paraformaldehyde, permeabilized, and processed for immunofluorescence as described,22 using rabbit affinity-purified antirat b5R antibodies cross-reactive with the human enzyme, followed by a fluorescein-labeled antirabbit IgG (Jackson Immunoresearch, West Grove, PA) as secondary reagent. Preparations were observed under an Axioplan microscope equipped for epifluorescence (Zeiss, Oberkochen, Germany). Mitochondria were stained with 100 nmol/L Mitotracker CMX Rose from Molecular Probes (Eugene, OR) as described.22
Cell fractionation.
Cells grown on 100-mm Petri dishes were washed free of medium with cold phosphate-buffered saline (PBS) containing 5 mmol/L EDTA, incubated in the same buffer for 5 minutes on ice, detached with a rubber policeman, and collected by centrifugation. The pellet was resuspended with cold hypotonic buffer (1 mmol/L EDTA, 15 mmol/L KCl, 30 mmol/L NaCl, 1 mmol/L Tris-HCl, pH 7.5) containing 1 mmol/L phenylmethylsulfonyl fluoride plus the cocktail of protease inhibitors described in Holt and Hart.24. A total of 300 μL of buffer for every 107 cells was used. After 5 minutes, cells were ruptured by passage through a cell cracker with a 0.0007-in clearance. The volume of the homogenate was measured and the same volume of cold two times concentrated isotonic buffer (0.5 mol/L sucrose, 0.2 mmol/L EDTA, 2 mmol/L Tris HCl, pH 7.4) was added. After sedimentation of nuclei, (500g for 10 minutes at 4°C), the resulting postnuclear supernatant (PNS) was centrifuged at 150,000g for 50 minutes at 4°C in a TLA 100.3 rotor (Beckman Instruments, Inc, Palo Alto, CA) to obtain a membrane pellet and a supernatant (cytosol) fraction. The pellet was resuspended with a small volume of isotonic buffer containing protease inhibitors.
Biochemical assays.
Reduced and alkylated pellet and supernatant fractions were analyzed by SDS-PAGE (11% gels) followed by blotting onto nitrocellulose and radioimmunostaining as described.25 The affinity-purified polyclonal antirat b5R antibodies described above were used at 2.5 μg/mL, followed by 125I-protein A (Amersham Life Science Ltd, Buckinghamshire, UK) at 200,000 dpm/mL.
Protein was assayed by the method of Lowry et al.26 b5R enzyme activity in cell fractions was determined by the rate of reduction of the ferrihemoglobin-ferrocyanide complex (MetHb-FeCN) according to Hegesh et al,27 slightly modified as described.28 Triton X-100, at a final concentration of 0.1%, was routinely included in the assay, a condition that allows measurement of the activity of both the membrane-bound and the soluble enzyme.28 29
RESULTS
Presence of b5R mRNAs with alternative first exons in human cells determined by RNase protection assay.
Basing ourselves on the published sequence of the b5R gene,14,15 we constructed probes for the 1M exon (the first exon of M-mRNA, which specifies the membrane-bound form of b5R) and for the recently identified 1S exon (the first exon of S-mRNA, which is thought to specify the soluble form). The probes, prepared by RT-PCR from polyA+ RNA purified from a human rhabdomyosarcoma cell line (TE671), were designed to contain, in addition to the first alternative exon, part of the downstream sequence common to the two transcripts (Figs 2A and3A). The M probe was obtained with 30 cycles of amplification, while for the S probe, two consecutive 30-cycle amplifications were required to obtain a product visible by ethidium bromide staining (see Materials and Methods), indicating that the expression of the 1S exon is much lower than that of the 1M exon in these cells. The sequences of three clones for each PCR-generated fragment were determined and found to be identical to the published ones,14,15 except for two differences, probably due to polymorphic replacements: (1) a silent C to G transversion at position 12,274,14 in exon 2, at position 3 of a val codon; this difference was seen also in a similar fragment obtained from HeLa cell RNA; (2) a G-T transversion in exon 1S (5′ UT region) at position 2206 (Tomatsu et al14 or position 2778 of the sequence in Du et al15).
Labeled antisense RNA probes, transcribed from the RT-PCR–generated fragments, were used in protection assays performed on RNA prepared from various human cell lines, as well as from primary erythroid cultures and from reticulocytes. The results obtained with the 1M-containing probe are shown in Fig 2. The presence of M-mRNA in the RNA samples was predicted to entirely protect the probe, with generation of a 225-nt–labeled fragment after digestion of the portions of the probe transcribed from the cloning vector (Fig 2A). In contrast, the presence of b5R mRNA containing any alternative first exon would result in only partial protection of the probe, with generation of a 174-nt–labeled fragment. As can be seen in Fig 2B, RNA from Hela cells (lane H), from K562 cells induced (lane Ki) or not induced (lanes Kc) to differentiate, as well as from TE671 cells (not shown) yielded a major species with the expected size for the fully protected probe (migrating approximately at the position of the 217 nt marker fragment). Some minor lower molecular weight fragments were also visible, including a band at approximately 174 nt with an intensity less than 10% that of the major 225 nt species, as determined by scanning analysis. This band was slightly more intense in the primary erythroid sample (lane E), however, its significance was difficult to evaluate because of the background in the lanes and because of its variability (compare the Kc lanes of the left and right panels). In any case, in these RNA samples, obtained from erythroid as well as nonerythroid cells, the major b5R mRNA was of the M type. A different situation was found for reticulocyte RNA (lane R). In this case, in addition to the fully protected species, a fragment at the position expected for the partially protected probe could clearly be seen (arrowhead). By scanning analysis, the intensity of this band was 80% that of the completely protected species. Taking into account the smaller size of the partially protected probe, this result suggests that in reticulocytes, a b5R mRNA with an alternative first exon is present in roughly equal amounts to the ubiquitous M-mRNA. It should be noted that the absolute level of expression of both b5R transcripts in reticulocytes was considerably lower than in the other tested cells; moreover, GAPDH mRNA, present at high levels in the other cells, was undetectable in the reticulocyte RNA preparation (row marked GAPDH in Fig 2B).
To determine whether the alternative transcript detected in reticulocytes contains the 1S exon, RNase protection assays were performed with the 1S probe, which is predicted to generate a 181-nt–labeled fragment when fully protected, and an 89 nt fragment if partially protected by a b5R mRNA not containing the 1S exon (Fig 3A). As can be seen in Fig 3B, by this assay, the S transcript was detectable not only in reticulocyte RNA (lanes R), but now, without the background problem, also in RNA from the terminal primary cultures of erythroid cells (lane E). The much higher ratio of M- to S-transcript in the primary cultured erythroid sample compared with reticulocytes can be explained by the presence in the cultures of nonerythroid cells (see Materials and Methods), which could contribute most of the M-transcript detected in this sample. In contrast to the situation with reticulocyte and erythroblast RNA, even prolonged exposure of the Hela (H), TE61 (T), induced and noninduced K562 (Ki and Kc) lanes failed to show any fully protected probe, indicating that levels of S transcript in these cells are below the limits of detection of the RNase protection assay. Scanning analysis of the reticulocyte lanes showed that the band intensity of the partially protected probe was 44% of that of the fully protected fragment. Again, taking into account the different size of the fully and partially protected probe, this result indicates that the two b5R transcripts are present in approximately equal amounts in reticulocytes, in agreement with the data in Fig 2.
Analysis of translation products of M- and S-b5R mRNAs in transfected cells.
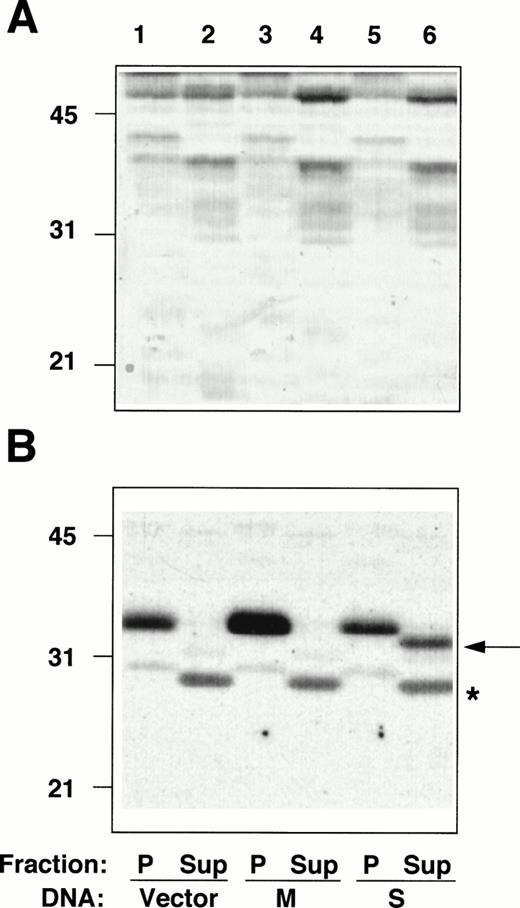
The entire coding sequences of M- and S-cDNAs, reconstructed by splicing the RT-PCR–generated fragments described above to an incomplete b5R cDNA clone, lacking the 5′ extremity,5were subcloned into a mammalian expression vector and transfected into HeLa cells. Membrane pellets (P), and cytosol fractions (Sup), prepared from the transfected cells, were analyzed by radioimmunoblotting (Fig 4). The amounts of protein of the pellet and supernatant fractions loaded on the gel were adjusted to correspond to the same number of cells. The pellet fraction of cells transfected with the vector alone (not containing a b5R insert) contained a polypeptide with an apparent Mr of ≈35 000, corresponding to b5R (calculated Mr = 34,232) (Fig 4B, lane 1), whereas no band at the position expected for soluble b5R (calculated Mr = 31,600) was detected in the cytosol fraction (Fig 4B, lane 2, Sup). This was true also for other cell lines that we analyzed (TE671 and K562 induced and not induced to differentiate, not shown), even with higher protein loads and prolonged exposure of the blots, consistently with the results of the RNase protection assays. A faint band comigrating with the membrane-bound form was detected after prolonged exposure (not shown), probably due to contamination of the cytosol fraction with small membrane fragments, which failed to sediment. In addition, the cytosol fraction from all cell lines analyzed contained a cross-reactive band at ≈28 kD (marked by asterisk in Fig 4B). The b5R band of the pellet fraction of cells transfected with M-cDNA was of increased intensity (Fig 4B, lane 3), while the cytosol fraction remained unaltered compared with the control (lane 4). Total protein load of the fractions of cells transfected with vector alone or with b5R-cDNA were roughly the same, as can be seen in Fig 4A. A striking result was obtained after transfection with S-cDNA: in this case, there was no increase in b5R in the pellet fraction, while a band at the position of the soluble enzyme (≈32 kD) appeared in the supernatant, representing about one half of the total b5R of the sample.
Radioimmunoblotting analysis of fractions from cells transfected with b5R cDNAs. Pellet (P) and supernatant (Sup) fractions prepared from PNS of Hela cells transfected with pCB6 (lanes 1 and 2, Vector), pCB6 containing M-cDNA (lanes 3 and 4, M) or S-cDNA (lanes 5 and 6, S) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by blotting onto nitrocellulose. (A) Shows the blot after staining with Ponceau S; (B) shows the autoradiogram of the same blot after radioimmunostaining with anti-b5R antibodies. A total of 55 μg protein of the pellet fractions (lanes 1, 3, and 5) and 57, 60, and 78 μg protein of the supernatant fractions from cells transfected with vector alone, M-, or S-cDNA, respectively, (lanes 2, 4, and 6) were loaded. Numbers on the left indicate positions and Mr (×10−3) of molecular mass markers. The arrow in (B) indicates the position of soluble b5R. The asterisk indicates the position of a cross-reactive band in the supernatant fraction, which remains unaltered regardless of the DNA used for the transfection.
Radioimmunoblotting analysis of fractions from cells transfected with b5R cDNAs. Pellet (P) and supernatant (Sup) fractions prepared from PNS of Hela cells transfected with pCB6 (lanes 1 and 2, Vector), pCB6 containing M-cDNA (lanes 3 and 4, M) or S-cDNA (lanes 5 and 6, S) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by blotting onto nitrocellulose. (A) Shows the blot after staining with Ponceau S; (B) shows the autoradiogram of the same blot after radioimmunostaining with anti-b5R antibodies. A total of 55 μg protein of the pellet fractions (lanes 1, 3, and 5) and 57, 60, and 78 μg protein of the supernatant fractions from cells transfected with vector alone, M-, or S-cDNA, respectively, (lanes 2, 4, and 6) were loaded. Numbers on the left indicate positions and Mr (×10−3) of molecular mass markers. The arrow in (B) indicates the position of soluble b5R. The asterisk indicates the position of a cross-reactive band in the supernatant fraction, which remains unaltered regardless of the DNA used for the transfection.
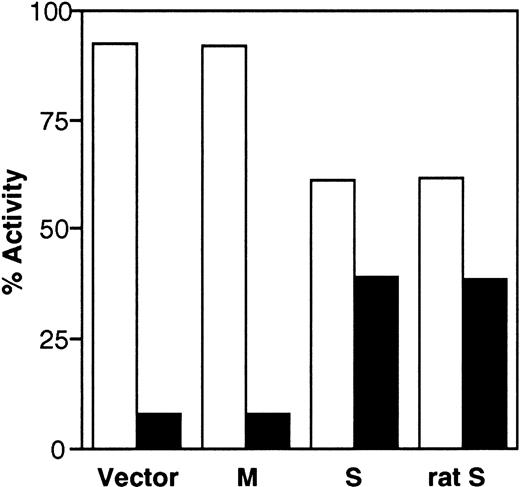
To investigate whether the transfected cDNAs generate enzymatically active b5R, we also determined enzyme activity in the cell fractions using an assay that detects the soluble, as well as the membrane-bound forms, of the enzyme.28,29 As can be seen in Fig 5, more than 90% of the enzyme activity of control cells was found in the pellet fraction. The small amount (≈8%) of activity assayed in the supernatant may be due to contaminating membrane fragments, as discussed above. The distribution of enzyme activity was unaffected by transfection of M-cDNA, whereas expression of S-cDNA resulted in a shift of the activity to the supernatant (39% of the total activity). Figure 5 also shows that rat S-cDNA, which is known to generate soluble b5R,10 when transfected into HeLa cells, yielded results indistinguishable from those obtained with the human clone (last set of columns in Fig 5). The results in Fig 5 are in good agreement with those obtained by radioimmunoblotting and demonstrate that the human S-cDNA generates enzymatically active soluble b5R.
Distribution of b5R enzyme activity between pellet (□) and supernatant (▪) fractions obtained from the PNS of cells transfected with pCB6 (Vector), M-cDNA (M), S-cDNA (S), or rat b5R S-cDNA (rat S). Enzyme activity in the fractions was assessed by the NADH-MetHb.FeCN reductase assay in the presence of detergent. Specific enzyme activities in the PNS fractions (nmol substrate reduced.min−1. mg protein) were: Vector, 49.9; M, 119; S, 55.2; rat S, 57.
Distribution of b5R enzyme activity between pellet (□) and supernatant (▪) fractions obtained from the PNS of cells transfected with pCB6 (Vector), M-cDNA (M), S-cDNA (S), or rat b5R S-cDNA (rat S). Enzyme activity in the fractions was assessed by the NADH-MetHb.FeCN reductase assay in the presence of detergent. Specific enzyme activities in the PNS fractions (nmol substrate reduced.min−1. mg protein) were: Vector, 49.9; M, 119; S, 55.2; rat S, 57.
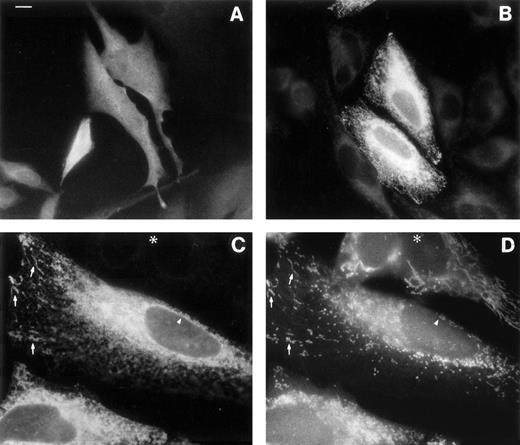
The rat M-mRNA generates a protein that associates both with endoplasmic reticulum (ER) and with mitochondrial outer membranes (MOM),30 and there is indirect evidence that this is the case also in man.2 We directly observed the subcellular localization of the products of b5R mRNAs by immunofluorescence analysis of the transfected HeLa cells (Fig6). Cells transfected with S-cDNA and stained with antireductase antibodies (at dilutions at which the endogenous b5R is undetectable) displayed uniform fluorescence throughout the cell, as expected for a soluble protein (Fig 6A). In contrast, cells expressing the M-cDNA product showed a reticular staining pattern (Fig 6B). The distribution of the membrane-bound form of b5R was compared with that of mitochondria in double-labeling experiments, using the mitochondrial fluorescent dye Mitotracker (Fig 6C and D). b5R was clearly present on mitochondria, as demonstrated by the coincident staining of antireductase antibodies and Mitotracker (arrows in Fig 6C and D show some of the most obvious identities). In addition, however, b5R showed a more widespread reticular staining and, importantly, was present also on the nuclear envelope (arrowhead in Fig 6C), indicating an ER localization. Thus, the human membrane-bound form of b5R localizes both to the ER and to MOM, like its rat homologue.
Immunofluorescence analysis of cells transfected with b5R S-cDNA (A) or M-cDNA (B through D). Transfection with S-cDNA results in the expression of a product distributed uniformly throughout the cell (A), while the product of M-cDNA is compartmentalized (B and C). In (B), cells expressing low levels of b5R (probably not transfected) are visible in the background. (C and D) Show the results of a double-labeling experiment, in which cells were labeled with the mitochondrial dye Mitotracker and with anti-b5R antibodies. The same field is shown viewed under the fluorescein filter for anti-b5R antibodies (C), or under the rhodamine filter for Mitotracker. The arrows point to some of the more striking identities between the two stains. The arrowhead indicates staining of the nuclear envelope by anti-b5R antibodies in (C) and absence of staining by Mitotracker in (D). The asterisk marks the position of an untransfected cell. Scale bar in (A) corresponds to 10 μm in (A) and (B) and to 6 μm in (C) and (D).
Immunofluorescence analysis of cells transfected with b5R S-cDNA (A) or M-cDNA (B through D). Transfection with S-cDNA results in the expression of a product distributed uniformly throughout the cell (A), while the product of M-cDNA is compartmentalized (B and C). In (B), cells expressing low levels of b5R (probably not transfected) are visible in the background. (C and D) Show the results of a double-labeling experiment, in which cells were labeled with the mitochondrial dye Mitotracker and with anti-b5R antibodies. The same field is shown viewed under the fluorescein filter for anti-b5R antibodies (C), or under the rhodamine filter for Mitotracker. The arrows point to some of the more striking identities between the two stains. The arrowhead indicates staining of the nuclear envelope by anti-b5R antibodies in (C) and absence of staining by Mitotracker in (D). The asterisk marks the position of an untransfected cell. Scale bar in (A) corresponds to 10 μm in (A) and (B) and to 6 μm in (C) and (D).
DISCUSSION
The molecular biology of b5R is of interest both because the gene offers an attractive model for the study of the regulation of alternative exon expression during erythroid maturation and because detailed knowledge of the biogenesis of the membrane-bound and soluble forms of b5R may contribute to the understanding and eventual treatment of hereditary methemoglobinemia caused by b5R deficiency. Although the mechanism of biogenesis of the two enzyme forms from one gene had been elucidated in the rat,10 less information was available in man. In the present study, we show that the basic features of the alternative promoter mechanism governing the production of the two enzyme forms in the rat are present in man, as well, although there are also some interesting differences between the two species.
Tissue specificity of the expression of alternative b5R exons in human cells.
In a recent report, Du et al15 identified an alternative first exon in the human b5R gene, called 1S, which was suggested to generate soluble b5R but which, because it was detected by RT-PCR in various nonerythroid cells, appeared not to be tissue-specific. Although we also could amplify S-cDNA from nonerythroid cells, our conclusion regarding the tissue specificity of exon 1S expression is different from that of Du et al.15 Using an RNase protection assay with antisense probes designed to simultaneously detect transcripts with alternative first exons in the same RNA sample, we found that the only cells expressing appreciable proportions of the S transcript were erythroid cells at late stages of maturation, ie, reticulocytes and terminally differentiated cultured erythroblasts. Indeed, with a 1S-containing antisense probe, the S-transcript was undetectable in nonerythroid cells, as well as in hemin-treated K562 cells, used as a model for erythroid cells at early stages of differentiation.31 Conversely, the use of a 1M-containing antisense probe demonstrated that in those same cells the major expressed transcript was of the M type. It must be noted that the presence of low concentrations of transcripts with alternative first exons different from 1S could not be ruled out from the experiments with the M probe because minor amounts of partially protected probe were difficult to evaluate over the background. In the rat, in addition to the 1M and 1S exons, two additional alternative first exons, apparently expressed ubiquitously at very low levels, have been described.32 We have not investigated whether these exons have counterparts in humans.
The discrepancy between the results with RT-PCR and RNase protection can, of course, be explained by the different sensitivities of the two methods. PCR can detect as little as 1 molecule of DNA, a sensitivity well above that of the RNase protection assay. Thus, although the 1S exon appears to be transcribed at very low levels in nonerythroid cells (detectable only by RT-PCR), its expression is upregulated specifically in developing red blood cells. It should be noted that “illegitimate” expression of various tissue-specific genes has been reported to occur at levels detectable by RT-PCR.33Thus, the presence of the S-transcript in nonerythroid cultured cells is of questionable biologic significance, considering also that these same cells do not have any soluble b5R detectable by radioimmunoblotting.
In addition to RT-PCR, Du et al15 also performed RNase protection experiments and detected low levels of the S-transcript in liver by this method. Their assay was not designed to compare the relative amounts of S and M transcript in the same sample, but because liver expresses high amounts of the membrane form of b5R,10 it is likely that the S transcript detected in liver represented a minor proportion of the total b5R transcripts. In rat liver, we previously also detected some S-transcript with a semiquantitative PCR assay, whereas none could be seen in skeletal muscle, heart, brain, or kidney.10 In that study, we suggested that the S-transcript detected in liver is due to the blood component and not to hepatocytes, however, it is also possible that hepatocytes do express slightly higher levels of the 1S exon than other nonerythroid cells.
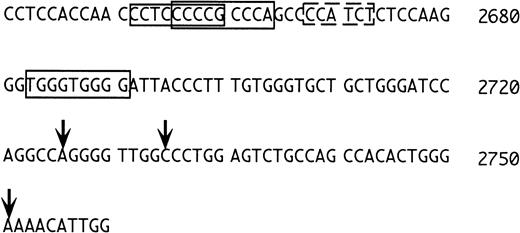
Because the 1S exon is expressed preferentially in erythroid cells, one would expect to find typical features of erythroid promoters in its 5′ flanking region. We were surprised that consensus sequences for the erythroid transcription factors NF-E2 and GATA-116are not present in this region, at variance with the situation in the rat.10 Closer inspection of the region (Fig 7) shows, however, that upstream to the transcriptional initiation sites determined by Du et al,15 there are three potential erythyroid Krüppel-like factor (EKLF) binding sites (Fig 7, regions boxed with continuous lines). EKLF is an erythroid cell-specific factor, which is required for expression of the β-globin gene, which binds the consensus CCNCNCCCN,34,35 and which interacts physically and synergistically with GATA-1.36 In this context, it is worth mentioning that a sequence with one mismatch to the consensus for GATA binding is present in between the two potential EKLF binding regions (dotted rectangle in Fig 7, AGATGG in reverse orientation, where the G in boldface replaces the A of the consensus). Binding of GATA-1 to a sequence not matching the consensus at position 5 has been reported,37thus, this sequence may also have functional significance. Clearly, further studies are required to understand the regulation of the 1S promoter.
5′ flanking region of the 1S exon. The numbering is as in Du et al15 and the vertical arrows mark the positions of the transcriptional initiation sites mapped by these investigators. The sequences boxed with the continuous lines correspond to putative EKLF binding sites. The sequence boxed by the dashed line is an inverted GATA consensus with one mismatch (see text).
5′ flanking region of the 1S exon. The numbering is as in Du et al15 and the vertical arrows mark the positions of the transcriptional initiation sites mapped by these investigators. The sequences boxed with the continuous lines correspond to putative EKLF binding sites. The sequence boxed by the dashed line is an inverted GATA consensus with one mismatch (see text).
S- and M-b5R mRNAs generate the soluble and membrane-bound forms of the enzyme, respectively.
Although the S-transcript was suggested to generate soluble b5R,15 until now, the translation products of the human M-and S-transcripts had not been directly characterized. The results of the present study demonstrate unequivocally that the M and S-transcripts, when expressed in HeLa cells, generate the membrane-bound and soluble forms of b5R, respectively. Considering also the results of RNase protection assays and of radioimmunoblotting experiments, which detected no S-transcript and soluble enzyme in nonerythroid cells, the S-transcript appears to be both necessary and sufficient for the generation of soluble b5R.
In the rat, the myristoylated product of the M-transcript specifically associates with two subcellular compartments: MOM and ER membranes.30 Immunofluorescence analysis of HeLa cells transfected with the human M-transcript showed that also the human enzyme is capable of associating with the ER and with mitochondria (presumably with MOM). One function of MOM-associated b5R is in ascorbate free radical reduction,2 3 but it probably has other roles, which remain to be investigated. The persistence of the M-transcript in reticulocytes may be related to the requirement for continued synthesis of MOM-associated b5R also at late stages of erythroid maturation.
The generation of the soluble form of b5R requires some further discussion. As explained earlier (Fig 1), in the rat, exon 1S has an in-frame initiation codon, which is used inefficiently. “Leaky scanning”11 allows use of a downstream AUG, contained in the second exon, with generation of soluble b5R.10 In humans, the 1S exon does not contain an initiation codon, however, the first amino acid of the stretch of 14 uncharged amino acids encoded in exon 2 is a methionine (instead of valine in the rat, see Fig 1). Therefore, also in humans, the potential initiation codon for soluble b5R is preceded by an upstream AUG. The results of this study strongly suggest that this upstream AUG allows leaky scanning, resulting in the use of the downstream AUG to produce soluble b5R. Initiation from the upstream AUG should have resulted in a protein capable of associating with the phospholipid bilayer through its N-terminal hydrophobic domain. Indeed, the additional hydrophobicity conferred by myristoylation is not required for association of b5R with artificial or natural membranes.30 38 Another hypothesis is that the soluble b5R in the transfected cells was generated by proteolysis of the translation product initiating from the upstream AUG. This possibility is very unlikely, because the product of the M transcript would contain the same hypothetical cleavage site at the end of the hydrophobic domain, yet it generated no detectable soluble b5R.
The first AUG of the human S transcript, although not preceded by a purine in position -3, is followed by a G in position +4, a context often found for AUGs, which function well as initiation codons.11 A recent study, however, shows that the enhancing effect of G at +4 is strongly diminished by a U in position +5.39 This is exactly the context of the first AUG of the human S-transcript and probably explains why it is used inefficiently.
In conclusion, although the position of the first AUG is shifted in man, the basic features of the S transcript described for the rat, ie, tissue specificity and generation of soluble b5R from an internal initiation codon, appear to be conserved. Whereas the importance of soluble b5R in MetHb reduction is well established, the biologic significance of the conserved and unusual features of its mode of production in erythrocyte precursors remains to be investigated.
ACKNOWLEDGMENT
We are grateful to T. Yubisui for his gift of a human b5R cDNA clone, to K. Shirabe for methodologic advice and for providing us with unpublished data, as well as with the entire human b5R gene sequence. We thank E. Battaglioli for helpful suggestions, S. Ottolenghi for fruitful discussion, and D. Fornasari and F. Clementi for critically reading the manuscript.
Supported by Grant No. E081 from Telethon, Rome, Italy (to N.B.).
Address reprint requests to Nica Borgese, PhD, C.N.R. Center for Cellular and Molecular Pharmacology, via Vanvitelli 32, 20129 Milano, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal