Abstract
The translocation t(8;14)(q24;q32) is the characteristic chromosomal aberration of Burkitt's-type lymphomas and leukemias (BLs). On the molecular level, the t(8;14) juxtaposes the c-myc gene in 8q24 next to the IgH locus in 14q32, resulting in overexpression of the transcription factor c-Myc. The detection of a t(8;14) is a major aim in the diagnostic process of all patients with high-grade B-cell lymphomas because treatment strategies differ between BL and other high-grade lymphomas. As chromosome analyses are sometimes hampered by the low yield or poor quality of metaphase spreads and as the application of molecular genetic techniques is limited by the distribution of the 8q24 breakpoints over a region of about some hundred kilobases, we set out to establish an interphase fluorescence in situ hybridization (FISH) assay for the detection of the t(8;14). A cosmid probe hybridizing to the IgH constant region in 14q32 was combined with a differently labeled probe of pooled cosmid clones spanning the c-myc locus in 8q24. Interphase nuclei lacking a t(8;14) show two separated signals corresponding to each probe, whereas interphase nuclei carrying a t(8;14) display a split of the c-myc probe and a colocalization of at least one of the splitted signals with the IgH probe. Based on the results of extensive control studies, the cutoff level for this stringent (type I) criteria was set at 2%. Additionally, colocalization of at least one c-myc signal with one IgH signal alone (without signal split for the c-myc probe) was used as a less stringent (type II) criteria with a cutoff limit of 11%. Nine BLs and one Burkitt-like lymphoma were investigated by this approach. Cytogenetically, all tumors contained a translocation t(8;14)(q24;q32) except for one BL, in which cytogenetic analysis had failed. In interphase FISH, all lymphomas and leukemias met the less stringent criteria for the diagnosis of the t(8;14). Additionally, in all tumors but the Burkitt-like lymphoma, a t(8;14) could be diagnosed according to the stringent criteria. The percentage of cells found to harbor the t(8;14) by FISH ranged from 4.3% to 100%. Comparison of cytogenetic and FISH results revealed a significantly lower percentage of t(8;14)+ interphase nuclei than metaphase cells (P = .004). In conclusion, the described FISH assay provides a feasible and sensitive tool for the routine detection of the translocation t(8;14) in interphase cells which might also offer new insights into the biology of high-grade B-cell lymphomas.
THE BURKITT translocation t(8;14)(q24;q32) was the first recurring chromosomal translocation shown to be associated with a lymphoproliferative disease.1Meanwhile, the pathogenetic role of the classical Burkitt translocation t(8;14) and its variants t(8;22)(q24;q11) and t(2;8)(p11;q24) in the tumorigenesis of Burkitt's lymphomas and its leukemic subtype, ie, acute lymphoblastic leukemia (ALL) FAB L3, has been well established.2-5 By cytogenetics, the translocation t(8;14) can be observed in 44% to 100% of Burkitt's lymphomas/leukemias (BLs), in up to 15% of other intermediate to high-grade B-cell lymphomas, and sporadically in low-grade B-cell lymphomas.2,3,6 7
On the molecular level, the t(8;14) juxtaposes the c-myc gene in chromosome region 8q24 next to the IgH locus in chromosome region 14q32, leading to deregulation of the transcription factor c-Myc. Although the t(8;14) translocation generally results in the overexpression of c-Myc protein, the location of the breakpoints within the c-myc gene region in 8q24 can vary greatly from patient to patient. Therefore, the t(8;14) translocations have been classified according to the position of the chromosomal breakpoints relative to the c-myc gene. Translocations with breakpoints in the first exon or intron of c-myc have been designated as class I, those with breakpoints immediately upstream of the gene as class II, and those with breakpoints distant as class III. In sporadic BL, class I (and II) translocations are predominant, whereas in endemic African cases, class III translocations with breakpoints dispersed over about 300 kb upstream of the gene are most frequent.8-12 On chromosome 14, the t(8;14) breakpoints are scattered between the variable and the constant part of the IgH locus, either 5′ of the intron enhancer in a joining or diversity segment, or 3′ of the intron enhancer in the μ switch region.10-14
As to the diagnostic and clinical relevance of the classical Burkitt translocation, the diagnosis of t(8;14) is an important aim in the management of patients with BL.6 So far, chromosomal analysis, Southern blot analysis, and polymerase chain reaction (PCR) analysis have been applied for the detection of a t(8;14). However, all these methods implicate technical limitations in the detection of this translocation. Cytogenetic analysis is hampered in about 10% to 20% of the specimens by a low mitotic index or a poor quality of metaphase spreads.15 The scattering of breakpoints in 8q24 and 14q32 and therefore the necessity of multiple probes and sequence specific primers render Southern blot analysis and PCR too time-consuming and unreliable for routine diagnosis. Fluorescence in situ hybridization (FISH) not only overcomes the limitations of cytogenetics and molecular genetics, but also provides a rapid and easy-to-handle technique for the detection of chromosomal abnormalities independent of the cycle status of cells. So far, the value of FISH for the detection of a t(8;14) translocation in interphase cells of BL has only been demonstrated in a small number of cell lines.11,16-18 No systematic data on the application of this method on primary tumor specimen have been published yet. To investigate the feasibility and sensitivity of FISH as a routinely applicant tool for the detection of a t(8;14), we established a FISH assay based on the approach of Joos et al11 using a cosmid pool spanning the breakpoint region in 8q24 and another cosmid probe flanking the centromeric border of the breakpoint region in the IgH locus in 14q32. Our results indicate that two-color FISH provides a highly sensitive tool for the detection of the classical Burkitt translocation in interphase nuclei of primary lymphoma/leukemia specimen.
MATERIALS AND METHODS
Patients
Samples from 10 patients ascertained during the last 8 years were analyzed by FISH for the presence of a translocation t(8;14). This included all patients in which cytogenetic analysis of a lymphoma or leukemia sample performed at our institution showed a t(8;14) and in which suitable cells for interphase FISH were still available. Additionally, a case of ALL-FAB L3 was included in the study, in which cytogenetic preparation failed because of bad metaphase quality (Table1). Biopsy specimens of all patients were obtained from affected tissues at the time of diagnosis. Mononuclear cells from six healthy individuals with a normal karyotype served as negative controls.
Clinico-Pathologic and Cytogenetic Features of the Lymphomas/Leukemias Investigated for the t(8;14) by FISH
| Case . | Sex . | Age (yr) . | Diagnosis . | Specimen Source . | Karyotype . | Percentage of t(8;14)+ Cells . | ||
|---|---|---|---|---|---|---|---|---|
| Cytogenetics . | FISH (Type I) . | FISH (Type II) . | ||||||
| BL1 | M | 28 | Burkitt NHL | Prostate | 46,XY,der(1)dup(1)(q42q21)add(1)(q21), t(8;14)(q24;q32)[18] | 100 | 100.0 | 100.0 |
| BL2 | M | 21 | Burkitt NHL | Bone marrow | 46,XY,t(8;14)(q24;q32)[6]/46,XY[15] | 29 | 36.5 | 49.5 |
| BL3 | M | 66 | Burkitt NHL | Bone marrow | 46,XY,t(8;14)(q24;q32),der(13)t(4;13) (q31;q34)[20]/47,idem,+12[4]/46,XY[2] | 92 | 15.5 | 37.5 |
| BL4 | M | 9 | Burkitt NHL | Peritoneal effusion | 46,XY,t(8;14)(q24;q32),add(13)(q24)[17] | 100 | 42.0 | 72.0 |
| BL5 | M | 50 | Burkitt NHL | Peritoneal effusion | 46,XY,dup(1)(q21q41),t(8;14)(q24;q32)[14] | 100 | 43.0 | 63.0 |
| BL6 | M | 12 | Burkitt NHL | Pleural effusion | 47,XY,t(8;14)(q24;q32),+12[2] | 100 | 33.3 | 88.0 |
| BL7 | M | 23 | Burkitt NHL | Lymph node | 46,inv(X)(p22q2?5),Y,t(8;14)(q24;q32)[18] | 100 | 4.3 | 14.3 |
| BL8 | M | 24 | Burkitt-like NHL | Lymph node | 51,XY,+5,+7,t(8;14)(q24;q32),+15, +19,+20[5]/46,XY[19] | 21 | 0.5 | 11.5 |
| BL9 | F | 76 | ALL-FAB L3 | Bone marrow | 46,XX,del(6)(q16q24),t(8;14)(q24;q32)[21]/46,idem,dup(1)(q32q22)[1] | 100 | 5.0 | 20.0 |
| BL10 | M | 9 | ALL-FAB L3 | Bone marrow | Failure | Failure | 54.5 | 58.1 |
| Case . | Sex . | Age (yr) . | Diagnosis . | Specimen Source . | Karyotype . | Percentage of t(8;14)+ Cells . | ||
|---|---|---|---|---|---|---|---|---|
| Cytogenetics . | FISH (Type I) . | FISH (Type II) . | ||||||
| BL1 | M | 28 | Burkitt NHL | Prostate | 46,XY,der(1)dup(1)(q42q21)add(1)(q21), t(8;14)(q24;q32)[18] | 100 | 100.0 | 100.0 |
| BL2 | M | 21 | Burkitt NHL | Bone marrow | 46,XY,t(8;14)(q24;q32)[6]/46,XY[15] | 29 | 36.5 | 49.5 |
| BL3 | M | 66 | Burkitt NHL | Bone marrow | 46,XY,t(8;14)(q24;q32),der(13)t(4;13) (q31;q34)[20]/47,idem,+12[4]/46,XY[2] | 92 | 15.5 | 37.5 |
| BL4 | M | 9 | Burkitt NHL | Peritoneal effusion | 46,XY,t(8;14)(q24;q32),add(13)(q24)[17] | 100 | 42.0 | 72.0 |
| BL5 | M | 50 | Burkitt NHL | Peritoneal effusion | 46,XY,dup(1)(q21q41),t(8;14)(q24;q32)[14] | 100 | 43.0 | 63.0 |
| BL6 | M | 12 | Burkitt NHL | Pleural effusion | 47,XY,t(8;14)(q24;q32),+12[2] | 100 | 33.3 | 88.0 |
| BL7 | M | 23 | Burkitt NHL | Lymph node | 46,inv(X)(p22q2?5),Y,t(8;14)(q24;q32)[18] | 100 | 4.3 | 14.3 |
| BL8 | M | 24 | Burkitt-like NHL | Lymph node | 51,XY,+5,+7,t(8;14)(q24;q32),+15, +19,+20[5]/46,XY[19] | 21 | 0.5 | 11.5 |
| BL9 | F | 76 | ALL-FAB L3 | Bone marrow | 46,XX,del(6)(q16q24),t(8;14)(q24;q32)[21]/46,idem,dup(1)(q32q22)[1] | 100 | 5.0 | 20.0 |
| BL10 | M | 9 | ALL-FAB L3 | Bone marrow | Failure | Failure | 54.5 | 58.1 |
The Burkitt translocation is indicated in bold face.
Abbreviations: M, male; F, female.
Preparations of Metaphase and Interphase Cells
Metaphase spreads and interphase nuclei of tumor cells were prepared from short-term cultures of bone marrow (3 cases), lymph nodes (2 cases), ascites (2 cases), pleural effusion (1 case), and prostate tissue (1 case) (Table 1). For chromosome banding, a fluorescence R-banding technique was applied. In the sample in which cytogenetic preparation failed, FISH was performed on an unstained bone marrow smear.
Probes
As probe for the IgH locus in chromosome region 14q32, the cosmid COS-cα1 spanning the IgH cα1 gene was used.11 To generate a probe spanning the entire c-myc breakpoint region, the cosmid clones COS-H4.1, COS-P380J9, and COS-MYC72 were pooled (Fig1A).11 The COS-cα1 probe was labeled by nick translation using digoxigenin-11-dUTP as described recently.19 The c-myc pool probe was labeled with biotin-16-dUTP by random primed labeling according to the manufacturer's instructions (Bioprime DNA labeling system; GIBCO-BRL, Life Technologies, Eggenstein, Germany).
(A) Schematic representation of the breakpoint regions in 8q24 and 14q32 and location of the respective FISH probes. (B) Ideogram representing the expected localization of signals by two-color FISH in t(8;14)− and + metaphase and interphase cells. Red color: C-myc-pool probe spanning the breakpoint region in 8q24; Green color: COS-cα1 hybridizing to the cα1-gene of the IgH locus in 14q32. T(8;14)+ interphase displaying the type I signal constellation show a split of one red (c-myc) signal and colocalization of one green and one red signal. Cells meeting type II criteria show a colocalization of one green and one red signal irrespective of a split of the c-myc signal.
(A) Schematic representation of the breakpoint regions in 8q24 and 14q32 and location of the respective FISH probes. (B) Ideogram representing the expected localization of signals by two-color FISH in t(8;14)− and + metaphase and interphase cells. Red color: C-myc-pool probe spanning the breakpoint region in 8q24; Green color: COS-cα1 hybridizing to the cα1-gene of the IgH locus in 14q32. T(8;14)+ interphase displaying the type I signal constellation show a split of one red (c-myc) signal and colocalization of one green and one red signal. Cells meeting type II criteria show a colocalization of one green and one red signal irrespective of a split of the c-myc signal.
FISH
FISH was performed on cells stored in Carnoy's fixative for up to 8 years as described recently.19 20 In case BL10, freshly prepared bone marrow smears were used. Briefly, 100 ng of each of the two differentially labeled DNA probes and 100 μg of unlabeled Cot1-DNA were coprecipitated and resolved in 10 μL of 50% formamide, 10% dextrane sulfate, and 2× SSC (pH 7.5). We placed 1.5 μL of this hybridization solution on the cell-containing area of the slides, covered it with a round 10-mm coverslip, and sealed it with rubber cement. The slides were denatured for 7 minutes at 75°C and hybridized overnight at 37°C. Posthybridization washes were performed three times in 0.1× SSC at 60°C for 5 minutes. The digoxigenin-labeled COS-cα1 probe was detected with a cascade using mouse anti-dogoxigenin antibody, digoxigenin-conjugated sheep anti-mouse antibody, and FITC-conjugated sheep anti-digoxigenin antibody (green fluorochrome; Boehringer, Mannheim, Germany). The biotin-labeled probe hybridizing to the c-myc breakpoint region was visualized by a cascade using Cy3-conjugated avidin (red fluorochrome; Jackson Immunoresearch Laboratories, West Grove, PA), biotinylated mouse anti-avidin antibody (Vector, Burlingame, CA), and again Cy3-conjugated avidin. Nuclei were counterstained with 4,6-diaminido-2-phenylindole dihydrochloride (DAPI).
Evaluation
Hybridization signals were analyzed by use of a Zeiss Axiophot fluorescence microscope (Zeiss, Oberkochen, Germany), with appropriate filtersets (Zeiss Nos. 02, 09, and 00), and documented using the ISIS imaging system (MetaSystems, Sandhausen, Germany). FISH experiments on lymphoma and control specimens were evaluated in blind fashion. In the mean, 185 interphase nuclei were investigated in each case.
RESULTS
Clinical Data and Cytogenetics
Of the nine patients in which cytogenetics showed a t(8;14)(q24;q32), 7 were diagnosed with sporadic Burkitt's type lymphoma, 1 with Burkitt-like non-Hodgkin's lymphoma (NHL), and 1 with ALL-FAB L3. The case in which cytogenetics failed was also diagnosed with ALL-FAB L3. In the two latter cases, the bone marrow was infiltrated by 70% and 95% of blasts displaying L3 morphology, respectively. Nine of the patients were male; 1 was female. The median age at diagnosis was 23.5 years (range, 9 to 76). In addition to a t(8;14), 3 samples carried a duplication of part of the long arm of chromosome 1. Clinical data and complete karytotypes are shown in Table 1.
FISH Analyses
Evaluation criteria.
As to the location of probes used in our assay, a translocation t(8;14) should lead to breakage within the region spanned by the c-myc pool probe and juxtapositioning of part of this probe to the IgH probe (Fig1). Nevertheless, dependent on the exact location of the breakpoint in 8q24, a split of the signal derived from the c-myc probe may be hard to detect in interphase cells because of the small size of one of the split signals. Therefore, we distinguished two types of hybridization patterns indicating a translocation t(8;14) in our experiments. The more stringent type I hybridization pattern was defined by (1) the presence of a split of one c-myc signal resulting in a total of at least three red signals, and (2) at least one colocalization of a red c-myc signal with a green signal of the IgH probe (Figs 1B and 2). The less stringent type II hybridization pattern was matched by each cell displaying at least one colocalization of a red c-myc pool signal and a green IgH probe signal (Fig 1B).
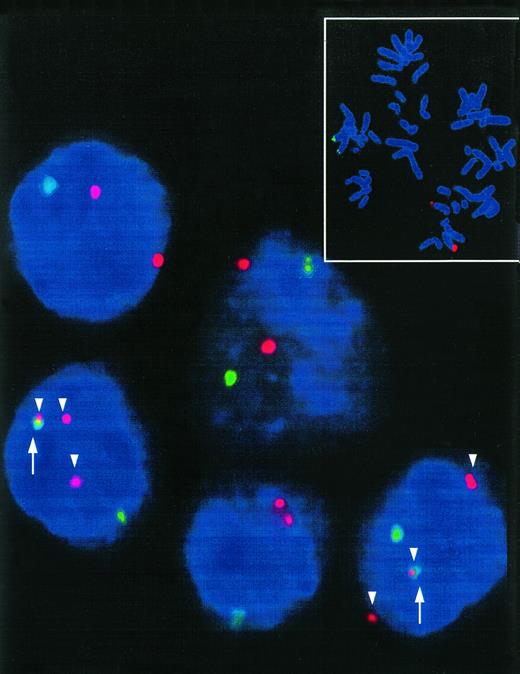
Interphase nuclei of a t(8;14)+ Burkitt's type ALL. Two interphase nuclei meeting the type I hybridization pattern show three red signals (c-myc-split, arrowhead) and colocalization (arrow) of one of these signals with a green COS-cα1 signal indicative for a t(8;14). Small picture: t(8;14)− control metaphase showing the location of the FISH probes. Distinct red (c-myc) and green (IgH) signals can be detected in the region 8q24 and 14q32, respectively.
Interphase nuclei of a t(8;14)+ Burkitt's type ALL. Two interphase nuclei meeting the type I hybridization pattern show three red signals (c-myc-split, arrowhead) and colocalization (arrow) of one of these signals with a green COS-cα1 signal indicative for a t(8;14). Small picture: t(8;14)− control metaphase showing the location of the FISH probes. Distinct red (c-myc) and green (IgH) signals can be detected in the region 8q24 and 14q32, respectively.
Controls.
To define the diagnostic thresholds of our assay for the detection of a t(8;14), 1,490 interphase nuclei prepared from six individuals were investigated. In these controls, 0% to 1.3% (mean, 0.3%; SD, 0.5%) of nuclei matched the more stringent type I hybridization pattern and 4% to 8.8% (mean, 5.6%; SD, 1.7%) of the nuclei matched the less stringent type II hybridization pattern. Analogous with previous studies,19 a t(8;14) was diagnosed if specimens exhibited highly significant percentages of cells with a distinct hybridization pattern. Therefore, the cutoff levels (mean + 3SD) for type I and type II hybridization patterns were set 2% and 11%, respectively (Fig 3).
Percentages of interphase nuclei meeting type I (grey) and type II (black) criteria for the detection of a t(8;14) by FISH in six controls (C1-C6) and 10 lymphoma/leukemia samples (BL1-BL10). The cutoff levels for type I (dotted line) and type II (black line) criteria were obtained from the statistical evaluation of the controls (mean percentage of false-positive nuclei +3SD).
Percentages of interphase nuclei meeting type I (grey) and type II (black) criteria for the detection of a t(8;14) by FISH in six controls (C1-C6) and 10 lymphoma/leukemia samples (BL1-BL10). The cutoff levels for type I (dotted line) and type II (black line) criteria were obtained from the statistical evaluation of the controls (mean percentage of false-positive nuclei +3SD).
Lymphoma and leukemia samples.
In all 10 patients investigated, a significant percentage of nuclei met the less stringent type II criteria. Thus, by use of the described interphase FISH assay, it was possible to diagnose the classical Burkitt translocation in all cases which were shown to carry a t(8;14) by chromosome analysis and in a sample of ALL-FAB L3, in which cytogenetic preparation failed. As shown in Fig 3, the percentage of cells displaying type II hybridization pattern ranged from 11.5% to 100% (mean, 51.4%; SD, 30.7%).
All samples except for the Burkitt-like lymphoma specimen also contained a significant proportion of cells displaying the stringent type I hybridization pattern. The percentage of cells meeting type I criteria varied between 4.3% and 100% (mean, 37.9%; SD, 29.4%) in the classical BL samples analyzed (Table 1, Fig 3).
Comparison of percentages of aberrant metaphases and interphases.
In the nine cases in which cytogenetic analysis was succesful, the mean percentage of metaphases carrying a t(8;14) was 82.4% (SD 32.7%). The mean percentage of interphase cells displaying type I and type II hybridization patterns as shown by FISH in these cases was 31.1% (SD 30.9%) and 50.6% (SD 32.4), respectively. Thus, even though the background rates of interphase cells carrying type I or II signal pattern by chance were not considered, there was a significantly lower percentage of type I or II positive interphase nuclei than t(8;14)+ metaphase cells (P = .004 andP = .03, respectively).
DISCUSSION
In the present study, we investigated the feasibility and sensitivity of two-color interphase FISH as a routine tool for the detection of the translocation t(8;14) in BLs. The assay described uses pooled cosmid clones covering the whole c-myc breakpoint region in 8q24 and a cosmid probe mapping proximal to the joining region of the IgH-locus in 14q32.11 As to the location of the applied probes, t(8;14)+ cells should display a signal constellation fullfilling two criteria: (1) split of the signal derived from one c-myc-probe, and (2) colocalization of one of these split signals with one IgH signal on the aberrant chromosome 14. Although a major advantage of the application of two criteria rather than of a single criterion for the diagnosis of a translocation in interphase FISH is the high specificity due to a low rate of false-positive results, the diagnosis of a t(8;14) according to this strict criteria may suffer from a low sensitivity due to high percentage of false-negative cells. Therefore, we also introduced the less specific but probably more sensitive type II criterion met by each cell displaying a colocalization of each one c-myc- and IgH-signal. Except for one case of Burkitt's lymphoma, in which all cells investigated met the type I criteria, all cases in our series contained a considerable percentage of cells (range, 3.6% to 55%) fullfilling type II but not type I criteria. This obvious lack of a detectable c-myc split signal in a high portion of interphase nuclei might be explained by breakpoints in 8q24 proximal to the COS-H4.1 probe. Nevertheless, c-myc breakpoints centromeric of the COS-H4.1 probe are uncommon in BL.11 12Thus, the lack of a c-myc split signal might be more likely caused by the small size of one c-myc signal, bad interphase morphology, or the loss of the derivative chromosome 8. Moreover, spatial orientation in the three-dimensional nucleus, chromosome decondensation, reactive lymphocytes, or a combination of these factors may play a role in the inability to detect the c-myc signal. Therefore, thorough “in-house” validation studies in each institution to determine the sensitivity and specificity of the probes used are necessary, especially if data provided by these FISH assays will be integrated in the management of the patients to guide therapy.
According to our extensive control studies, the diagnostic threshold for the type I signal constellation was set at 2%. The cutoff level of the type II signal constellation in our assay is 11%. Comparably high thresholds have been determined for other two-color FISH systems, in which positive cells are also indicated solely by the colocalization of two probes, like those for the detection of the t(14;18), t(11;14) or t(9;22).21-23
All cases investigated were positive for t(8;14), proving the principal validity of our interphase FISH assay in primary tumor specimen. In all classical BLs shown to carry this aberration by chromosome analysis, the thresholds for the diagnosis of a translocation t(8;14) were reached for both type I and type II criteria. These data show that both criteria should be equally valid for the diagnosis of a t(8;14) by interphase FISH in classical BL. According to type I criteria, we could not diagnose a t(8;14) in the only case of a Burkitt-like lymphoma, which was shown to contain this translocation by chromosome analysis. Nevertheless, the type II signal pattern was diagnostic for a t(8;14) and indicated a lower percentage of interphase cells carrying this translocation in this case as compared with the classical BL. These data not only provide evidence for the higher sensitivity of the type II criteria but also presumably mean that a higher fraction of cells in the tumor are nonmalignant in Burkitt-like lymphoma than in classical BL. The difference in the portion of t(8;14)+ cells between classical BL and Burkitt-like lymphoma might be hard to detect by chromosome analysis, because as indicated by the significantly higher percentage of t(8;14)+ metaphase than interphase cells reported above, t(8;14)+ cells might have a growth advantage in vivo. As Burkitt-like lymphoma is as yet only a provisionally defined entity,5 the presented interphase FISH assay should contribute to the biologic characterization of this newly described subtype of high-grade B-cell lymphomas.
To date only very few assays for the detection of the t(8;14) by interphase FISH have been published, and none of them have been applied to a substantial series of primary lymphoma specimen. Veronese et al18 reported a FISH system for the detection of the t(8;14), in which the diagnosis of this aberration also relies on two criteria, namely, a split of a signal derived from two YAC-probes spanning the c-myc breakpoint region, and colocalization of at least one of these signals with a probe specific for the telomeric or centromeric part of chromosome 14. By this FISH approach, they were able to diagnose a t(8;14) in metaphase and interphase cells of four BL cell lines and five primary lymphomas shown to carry this translocation by classical cytogenetics. Unfortunately, the study suffers from the lack of control data. Therefore, neither the false-positive rate nor the diagnostic threshold of this approach can be determined. Additionally, there might be a considerable spatial separation of the breakpoint region in 14q32 from the centromeric or telomeric region of chromosome 14 in interphase nuclei because of the decondensation of the chromosome. As this may render the detection of colocalized signals difficult, and in some cases, nearly impossible, caution has to be warranted to apply this assay for routine use.
A recently described three-color FISH approach for the detection of breakpoints in 8q24 uses a chromosome 8 painting probe in combination with differently labeled bacteriophage clones flanking the c-myc locus.16 Breakage in 8q24 should lead to both dissociation of the c-myc signals and to a third chromosome 8 domain containing one c-myc signal. The validity of this assay was confirmed on one normal control and one Burkitt lymphoma cell line. Nevertheless, the necessity of three-color FISH and the difficulties associated with evaluation of chromosome territories in interphase cells render its routine applicatibility limited. Moreover, in the routine diagnostic scenario, an additional hybridization for the identification of the partner chromosome to which a fragment of chromosome 8 is translocated must be demanded. This is also true for FISH assays, in which two differently labeled probes hybridizing to either side of the IgH-locus are used to screen for all 14q32 aberrations including the t(8;14).24 25
Another two-color FISH system for the detection of the t(8;14) uses a 3.5-kb plasmid probe for the joining region of the IgH locus and a 9-kb genomic fragment containing the second and third exons of the c-myc gene.17 In this assay, each interphase cell containing a t(8;14) shows a colocalization of one signal. The signal constellation indicative for a t(8;14) was found in 100% of 30-65 interphase cells from five t(8;14)+ BL cell lines and one primary lymphoma, but in only 1.4% of 68 interphase cells of a t(8;14)− cell line. As a result of the extraordinarily small number of nuclei evaluated in each case and the lack of sufficient controls in this study, the validity and sensitivity of this assay can not be determined. Furthermore, due to the tiny size of the applied probes, the routine detection of the signals may not be reliable.26 This especially might be the case in those BLs in which breakage in the joining region of the IgH-locus leads to disruption of the corresponding plasmid probe.
In summary, in the present study we have successfully proved the feasibility, sensitivity, and validity of two-color FISH for the detection of the t(8;14) in interphase cells of routinely ascertained lymphoma samples. The easy-to-handle assay described allows the reliable diagnosis of this translocation on fresh and fixed tumor samples within 24 hours without the necessity of time-consuming cell culturing and metaphase preparation or the limitations of molecular genetics. Therefore, this interphase FISH system not only allows a rapid biologic classification of high-grade B-cell lymphomas, but also provides the opportunity for rational therapy regimens adjusted a priori for genetic characteristics.
ACKNOWLEDGMENT
The authors are grateful to Prof Dr A. Feller (Department of Pathology, University of Lübeck, Germany) and Prof Dr H.-K. Müller-Hermelink (Department of Pathology, University of Würzburg, Germany) for providing histopathologic diagnoses.
R.S. and P.M. contributed equally to this work.
Supported by the Wilhelm-Sander-Stiftung (95.003.1), and the Deutsche Krebshilfe (10-0992-Schl3). B.S. holds a Hermann and Lilly Schilling-Stiftungsprofessur.
Address reprint requests to Reiner Siebert, MD, Department of Human Genetics, University of Kiel, Schwanenweg 24, 24105 Kiel, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal