Abstract
The consequences of assembling the contact system of proteins on the surface of vascular cells has received little study. We asked whether assembly of these proteins on the surface of cultured human endothelial cells (HUVECs) results in the activation of prekallikrein (PK) and its dependent pathways. Biotinylated PK binds specifically and reversibly to HUVECs in the presence of high molecular weight kininogen (HK) (apparent Kd of 23 ± 11 nmol/L,Bmax of 1.7 ± 0.5 × 107 sites per cell [mean ± SD, n = 5 experiments]). Cell-associated PK is rapidly converted to kallikrein. Surprisingly, the activation of cell-associated HK•PK complexes is entirely independent of exogenous factor XII (Km = 30 nmol/L,Vmax = 12 ± 3 pmol/L/min in the absencevKm = 20 nmol/L,Vmax = 9.2 ± 2.1 pmol/L/min in the presence of factor XII). Rather, kallikrein formation is mediated by an endothelial cell-associated, thiol protease. Cell-associated HK is proteolyzed during the course of prekallikrein activation, releasing kallikrein from the surface. Furthermore, activation of PK bound to HK on HUVECs promotes kallikrein-dependent activation of pro-urokinase, resulting in the formation of plasmin. These results indicate the existence of a previously undescribed, factor XII-independent pathway for contact factor activation on HUVECs that regulates the production of bradykinin and may contribute to cell-associated plasminogen activation in vivo.
THE BIOLOGIC FUNCTION(S) of the plasma proteins of the contact system in hemostasis has been uncertain. Deficiencies of these proteins are not associated with clinical bleeding despite marked prolongation of in vitro surface-activated coagulation times. Paradoxically, studies suggest a role for contact system proteins in fibrinolysis. Patients deficient in individual contact factor proteins may be at increased risk for thrombosis.1-7 Activation of the contact factor pathway promotes plasma fibrinolytic activity8 and antibodies to tissue-type and urokinase plasminogen activators neutralize only 75% of plasma fibrinolytic activity after stimulation with 1-desamino-8-D-arginine vasopressin.9 However, direct plasminogen activator activity of kallikrein, activated factor XII, or factor XIa are only 1/10,000 to 1/40,000 of that of tissue-type and urokinase-type plasminogen activators.10-14 Alternatively, kallikrein has been reported to cleave pro-urokinase (Pro-UK) at a rate that suggests a potential physiologic role.15 Recent observations also indicate that binding of contact factor proteins to cell surfaces accelerated the formation of two-chain urokinase (tcuPA) and plasmin,16-18 suggesting a potential mechanism by which these reactions may occur in vivo.
The mechanism by which contact system proteins are assembled and activated on cell surfaces has received little attention. We and others have previously reported that platelets and endothelial cells express binding sites for high molecular weight kininogen (HK),19-22 a multidomain protein that is both a binding site and a cofactor for the activation of prekallikrein (PK) and factor XI. HK binds to cells through sites present in domains 3, 4, and 523-28 and binds to PK through a site on domain 6.29-31 Because endothelial cells also have the capacity to bind factor XII (FXII),32 it has been assumed that activation of the contact pathway on endothelium proceeds similarly to that which occurs in plasma on artificial surfaces.16-18The present study indicates that assembly of the contact factor proteins on endothelial cells results in PK activation. Activation of PK results in proteolysis of HK with probable liberation of bradykinin and stimulation of plasminogen activator activity. However, to our surprise, PK activation on endothelial cells is independent of FXII or its activated forms.
MATERIALS AND METHODS
Proteins.
HK was purified from plasma using sequential carboxymethyl-papain-Sepharose (CM-papain-Sepharose) and Blue-Sepharose affinity chromatography as previously reported.28,29 HK migrated as a 120-kD protein on sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-PAGE) after reduction with 2% β-mercaptoethanol. HK had a specific activity of 12 to 20 U/mg. Purified HK was iodinated with IODOGEN (Pierce, Rockford, IL) as previously reported.20 Human PK was purchased from Enzyme Research Laboratories (South Bend, IN). The protein migrated as a doublet at 88 and 85 kD on 10% SDS-PAGE under reduced conditions and expressed approximately 1% to 3% of the amidolytic activity of kallikrein.33 No FXII or its activated forms were found in the HK or PK preparations by immunoblotting using a monospecific goat antisera to human FXII. PK was also iodinated with IODOGEN using identical techniques previously reported for HK.20 23Iodinated PK was a doublet at 88 and 85 kD on 10% SDS-PAGE under reducing conditions. FXII, purchased from Enzyme Research Laboratories, migrated predominantly as a single band at 80 kD on 10% SDS-PAGE under reduced conditions and expressed less than 1% of the amidolytic activity of activated FXII. Activated factor XII (αFXIIa) was purchased from Enzyme Research Laboratories. αFXIIa migrated as two bands at 50 and 28 kD on 10% SDS-PAGE under reduced conditions. Factor XIIa fragment (βFXIIa; a generous gift from Dr Robin Pixley, Temple University School of Medicine, Philadelphia, PA) migrated predominantly as a single band at 28 kD on 10% SDS-PAGE under reduced conditions. PK was converted to kallikrein by adding βFXIIa at a molar ratio of 1:74 (βFXIIa:PK). On reduced 10% SDS-PAGE, kallikrein migrated at three bands at 51, 37, and 34 kD.
Pro-UK and tcuPA were purchased from American Diagnostica (Greenwich, CT) or were the generous gifts of Dr Jack Henkin (Abbott Laboratories, Abbott Park, IL). Pro-UK migrated as a single band at 56 kD on 10% SDS-PAGE under reduced conditions. Glu-plasminogen was purchased from American Diagnostica. Fibrinogen, factor XI, lys-plasminogen, and plasmin were purchased from Enzyme Research Laboratories. C1-inhibitor was purified from plasma and antibodies to it were purchased as previously reported.34 SDD31 (SDDDWIPDIQTDPNGLSFNPISDFPDTTSPK), a 31-amino acid peptide that constitutes the PK binding region on HK,35 was synthesized at the University of Michigan Protein and Carbohydrate Structure Facility. This peptide, which spans amino acids 565 to 595 in the mature sequence of HK, is named by using the first three letters of its sequence followed by the number of total amino acids in the peptide. Plasminogen activator inhibitor-1 and rabbit antisera to it were generously supplied by Dr David Ginsburg (University of Michigan, Ann Arbor, MI). Monoclonal antibodies (MoAbs) HKL13, which is directed to domain 5 of HK; HKL16, which is directed to the PK binding site on HK; and PK6, which is directed to the HK binding site on PK, were generously provided by Dr Werner Müller-Esterl (Johannes Gutenberg University, Mainz, Germany).35,36 Goat antisera to human FXII (generously provided by Dr Robin Pixley, Temple University) was adsorbed with FXII-deficient plasma to make it monospecific before use. Polyclonal antibodies to factor XII were also purchased from Enzyme Research Laboratories and Haematologic Technologies (Essex Junction, VT). Rabbit antisera to human PK was prepared as previously reported,37 and antibodies were purified on kallikrein-Sepharose. Pooled normal human plasma and FXII-deficient plasma were purchased from George King Biochemicals, Inc (Overland Park, KS). Prekallikrein-deficient human plasma was from a patient who was characterized to have less than 1% prekallikrein activity and antigen levels.
Functional and immunochemical assays.
HK and FXII procoagulant activities were measured using a one-stage, kaolin-activated coagulant assay38 with total kininogen-deficient and FXII-deficient (George King Biochemicals, Inc) plasmas as the substrate, respectively. Total kininogen-deficient plasma was donated by the late Mayme Williams (Philadelphia, PA). One unit of HK and FXII procoagulant activity is equal to that found in 1 mL of normal plasma. The amidolytic activity of plasma kallikrein and activated FXII were measured using 0.4-1 mmol/L chromogenic substrate, H-D-Pro-Phe-Arg-pNA · 2HCl (S-2302; Pharmacia, Franklin, OH) as previously reported.33 One unit of activity (40 μg/mL) is equal to 2.47 μmol of substrate hydrolyzed/min/mL plasma.33 Immunoblotting of HK, PK, FXII, plasminogen activator inhibitor-1, and C1 inhibitor was performed using the chemiluminesence system of Amersham (Arlington Park, IL), as previously reported.39
Biotinylation of purified PK.
PK was biotinylated as previously reported for HK.25,26 The protein concentration in each fraction of gel-filtered biotinylated-PK (biotin-PK) was determined by its absorbance at 280 nm using an extinction coefficient of 11.7.40 Incorporation of biotin into PK was determined by adding 2-(4′-hydroxyazobenzene)benzoic acid41according to the manufacturer's instructions (Pierce). Each molecule of PK was labeled with 1 to 3 molecules of biotin without causing its activation as determined either by a change in migration on SDS-PAGE or by the development of amidolytic activity. Biotin-PK could be completely converted to biotin-kallikrein by βFXIIa.
Endothelial cell culture.
Cultures of human umbilical vein endothelial cells (HUVECs) were established as previously described.21 Primary cultured cells were passaged twice and then frozen in liquid nitrogen at 1 to 3 × 106 cells/mL. Frozen cells (1.0 mL) were thawed quickly and resuspended in 10.0 mL Endothelial Cell Growth Medium-Modified MCDB 131 (Clonetics, San Diego, CA), which contained 2% fetal calf serum and which was supplemented with Bovine Brain Extract (Clonetics) and centrifuged at 300g for 5 minutes at room temperature. The pellet was resuspended in 15.0 mL of the medium, and the cell suspension was grown to confluence on fibronectin (20 μg/mL) -coated flasks (Corning Inc, Corning, NY).
Binding of biotin-PK to HUVECs.
All binding experiments were performed at 37°C, unless otherwise stated, on fibronectin-coated, 96-well microtiter plates (Nunclon; Thomas Scientific, Swedesboro, NJ) using cells passaged three or four times as previously published.25,26 HUVECs were always used within 24 hours of reaching confluence. Each well contained 3 to 4 × 104 cells. All incubation and washing steps were performed using HEPES-Tyrode's binding buffer [0.135 mol/L NaCl, 2.7 mmol/L KCl, 11.9 mmol/L NaHCO3, 0.36 mmol/L NaH2PO4, 14.7 mmol/L HEPES (N-2-hydroxyethylpiperazine-N,-2-ethanosulfonic acid)] containing 50 μmol/L Zn+2, 1 mmol/L Mg+2, 3.5 mg/mL bovine serum albumin, 3.5 mg/mL dextrose, pH 7.35, in the presence of 2 mmol/L Ca+2, unless otherwise stated. HUVECs were washed five times before performing all binding studies. In most experiments, HUVECs were incubated with 20 nmol/L HK in 100 μL for 1 hour, which is sufficient to saturate their specific binding sites,21,25 and the unbound HK was removed, whereas in other experiments the cells were washed three times. The cells were then incubated with various concentrations of biotin-PK in a 100 μL reaction volume for 1 hour at 37°C unless otherwise stated. Binding of biotin-PK to HUVECs was the same whether unbound HK was removed by aspiration alone or by washing. Nonspecific binding, unless otherwise stated, was determined by measuring binding in the presence of 50-fold molar excess unlabeled PK. Specific binding was determined by subtracting nonspecific binding from total binding. Cell-associated biotin-PK was measured using ImmunoPure streptavidin horseradish peroxidase conjugate (Pierce) and the peroxidase-specific fast-reacting substrate, turbo-3,3′,5,5′-tetramethylbenzidine dihydrochloride (turbo-TMB; Pierce), as previously reported.25 Bound biotin-PK was measured by the absorbance at OD450nm using a Microplate auto reader EL 311 (Bio-Tek Instrument, Winooski, VT).
In certain experiments, binding of 125I-PK to HUVECs in suspension was measured. Briefly, HUVECs were removed from culture dishes with 0.05% trypsin, 0.53 mmol/L EDTA solution (Life Technologies, Grand Island, NY), which was immediately inactivated with trypsin neutralizing solution (Clonetics), and the cells were washed twice by centrifugation using HEPES-Tyrode's binding buffer and brought to a final cell suspension of 3.5 × 105cells/mL. The suspended cells were preincubated with 20 nmol/L HK and125I-PK (20 nmol/L) was added in the absence or presence of 50-fold molar excess PK. After 5 to 40 minutes, 50-μL aliquots of the cell suspension were centrifuged at 10,000g for 2 minutes in a Beckman Microcentrifuge Model E (Fullerton, CA) over a 200-μL oil gradient consisting of 1 part Apiezon (Biddle Instruments, Blue Bell, PA) and 9 parts N-butylphthalate (Fisher Scientific, King of Prussia, PA). The tips of the tapered centrifuge tubes were amputated and counted in a gamma counter. The amount of bound 125I-PK was calculated based on the specific radioactivity of the ligand.
Additional studies were performed to evaluate the reliability of using biotin-PK to measure binding to cells. Although four additional wash steps were used when biotin-PK was used as the ligand than when125I-PK was used, the percentage of specific binding of biotin PK (0.5% ± 0.14%) and 125I-PK (0.62% ± 0.12%) were not significantly different (P > .05). These data indicated that, like biotin-HK and125I-HK,25 biotin-PK and 125I-PK bound with sufficient affinity to cell-associated HK to permit them to be used interchangeably.
Quantification of HUVEC-bound biotin-PK.
To convert the color reaction of bound biotin-PK to pmoles PK bound, standard curves for each batch of biotin-PK were developed as previously reported for biotin-HK binding.25 Kaolin (200 mg/mL) in HEPES-Tyrode's binding buffer without added divalent cations was preincubated with 60 nmol/L HK for 1 hour at 37°C with constant mixing. After removing unbound HK by centrifugation at 3,000gfor 30 seconds, biotin-PK (0.02 to 3.0 pmol) was incubated with the HK-treated kaolin suspension in triplicate for 10 minutes at 37°C with constant mixing. The kaolin suspension was then blocked by adding 1% bovine serum albumin and then incubated with a 1:500 dilution of streptavidin horseradish peroxidase conjugate for 1 hour. The relative amount of streptavidin horseradish peroxidase conjugate bound to the centrifuged and resuspended kaolin was determined as described for biotin-HK binding to HUVECs.25 After subtracting the absorbance of kaolin-HK alone, a standard curve from three or more identical experiments was generated relating the absorbance at each amount of biotin-PK to its concentration by linear regression.
Characterization of PK binding to HUVECs.
The apparent affinity (Kd) and number of binding sites (Bmax) for specific biotin-PK binding to HUVECs, preincubated with HK (20 nmol/L), were determined by the method of Scatchard.42 The apparent affinity of biotin-PK binding for HUVECs was also assessed by determining the capacity of unlabeled PK to inhibit the binding of biotin-PK. Biotin-PK (20 to 40 nmol/L) was incubated with 0- to 100-fold molar excess PK for 1 hour with confluent HUVECs that had been preincubated in the presence (20 nmol/L) or absence of HK. The 50% inhibitory concentration (IC50) was determined, and the apparent Ki of the competitor was calculated by the technique of Müller43 as previously reported,20 using the formula Ka = 8/3 ([It] − [Tt]), where [It] equals the molar concentration of the IC50 of the competitor and [Tt] is molar concentration of the added biotin-PK.
PK activation on endothelial cells.
Activation of PK bound to HUVECs was measured three ways. First, confluent monolayers of HUVECs in microtiter plate wells were preincubated with buffer containing 2% radioimmunoassay grade bovine serum albumin (Sigma, St Louis, MO) for 1 hour at 37°C. PK in HEPES-Tyrode's binding buffer containing 50 μmol/L Zn+2 was incubated with cells preincubated with either saturating concentrations of HK (20 nmol/L) or buffer. Kallikrein activity was then measured as the hydrolysis of 0.4 mmol/L S2302 over 1 hour (Pharmacia). In certain experiments, FXII (20 nmol/L), αFXIIa (3.4 nmol/L), or βFXIIa (3.4 nmol/L) was added along with the chromogenic substrate. In other experiments, plasma kallikrein (20 nmol/L) was substituted for PK. In other experiments, the activation of PK bound to HK on HUVECs was measured in the presence of a twofold neutralizing concentration of an antibody to FXII. To determine this, 20 nmol/L αFXIIa was incubated with increasing concentrations of anti-factor XII IgG (1.0 to 1,000 μg/mL). It was found that 0.2 mg/mL antibody from two sources inhibited greater than 95% of the hydrolytic activity of αFXIIa for S2302. Therefore, 0.4 mg/mL of anti-FXII IgG was added to 4 × 104 HUVECs pretreated with 20 nmol/L HK in the presence of 20 nmol/L PK and the amidolytic activity was determined relative to the activity generated in the absence of antibody. In different experiments, endothelial cell-bound PK activation was measured when the source of PK were different plasmas. Briefly, HUVECs, preincubated with 20 nmol/L HK, were incubated with NHP, FXII-deficient plasma, or PK-deficient plasma for 1 hour at 37°C. After washing the cells with HEPES-Tyrode's buffer containing 50 μmol/L Zn+2, 0.4 mmol/L S2302 was added and the extent of hydrolysis of the chromogenic substrate was measured over the next 1 hour.
Second, the kinetics of PK activation on HUVEC monolayers were determined in the absence or presence of exogenous FXII.20The cells were preincubated with HK (20 nmol/L) for 1 hour and PK (1 to 100 nmol/L) was added for an additional 1 hour. The wells were washed three times with HEPES-Tyrode's binding buffer containing 50 μmol/L Zn+2, after which FXII (20 nmol/L) and S2302 (0.4 mmol/L) were added and hydrolysis of the substrate was measured over the next 1 hour. In certain experiments, FXII was omitted. The amount of kallikrein formed was determined using a standard curve generated by adding known amounts of soluble plasma kallikrein under identical conditions. The Km and Vmax of PK activation on HUVECs were determined from double reciprocal plots.
Third, experiments were performed to determine if cell-bound PK was activated to kallikrein or whether cell-associated PK expressed intrinsic activity. HUVECs were incubated for 1 hour with 20 nmol/L HK, unbound HK was removed, and the cells were incubated with 20 nmol/L biotin-PK in HEPES-Tyrode's buffer containing 50 μmol/L Zn+2 for variable lengths of time. The reaction was stopped by washing the cells three times, after which the contents of the wells were solubilized by adding electrophoresis sample buffer containing 2% β-mercaptoethanol. The proteins were then boiled, separated on 10% SDS-PAGE, electroblotted onto a nitrocellulose membrane, and blocked with Blotto44 and streptavidin-horseradish peroxidase (1:500; Pierce) was added. Biotin-PK bound to the nitrocellulose was detected by measuring chemiluminesence (Amersham). The extent to which the biotin-PK was cleaved was determined by densitometer scanning of the blot using a transmittance/reflectance scanner (Model GS 300; Hoefer Scientific Instruments, San Francisco, CA) in the transmittance mode.
Determination of the protease class of the cell-associated PK activating enzyme.
Two sets of experiments were performed to determine the biochemical features of the protease(s) that activated PK bound to HK on HUVECs. First, studies were performed to determine if contaminating factor XIIa, kallikrein, or another serine protease could be responsible for PK activation. To do this, we determined if neutralizing concentrations (0.4 mg/mL) of two antibodies to FXII, phenylmethyl sulfonylfluoride (PMSF; 1 mmol/L), SBTI (1 mg/mL), Pro-Phe-Arg-chloromethylketone (10 μmol/L), or benzamidine (1 mmol/L) would block activation of HK-bound PK on HUVECs. Activation was determined by detecting cleavage of PK (88,85-kD doublet) to kallikrein on reduced 10% SDS-PAGE, which mostly appeared as the 51-kD heavy chain of kallikrein. Pro-Phe-Arg-chloromethylketone was the generous gift of Dr Charles Kettner (Dupont-Merck Pharmaceutical Co, Wilimington, DE). All other inhibitors were purchased from Sigma. Each of these protease inhibitors were titered against 20 nmol/L factor XIIa or kallikrein to determine the minimal concentration that would abolish the enzymes' hydrolytic activity against S2302. This inhibitory concentration was the one used in the experiments.
Second, studies were performed to determine the biochemical requirements of the membrane-associated, PK activating enzyme. In these experiments, the minimal concentration of the inhibitor that produced maximal inhibition was used. Furthermore, each inhibitor was examined for its ability to directly inhibit kallikrein itself. In these experiments, HK-saturated HUVECs were incubated with PK in the absence or presence of antipain (100 μmol/L), cysteine (10 mmol/L), HgCl2 (1 mmol/L), glutathione (100 μmol/L), calpain inhibitor (10 mmol/L; Calbiochem, La Jolla, CA), E64 (10 mmol/L), hydroxy-mercuribenzoic acid (10 mmol/L), iodoacetamide (10 mmol/L), iodoacetic acid (10 mmol/L), dithiothreitol (DTT; 10 mmol/L), 2-mercaptoethanol (5%), TIMP-1 and TIMP-2 (20 μg/mL; Calbiochem), BB94 (15 μmol/L), Cystatin (1 mmol/L), pepstatin A (10 mmol/L), EDTA (10 mmol/L), EGTA (10 mmol/L), 1,10 phenanthroline (10 mmol/L), phosphoramidon (10 mmol/L), zincov (100 μmol/L), enalapril (10 mmol/L), lisinopril (10 mmol/L), bathophenanthroline (100 μmol/L), or sodium hydrosulfite (10 mmol/L). After incubation, S2302 (0.4 mmol/L) was added and the extent of hydrolysis of the substrate was determined. BB94 was a generous gift of Dr Steve Weiss (University of Michigan). Unless otherwise stated, the remaining inhibitors were purchased from Sigma.
Cleavage of HUVEC-bound HK by activated PK.
Experiments were performed to determine whether activation of PK was accompanied by cleavage of the HK to which it was bound. HUVECs were incubated with 20 nmol/L 125I-HK for 1 hour. Unbound ligand was removed and the cells were incubated with 20 nmol/L PK or its buffer for variable lengths of time. The reaction was stopped by washing the cells three times, after which the contents of the wells were solubilized by adding electrophoresis sample buffer containing 2% β-mercaptoethanol. The samples were boiled, separated on 10% SDS-PAGE, and analyzed by autoradiography and scanning densitometry as described above.
Measurement of Pro-UK activation on endothelial cells.
Pro-UK activation on HUVECs was measured. Activation of Pro-UK (20 nmol/L) on HUVECs incubated with PK (20 nmol/L) or sequentially with HK (20 nmol/L) and PK (20 nmol/L) was determined. HUVECs were incubated with 20 nmol/L HK in 100 μL HEPES-Tyrode's binding buffer containing 50 μmol/L Zn+2 for 1 hour at 37°C. Unbound HK was removed, 20 nmol/L PK was incubated for an additional 1 hour, and the cells were washed three more times. Pro-UK (20 nmol/L) and 0.6 mmol/L Glu-Gly-Arg-pNA. HCL (S2444; Pharmacia) were added and the hydrolysis of the substrate was monitored for 75 minutes at 37°C. In certain of these experiments, a neutralizing concentration (0.4 mg/mL) of anti-FXII antibody was added. The kinetics of Pro-UK activation (5 to 1,000 nmol/L) on HUVECs in the presence or absence of HK (20 nmol/L), PK (20 nmol/L), or FXII (20 nmol/L) was studied as well. Formation of tcuPA on HUVECs was also determined with reference to a standard curve generated by adding known amounts of soluble tcuPA to S2444. The Km and Vmax of two-chain urokinase formation was determined from double reciprocal plots.
Measurement of plasminogen activation on endothelial cells.
Plasminogen (1 μmol/L) activation in plastic wells and on confluent HUVECs in microtiter plate wells was determined by measuring the hydrolysis of 0.3 mmol/L Val-Leu-Lys-pNA.HCl (S2251; Pharmacia). The contribution of Pro-UK (2 nmol/L) to plasminogen activation under the same condition was determined. In other experiments, HUVECs were preincubated with HK (20 nmol/L) for 1 hour, unbound HK was removed, PK or kallikrein (20 nmol/L) was added for second hour, and plasminogen (1 μmol/L) was added for a third hour, before Pro-UK (2 nmol/L) and S2251 (0.3 mmol/L) were added to start the reaction. In other experiments, Pro-UK was added to the wells in the presence of a twofold neutralizing concentration of antibody to FXII (0.4 mg/mL), 20 nmol/L FXII, or 3.4 nmol/L β-FXIIa. Under all experimental conditions, the hydrolysis of chromogenic substrate was monitored continuously for 210 minutes at 37°C. The amount of plasmin formed from plasminogen was determined using a standard curve generated by adding known amounts of soluble plasmin.
Statistics.
Significant differences were measured using the t-test for groups of unpaired data.
RESULTS
Binding of biotin-PK to HUVECs.
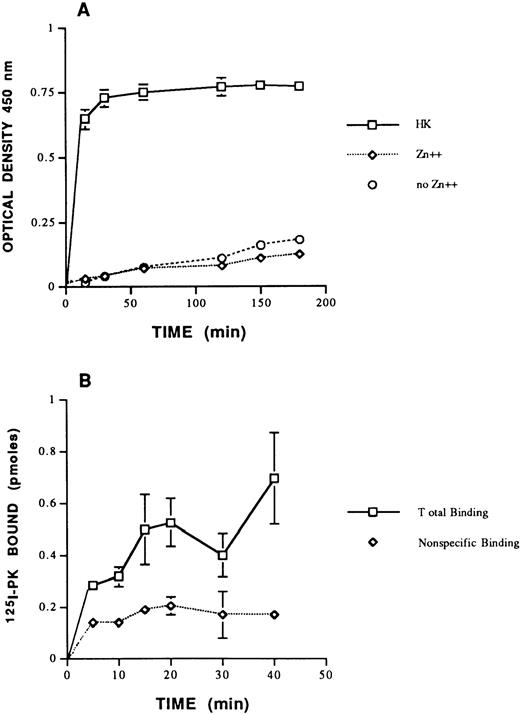
In view of the fact that PK circulates in plasma predominantly complexed with HK,29-31 initial experiments were performed to determine if PK bound to HUVECs through HK. Biotin-PK bound specifically to HUVEC monolayers preincubated with HK and Zn+2 (Fig1A). Zinc ion alone did not support binding of biotin-PK in the absence of HK. The presence of 2 mmol/L Ca+2 in the buffers had no effect on binding (data not shown). Additional studies showed that125I-PK also bound specifically to suspensions of HUVECs preincubated with HK (Fig 1B). These combined data indicated that the labeled PKs bound to HK on HUVECs, irrespective of whether the cells were in monolayers or suspension.
Binding of PK to HUVEC. (A) The effect of HK and Zn+2 on biotin-PK binding to HUVECs. HUVECs were incubated with 20 nmol/L HK (□) for 1 hour at 37°C in the presence of 50 μmol/L Zn+2. Unbound HK was removed and biotin-PK (20 nmol/L) was added in HEPES-Tyrode's binding buffer containing Zn+2 for variable times. In other wells, HUVECs were incubated with biotin-PK (20 nmol/L) for variable times at 37°C in the absence of HK, but in the presence (⋄) or absence (○) of Zn+2. The specific binding of biotin-PK bound is shown. The data presented represent the mean ± SEM of three experiments. (B) Binding of 125I-PK (20 nmol/L) to HUVECs in suspension in HEPES-Tyrode's binding buffer containing 50 μmol/L Zn+2 in the absence (total binding; □) or presence (nonspecific binding; ⋄) of 50-fold molar excess PK. The points presented represent the mean ± SEM of three experiments. The absence of standard error bars at some points indicates that the variation was too low to indicate visually.
Binding of PK to HUVEC. (A) The effect of HK and Zn+2 on biotin-PK binding to HUVECs. HUVECs were incubated with 20 nmol/L HK (□) for 1 hour at 37°C in the presence of 50 μmol/L Zn+2. Unbound HK was removed and biotin-PK (20 nmol/L) was added in HEPES-Tyrode's binding buffer containing Zn+2 for variable times. In other wells, HUVECs were incubated with biotin-PK (20 nmol/L) for variable times at 37°C in the absence of HK, but in the presence (⋄) or absence (○) of Zn+2. The specific binding of biotin-PK bound is shown. The data presented represent the mean ± SEM of three experiments. (B) Binding of 125I-PK (20 nmol/L) to HUVECs in suspension in HEPES-Tyrode's binding buffer containing 50 μmol/L Zn+2 in the absence (total binding; □) or presence (nonspecific binding; ⋄) of 50-fold molar excess PK. The points presented represent the mean ± SEM of three experiments. The absence of standard error bars at some points indicates that the variation was too low to indicate visually.
Biotin-PK binding to HUVECs in the presence of added HK.
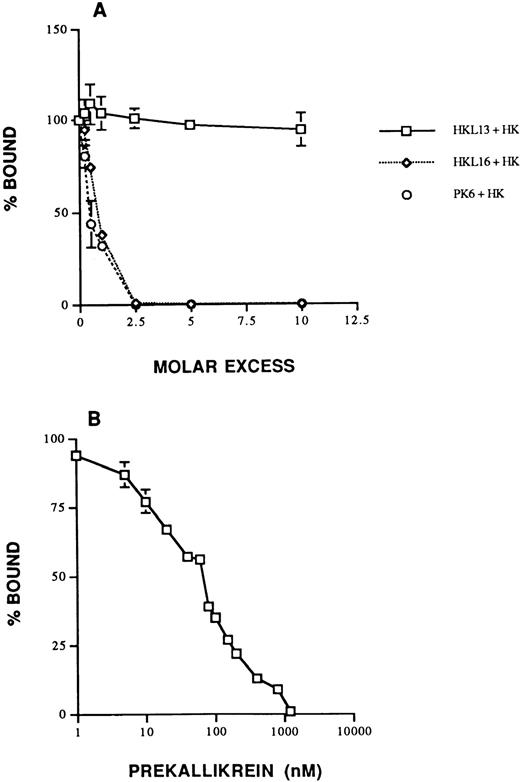
The specificity of biotin-PK binding to HK on HUVECs was shown several ways. First, biotin-PK binding to HUVECs preincubated with 20 nmol/L HK was completely inhibited by 50-fold molar excess PK, factor XI, and the peptide SDD31 (data not shown). In contrast, 100-fold molar excess Lys-plasminogen, C1-inhibitor, fibrinogen, and Glu-plasminogen inhibited biotin-PK binding by 34%, 21%, 15%, and 8%, respectively (P ≤ .05) compared with the absence of inhibitor (data not shown). FXII appeared to increase biotin-PK binding, but this difference was not statistically significant. Second, binding of PK in the presence of added HK was 100% inhibited by 2.5 molar excess MoAbs HKL16, which is known to recognize the PK binding site on HK, and PK6, which is directed to the HK binding site on PK, whereas HKL13, which is directed to domain 5 on HK, had no effect (Fig 2A). Taken together, the data suggested that PK bound to a site on domain 6 of cell-associated HK that was shared with factor XI.31 PK also inhibited biotin-PK binding to HUVECs preincubated with HK in a concentration-dependent fashion with an IC50 of 80 nmol/L (apparent Ki = 23 nmol/L; Fig 2B), suggesting that biotin-PK and PK compete for the same site. When HUVECs were preincubated with biotin-PK for 60 minutes, 86% of the binding was reversed by adding 100-fold molar excess PK (data not shown). Similarly, when HUVECs were preincubated for ≤40 minutes with biotin-PK, binding was 100% reversible (data not shown). We next determined the affinity and number of saturable biotin-PK binding sites expressed by HUVECs under calculated equilibrium conditions. Binding of biotin-PK saturated at a concentration of ≥20 nmol/L. Assuming a 1:1 stoichiometry between PK and cell-bound HK as exists in plasma,29-31 biotin-PK bound with an apparentKd = 23 ± 11 nmol/L and aBmax of 1.7 ± 0.5 × 107sites/cell (mean ± SD of 5 individual experiments). However, these data need to be interpreted as descriptive, because the interaction of biotin-PK binding with HK on HUVECs did not fulfill the criteria for true equilibrium conditions (see below).
Specificity of biotin-PK binding to HUVECs in the presence of added HK. (A) The effect of MoAbs to HK and PK on the binding of biotin-PK to HUVECs preincubated with 20 nmol/L HK. Binding of biotin-PK to HUVECs was measured in the presence of 1- to 10-fold molar excess of the MoAbs, HKL13 (□), HKL16 (⋄), or PK6 (○). Binding of biotin-PK in the presence of the antibodies is expressed as a percentage of the binding in their absence. The data presented are the mean ± SEM of three experiments. (B) PK and biotin-PK compete for binding to HUVECs. HUVECs pretreated with 20 nmol/L HK were incubated with 20 nmol/L biotin-PK in the presence of 10 to 2,000 nmol/L PK. Binding of biotin-PK in the presence of PK is expressed as a percentage of its binding in the absence of PK. The data shown are the mean ± SEM of three experiments. The absence of standard error bars at some points indicates that the variation was too low to indicate visually.
Specificity of biotin-PK binding to HUVECs in the presence of added HK. (A) The effect of MoAbs to HK and PK on the binding of biotin-PK to HUVECs preincubated with 20 nmol/L HK. Binding of biotin-PK to HUVECs was measured in the presence of 1- to 10-fold molar excess of the MoAbs, HKL13 (□), HKL16 (⋄), or PK6 (○). Binding of biotin-PK in the presence of the antibodies is expressed as a percentage of the binding in their absence. The data presented are the mean ± SEM of three experiments. (B) PK and biotin-PK compete for binding to HUVECs. HUVECs pretreated with 20 nmol/L HK were incubated with 20 nmol/L biotin-PK in the presence of 10 to 2,000 nmol/L PK. Binding of biotin-PK in the presence of PK is expressed as a percentage of its binding in the absence of PK. The data shown are the mean ± SEM of three experiments. The absence of standard error bars at some points indicates that the variation was too low to indicate visually.
Biotin-PK binding to HUVECs in the absence of added HK.
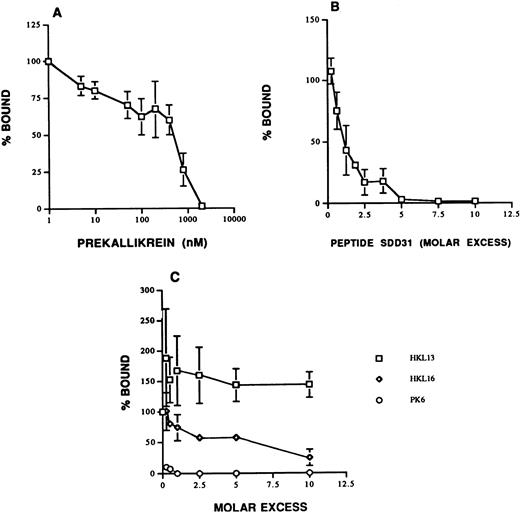
The results of the binding experiments suggested that HUVECs expressed more binding sites for biotin-PK than the actual number of molecules of HK bound (1.0 ± 0.02 × 107sites/cell).25 Several additional experiments were performed to address the possibility that there may be additional PK binding sites on HUVECs independent of those provided by added HK. Biotin-PK binding to HUVECs in the absence of added HK was blocked by PK with an IC50 of 800 nmol/L (apparentKi = 285 nmol/L; Fig3A). These data suggested that, in the absence of added HK, a lower affinity, specific binding site(s) for biotin-PK was present on these cells. SDD31, a peptide corresponding to the PK binding site on HK, also blocked biotin-PK binding to HUVECs in the absence of added HK with an IC50 of 40 nmol/L (Fig 3B). In support of this finding, MoAb PK6, which is directed to the region on PK that binds to HK, blocked binding completely (Fig 3C). Taken together, these data indicated that all of the binding of PK to HUVECs is mediated by the same region on PK. We then asked whether endogenous HUVEC HK21 could account for some of this biotin-PK binding in the absence of added HK. MoAb HKL16, which is directed to the PK binding site on HK, blocked biotin-PK binding to HUVECs in the absence of added HK (Fig 3C). This finding suggested that HUVECs did express endogenous HK that was available to bind PK. We then asked whether the PK binding site was human HK derived from the HUVECs21 or heterologous HK adsorbed from bovine serum. In the support of the former possibility, HKL16 did not recognize bovine plasma HK on immunoblot, but it did detect human plasma HK (data not shown). However, it was noted that binding of biotin-PK was inhibited only 75% by 10-fold molar excess HKL16 in the absence of added HK. This finding indicated that about 25% of PK binding may involve an additional, uncharacterized site(s) (Fig 3C). However, because virtually all plasma PK circulates as a complex with HK,29 30 we focused the remaining investigations on characterizing the biologic consequences of forming HK and PK complexes on the endothelial cells.
Specificity of biotin-PK binding to HUVECs in the absence of added HK. (A) PK and biotin-PK compete for binding to HUVECs. HUVECs were incubated with 40 nmol/L biotin-PK and 10 to 2,000 nmol/L PK in the absence of added HK. Binding of biotin-PK in the presence of PK is expressed as a percentage of its binding in the absence of PK. The data shown are the mean ± SEM of three experiments. (B) Binding of 40 nmol/L biotin-PK to HUVECs was measured in the absence of added HK but in the presence of increasing concentrations of the peptide SDD31, which corresponds to the PK binding site on HK. The data shown are the mean ± SEM of three experiments. (C) The effect of MoAbs to HK and PK on the binding of biotin-PK to HUVECs in the absence of added HK. Binding of biotin-PK to HUVECs was measured in the presence of 1- to 10-fold molar excess of the MoAbs, HKL13 (□), HKL16 (⋄), and PK6 (○). Binding of biotin-PK in the presence of the antibodies is expressed as percent of the binding in their absence. The data presented are the mean ± SEM of three experiments.
Specificity of biotin-PK binding to HUVECs in the absence of added HK. (A) PK and biotin-PK compete for binding to HUVECs. HUVECs were incubated with 40 nmol/L biotin-PK and 10 to 2,000 nmol/L PK in the absence of added HK. Binding of biotin-PK in the presence of PK is expressed as a percentage of its binding in the absence of PK. The data shown are the mean ± SEM of three experiments. (B) Binding of 40 nmol/L biotin-PK to HUVECs was measured in the absence of added HK but in the presence of increasing concentrations of the peptide SDD31, which corresponds to the PK binding site on HK. The data shown are the mean ± SEM of three experiments. (C) The effect of MoAbs to HK and PK on the binding of biotin-PK to HUVECs in the absence of added HK. Binding of biotin-PK to HUVECs was measured in the presence of 1- to 10-fold molar excess of the MoAbs, HKL13 (□), HKL16 (⋄), and PK6 (○). Binding of biotin-PK in the presence of the antibodies is expressed as percent of the binding in their absence. The data presented are the mean ± SEM of three experiments.
Activation of PK on HUVECs.
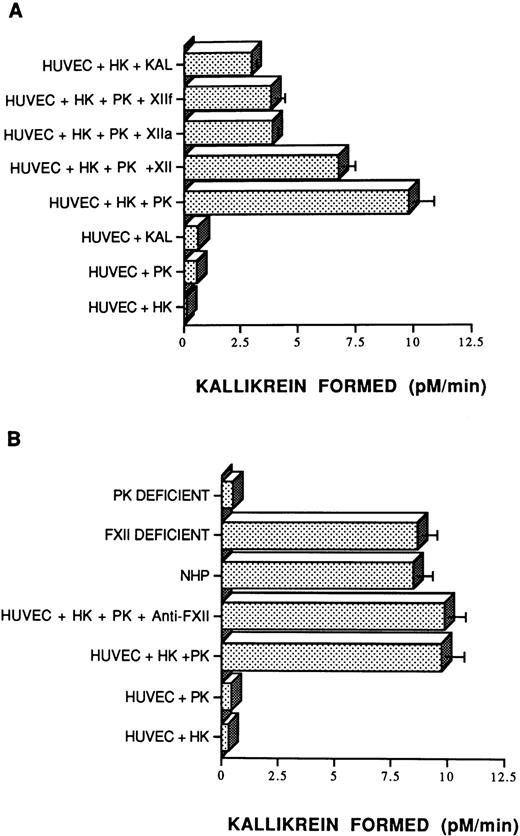
Investigations were performed to determine if PK bound to HK on the surface of HUVECs could be activated through mechanisms similar to those known to occur in plasma and on artificial surfaces. HUVECs did not hydrolyze the plasma kallikrein chromogenic substrate in the absence of added contact proteins (data not shown). These data suggested that washed HUVECs have little if any kallikrein- and/or activated FXII-like activity tightly bound and nonexchangeable to be measured in this system. Further, little enzymatic activity was seen when HK, PK, or kallikrein alone was permitted to bind to HUVECs (Fig 4A). However, when HUVECs were incubated sequentially with HK and then PK, chromogenic activity was readily detected (Fig 4A). Indeed, when equal amounts of PK and plasma kallikrein were added to HUVECs preincubated with HK, significantly more activity (P < .001) was detected from the assembly of the HK•PK complex than from the HK•kallikrein complex. Furthermore, the addition of FXII, αFXIIa, or βFXIIa to the HK•PK complex on HUVECs did not increase the extent of chromogenic activity above that which could be accounted for by activating PK bound to HK on HUVECs alone. In fact, the addition of FXII, αFXIIa, or βFXIIa resulted in significantly less measured enzymatic activity (P ≤ .005) than that seen with the HK•PK complex alone, similar to what was seen when kallikrein was substituted for PK (Fig 4A).
Activation of PK on HUVECs. (A) Endothelial cell monolayers (HUVECs) were preincubated with 200 μL of a solution containing 2% bovine serum albumin. HK (20 nmol/L) or buffer was then added for 1 hour at 37°C, the unbound protein was removed by washing, and 20 nmol/L PK or 20 nmol/L plasma kallikrein (Kal) was incubated for an additional 1 hour. The wells were then washed and 0.4 mmol/L S2302 was added in the absence or presence of 20 nmol/L FXII (XII), 3.4 nmol/L αFXIIa (XIIa), or 3.4 nmol/L βFXIIa (XIIf), as indicated. The data presented are the mean ± SEM of three experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually. (B) Endothelial cell monolayers (HUVECs) were preincubated with 200 μL of a solution containing 2% bovine serum albumin. HK (20 nmol/L) or buffer were then added for 1 hour at 37°C, the unbound protein was removed by washing, and 20 nmol/L PK was incubated for 1 additional hour in the absence or presence of 0.4 mg/mL of an anti-FXII antibody (Anti-FXII). In other experiments, HUVECs saturated with HK (20 nmol/L) were incubated with 50 μL of pooled normal plasma (NHP), FXII-deficient plasma, or PK-deficient plasma for 1 hour at 37°C. After washing, 0.4 mmol/L S2302 was added and hydrolysis was monitored for 1 hour. The data presented are the mean ± SEM of three experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually.
Activation of PK on HUVECs. (A) Endothelial cell monolayers (HUVECs) were preincubated with 200 μL of a solution containing 2% bovine serum albumin. HK (20 nmol/L) or buffer was then added for 1 hour at 37°C, the unbound protein was removed by washing, and 20 nmol/L PK or 20 nmol/L plasma kallikrein (Kal) was incubated for an additional 1 hour. The wells were then washed and 0.4 mmol/L S2302 was added in the absence or presence of 20 nmol/L FXII (XII), 3.4 nmol/L αFXIIa (XIIa), or 3.4 nmol/L βFXIIa (XIIf), as indicated. The data presented are the mean ± SEM of three experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually. (B) Endothelial cell monolayers (HUVECs) were preincubated with 200 μL of a solution containing 2% bovine serum albumin. HK (20 nmol/L) or buffer were then added for 1 hour at 37°C, the unbound protein was removed by washing, and 20 nmol/L PK was incubated for 1 additional hour in the absence or presence of 0.4 mg/mL of an anti-FXII antibody (Anti-FXII). In other experiments, HUVECs saturated with HK (20 nmol/L) were incubated with 50 μL of pooled normal plasma (NHP), FXII-deficient plasma, or PK-deficient plasma for 1 hour at 37°C. After washing, 0.4 mmol/L S2302 was added and hydrolysis was monitored for 1 hour. The data presented are the mean ± SEM of three experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually.
Additional studies showed that PK activation when bound to HK on HUVECs occurred through an FXII-independent mechanism. First, the presence of neutralizing quantities of antibody to FXII did not inhibit the extent of PK activation on HUVECs (Fig 4B). Second, PK from normal plasma and FXII-deficient plasma was activated comparably, whereas no activation occurred when PK-deficient plasma was used (Fig 4B). Lastly, theKm and Vmax of PK activation bound to HK on HUVECs in the absence of FXII (Km = 20 ± 8 nmol/L; Vmax = 12 ± 3 pmol/L/min) was virtually the same as that generated in the presence of FXII (Km = 30 ± 4.2 nmol/L; Vmax= 9.2 ± 2.1 pmol/L/min). These data indicated that exogenous FXII did not contribute to the rate of PK activation when bound to HK on HUVECs.
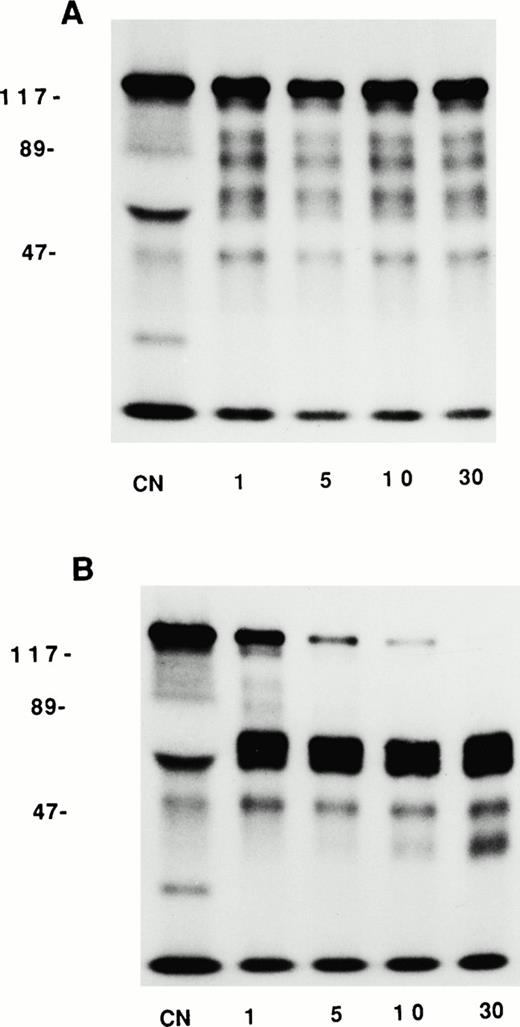
Studies were next performed to determine if the chromogenic activity was attributable to the enzymatic conversion of PK to kallikrein, to a distinct enzyme with kallikrein like-activity, or to a conformational change in PK that occurred upon binding that exposed its catalytic site (Fig 5). Biotin-PK predominantly migrated as a doublet at 88 and 85 kD on SDS-PAGE under reduced conditions. When βFXIIa was added, biotin-PK was cleaved into a heavy chain of 51 kD and two light chains at 37 and 34 kD. A fourth band also was seen at 40 kD (Fig 5A and B). When biotin-PK was incubated with HUVECs in the absence of HK for up to 120 minutes, biotin-PK predominantly migrated at 85 and 88 kD (Fig 5A). However, a new band appeared at 116 kD within the first minutes, consistent with a SDS-stable complex having formed between biotin-PK or biotin-kallikrein and another protein. This band did not increase over time and constituted approximately 28% of the total biotin-PK (average of all lanes from 1 to 120 minutes) on densitometer scan. Furthermore, threefold molar excess HKL16 did not block the formation of the 116-kD complex on HK-treated cells, indicating that this other PK-binding protein was not HK (data not shown). This higher molecular mass band also was not found to be a complex between kallikrein and plasminogen activator inhibitor-1 or C1-inhibitor on immunoblot (data not shown). By 60 to 120 minutes, small amounts of cleaved forms of biotin-PK (<5% of the total protein) were seen at 51 and 40 kD (Fig 5A). Thus, little cleavage of PK occurred when it was incubated with HUVECs in the absence of added HK. In contrast, when biotin-PK was incubated with HK prebound to HUVECs, changes in PK structure occurred more rapidly and extensively (Fig 5B). The 116-kD band consisted of only 5% of the total protein over 120 minutes. Cleaved products of biotin-PK at 51, 40, and 37 kD appeared at 1 to 5 minutes and increased in intensity over the ensuing 120 minutes (Fig 5B). By 60 minutes, the 51-kD band constituted 46% of the total protein. These data indicated first that the chromogenic activity described in Fig 4 was due to the conversion of PK to kallikrein; second, the conversion occurred more rapidly in the presence of HK; and third, the generation of kallikrein on HUVECs did not require an exogenous source of activated FXII. In experiments not shown, the ability of PK bound to HK on HUVECs to become activated occurred regardless of whether the PK was incubated in plasma or buffer. Lastly, PK (or kallikrein) formed an SDS-stable 116-kD complex with an endogenous HUVEC protein other than HK.
Conversion of HUVEC-bound PK to kallikrein. HUVECs were incubated with buffer (A) or with 20 nmol/L HK (B) for 1 hour at 37°C. Unbound HK was removed and 20 nmol/L biotin-PK was added for 1 to 120 minutes at 37°C. After washing, the cells were solubilized by adding electrophoresis sample buffer containing 2% β-mercaptoethanol. The proteins were boiled and then separated on 10% SDS-PAGE and electroblotted onto nitrocellulose, and biotin-PK was detected by adding streptavidin-horseradish peroxidase. Photographs of ligand blots on nitrocellulose are shown. The numbers on each side of the photographs are molecular mass standards in kilodaltons. The numbers between the photographs show the time of incubation with PK. CN represents soluble biotin-PK starting material and XIIf represents βFXIIa-cleaved biotin-PK analyzed by SDS-PAGE in the absence of cells.
Conversion of HUVEC-bound PK to kallikrein. HUVECs were incubated with buffer (A) or with 20 nmol/L HK (B) for 1 hour at 37°C. Unbound HK was removed and 20 nmol/L biotin-PK was added for 1 to 120 minutes at 37°C. After washing, the cells were solubilized by adding electrophoresis sample buffer containing 2% β-mercaptoethanol. The proteins were boiled and then separated on 10% SDS-PAGE and electroblotted onto nitrocellulose, and biotin-PK was detected by adding streptavidin-horseradish peroxidase. Photographs of ligand blots on nitrocellulose are shown. The numbers on each side of the photographs are molecular mass standards in kilodaltons. The numbers between the photographs show the time of incubation with PK. CN represents soluble biotin-PK starting material and XIIf represents βFXIIa-cleaved biotin-PK analyzed by SDS-PAGE in the absence of cells.
Characterization of the FXII-independent, HUVEC-mediated PK activation.
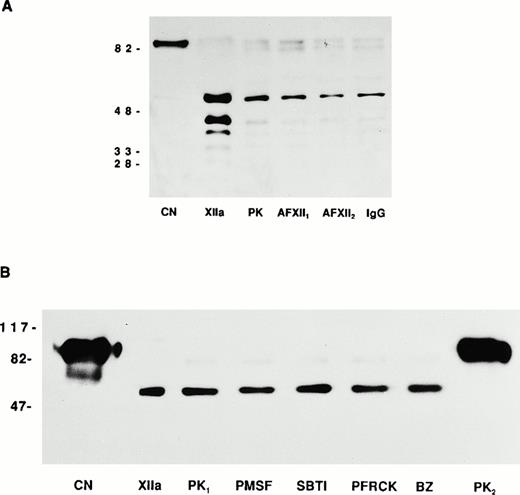
Investigations were then performed to determine the mechanism by which kallikrein was generated from the cell-associated HK•PK complex. Studies were first performed to determine if residual tissue culture media, which contained 2% fetal calf serum, was a source of activated FXII. Tissue culture media contained less than 0.0001 U/mL activated FXII coagulant activity relative to pooled normal plasma. A second set of investigations was performed to determine the chemical class of protease inhibitor(s) capable of blocking the generation of kallikrein. Initial studies were directed at determining if factor XIIa, kallikrein, or another serine protease could be responsible for the PK activation seen when bound to HK on HUVECs. In view of the fact that serine protease inhibitors would directly block kallikrein amidolytic activity, these experiments determined if serine protease inhibitors could block the generation of kallikrein, as indicated by a change in the conversion of PK to kallikrein assessed by SDS-PAGE, an event that is independent of kallikrein amidolytic activity (Fig 6). Biotin-PK migrated a thick band between 88 and 85 kD (Fig 6A). αFXIIa alone produced multiple breakdown products of biotin-PK that migrated at 51, 40, 37, and 34 kD (Fig 6A). When biotin-PK was bound to HK on HUVECs, activated PK migrated predominantly at the 51-kD band with minor bands seen at 40, 37, and 34 kD (Fig 6A). No change in the migration of endothelial cell-associated biotin-PK was seen in the presence of neutralizing concentrations of two antibodies to FXII or IgG (Fig 6A). These data indicated that the PK activating enzyme(s) was not due to a protein antigenically related to plasma factor XIIa that might have been present either in the PK preparation or associated with endothelial cells.
Endothelial cell prekallikrein activation. (A) CN represents biotin-PK directly added to the SDS-PAGE. XIIa represents factor XIIa-cleaved soluble biotin-PK. PK represents the form of 20 nmol/L biotin-PK bound to HK on HUVECs. In this experiment, HUVECs were incubated with 20 nmol/L HK for 1 hour at 37°C. After washing the cells, 20 nmol/L biotin-PK was added in the absence (PK) or presence of two neutralizing antibodies to FXII (AFXII1 and AFXII2) or normal goat IgG (IgG). The biotin-PK was detected by chemilluminesence. The figure is a photograph of a 10% SDS-PAGE after the proteins were reduced with 5% β-mercaptoethanol and boiling. (B) CN represents biotin-PK directly added to the SDS-PAGE. XIIa represents factor XIIa-cleaved soluble biotin-PK. In this experiment, HUVECs were treated with 20 nmol/L HK for 1 hour at 37°C. After washing the cells, 20 nmol/L biotin-PK was added in the absence (PK1) or presence of PMSF (1 mmol/L), SBTI (2 μg/mL), Pro-Phe-Arg-chloromethylketone (PFRCK; 100 μmol/L), or benzamidine (BZ; 1 mmol/L). PK2 was 40 nmol/L biotin-PK bound to HUVECs in the absence of HK. The biotin-PK was detected by chemilluminesence. The figure is a photograph of a 7% SDS-PAGE after the proteins were reduced with 5% β-mercaptoethanol and boiling.
Endothelial cell prekallikrein activation. (A) CN represents biotin-PK directly added to the SDS-PAGE. XIIa represents factor XIIa-cleaved soluble biotin-PK. PK represents the form of 20 nmol/L biotin-PK bound to HK on HUVECs. In this experiment, HUVECs were incubated with 20 nmol/L HK for 1 hour at 37°C. After washing the cells, 20 nmol/L biotin-PK was added in the absence (PK) or presence of two neutralizing antibodies to FXII (AFXII1 and AFXII2) or normal goat IgG (IgG). The biotin-PK was detected by chemilluminesence. The figure is a photograph of a 10% SDS-PAGE after the proteins were reduced with 5% β-mercaptoethanol and boiling. (B) CN represents biotin-PK directly added to the SDS-PAGE. XIIa represents factor XIIa-cleaved soluble biotin-PK. In this experiment, HUVECs were treated with 20 nmol/L HK for 1 hour at 37°C. After washing the cells, 20 nmol/L biotin-PK was added in the absence (PK1) or presence of PMSF (1 mmol/L), SBTI (2 μg/mL), Pro-Phe-Arg-chloromethylketone (PFRCK; 100 μmol/L), or benzamidine (BZ; 1 mmol/L). PK2 was 40 nmol/L biotin-PK bound to HUVECs in the absence of HK. The biotin-PK was detected by chemilluminesence. The figure is a photograph of a 7% SDS-PAGE after the proteins were reduced with 5% β-mercaptoethanol and boiling.
Additional experiments were performed to exclude the possibility of factor XIIa, kallikrein, or another serine protease contaminating the preparations to account for PK activation. Biotin-PK migrated as a thick band at about 88-85 kD and contained a minor fragment at about 66 kD (Fig 6B). When the biotin-PK was activated with αFXIIa, greater than 95% of the labeled PK now migrated at the 51-kD band identical to the migration of biotin-PK bound to HK on HUVECs (designated PK1; Fig 6B). This finding was distinctly different from the migration of PK bound to HUVECs in the absence of HK, which remained greater than 95% as a doublet of 88-85 kD (designated PK2; Fig 6B). When the HK and biotin-PK assembled on HUVECs were incubated with either 1 mmol/L PMSF, 1 mg/mL of SBTI, 10 μmol/L Pro-Phe-Arg-chloromethylketone, or 1 mmol/L benzamidine, biotin-PK bound to HUVECs also migrated greater than 95% as a 51-kD protein (Fig6B). These latter data showed that serine protease inhibitors in concentrations sufficient to inhibit 20 nmol/L αFXIIa or kallikrein, or other serine proteases, were not able to prevent PK activation. These data strongly suggested that the PK activating enzyme was not a serine protease.
Additional investigations were performed to determine the class of protease inhibitor that blocked PK activation when bound to HK on HUVECs. Antipain, cysteine, HgCl2, and 2-mercaptoethanol were potent inhibitors of the membrane-associated, PK-activating enzyme, suggesting that the enzyme was a thiol protease (Table 1). Accordingly, both DTT and glutathione also inhibited the enzyme, but these compounds also decreased kallikrein activity itself (Table 1). Z-Phe-OH and one preparation of a calpain inhibitor also blocked the enzyme, but the enzyme itself cannot be calpain because E64 did not inhibit its activity (Table 1). However, the enzyme(s) differs from other thiol proteases, because iodoacetamide, iodoacetic acid, N-ethylmalimide, cystatin, and hydroxy-mercuribenzoic acid did not block the membrane-associated, PK-activating enzyme (Table 1).
Inhibition Profile of Endothelial Cell Prekallikrein-Activating Enzyme(s)
| Inhibitor Class . | Inhibitor . | [Inhibitor]* . | % Inhibition† . | % Kallikrein Inhibition‡ . |
|---|---|---|---|---|
| Cysteine protease inhibitors | Antipain | 100 μmol/L | 92 ± 7.1 | ≤5% |
| Cysteine | 10 mmol/L | 99 ± 3.4 | ≤5% | |
| HgCl2 | 1 mmol/L | 96 ± 3.6 | ≤5% | |
| DTT | 10 mmol/L | 99 ± 3.5 | 28.6% | |
| 2-Mercaptoethanol | 5% | 98 ± 2.3 | ≤5% | |
| Glutathione | 100 μmol/L | 62 ± 4.5 | 10.4% | |
| Calpain Inhibitor | 10 mmol/L | 46 ± 3.6 | ≤5% | |
| Z-Phe-OH | 1 mmol/L | 45 ± 2.4 | ≤5% | |
| E64 | 10 mmol/L | 2.5 ± 3.8 | ≤5% | |
| Iodoacetamide | 10 mmol/L | 2.5 ± 1.5 | ≤5% | |
| Iodoacetic Acid | 10 mmol/L | 3.1 ± 1.8 | ≤5% | |
| N-ethylmalimide | 3 mmol/L | 0.3 ± 0.1 | Not done | |
| Cystatin | 1 mmol/L | 1.0 ± 0.3 | ≤5% | |
| OH-Mercuribenoic Acid | 10 mmol/L | 1.2 ± 0.1 | ≤5% | |
| Metalloprotease inhibitors | Zincov | 100 μmol/L | 20 ± 2.5 | ≤5% |
| TIMP-1 | 20 μg/mL | 15 ± 2.9 | ≤5% | |
| TIMP-2 | 20 μg/mL | 0.4 ± 0.2 | ≤5% | |
| BB94 | 15 μmol/L | 0.1 ± 0.0 | ≤5% | |
| Angiotensin converting enzyme inhibitors | Lisinopril Enalapril | 10 mmol/L 10 mmol/L | 1.1 ± 0.4 0.4 ± 0.2 | ≤5% ≤5% |
| Neutral endopeptidase inhibitor | Phosphoramidon | 10 mmol/L | 16 ± 3.2 | ≤5% |
| Aspartic protease inhibitor | Pepstatin A | 10 mmol/L | 0.2 ± 0.1 | ≤5% |
| Inhibitor Class . | Inhibitor . | [Inhibitor]* . | % Inhibition† . | % Kallikrein Inhibition‡ . |
|---|---|---|---|---|
| Cysteine protease inhibitors | Antipain | 100 μmol/L | 92 ± 7.1 | ≤5% |
| Cysteine | 10 mmol/L | 99 ± 3.4 | ≤5% | |
| HgCl2 | 1 mmol/L | 96 ± 3.6 | ≤5% | |
| DTT | 10 mmol/L | 99 ± 3.5 | 28.6% | |
| 2-Mercaptoethanol | 5% | 98 ± 2.3 | ≤5% | |
| Glutathione | 100 μmol/L | 62 ± 4.5 | 10.4% | |
| Calpain Inhibitor | 10 mmol/L | 46 ± 3.6 | ≤5% | |
| Z-Phe-OH | 1 mmol/L | 45 ± 2.4 | ≤5% | |
| E64 | 10 mmol/L | 2.5 ± 3.8 | ≤5% | |
| Iodoacetamide | 10 mmol/L | 2.5 ± 1.5 | ≤5% | |
| Iodoacetic Acid | 10 mmol/L | 3.1 ± 1.8 | ≤5% | |
| N-ethylmalimide | 3 mmol/L | 0.3 ± 0.1 | Not done | |
| Cystatin | 1 mmol/L | 1.0 ± 0.3 | ≤5% | |
| OH-Mercuribenoic Acid | 10 mmol/L | 1.2 ± 0.1 | ≤5% | |
| Metalloprotease inhibitors | Zincov | 100 μmol/L | 20 ± 2.5 | ≤5% |
| TIMP-1 | 20 μg/mL | 15 ± 2.9 | ≤5% | |
| TIMP-2 | 20 μg/mL | 0.4 ± 0.2 | ≤5% | |
| BB94 | 15 μmol/L | 0.1 ± 0.0 | ≤5% | |
| Angiotensin converting enzyme inhibitors | Lisinopril Enalapril | 10 mmol/L 10 mmol/L | 1.1 ± 0.4 0.4 ± 0.2 | ≤5% ≤5% |
| Neutral endopeptidase inhibitor | Phosphoramidon | 10 mmol/L | 16 ± 3.2 | ≤5% |
| Aspartic protease inhibitor | Pepstatin A | 10 mmol/L | 0.2 ± 0.1 | ≤5% |
*The values given represent the lowest concentration that gave the maximal level of inhibition of kallikrein-generating activity.
The values given represent the mean ± SD of inhibition of the prekallikrein-activating enzyme associated with HUVECs.
The values given represent the percentage of inhibition of kallikrein itself by the protease inhibitor.
In addition to thiol protease inhibitors, certain metal ion chelators also inhibited PK activation when bound to HK on HUVECs. EDTA, EGTA, 1,10-phenanthroline, the impermeable bathophenanthroline, or sodium hydrosulfite inhibited the PK-activating enzyme ≥86% (data not shown). The metalloprotease inhibitor zincov also decreased activity of the PK-activating enzyme (Table 1). However, the enzyme cannot be a known matrix metalloprotease because TIMP-1, TIMP-2, and BB94 were not inhibitory (Table 1). In view of the observations that PK is not activated unless bound to HK on HUVECs and that Zn+2 is essential for HK binding to HUVECs, the inhibitory effect of the metal ion chelators is likely attributable to chelation of the Zn+2 and removal of the HK•PK complex from the HUVEC membrane, rather than to a direct inhibitory effect on the PK-activating enzyme itself.
The angiotensin converting enzyme inhibitors, lisinopril and enalapril, had no inhibitory effect (Table 1). Likewise, phosphoramidon, an inhibitor of neutral endopeptidase, and pepstatin A, an aspartic protease inhibitor, did not interfere with the activity of the PK-activating enzyme (Table 1). Thus, our data indicate that the HUVEC PK-activating enzyme may be a thiol protease whose activity is blocked with when the divalent cations necessary for the HK•PK complex are chelated by certain metalloprotease inhibitors.
Effect of PK activation on cleavage of HK.
We then asked whether kallikrein formed on HUVECs was able to cleave its natural substrate, HK (Fig 7). Soluble125I-HK migrated predominantly as a 120-kD protein under reduced conditions. When incubated with HUVECs for 30 minutes,125I-HK (120 kD) was not cleaved to any great extent. A few lower molecular weight bands were evident by 1 minute; however, these did not increase in intensity over the next 30 minutes (Fig 7A). These data are consistent with our previous findings that endothelial cell bound exogenous HK is not substantially cleaved upon binding.21 In contrast, when HUVEC-bound125I-HK was incubated with PK (20 nmol/L), the intensity of the 120-kD band was reduced by 75% and new bands appeared at 64-55 and 46 kD within the first minutes that constituted 61% and 14% of the protein, respectively (Fig 7B); by 30 minutes, the 120-kD band had disappeared completely and a new 40-kD HK band progressively increased in intensity ultimately accounting for 14% of the total protein. These data indicated that the kallikrein formed on HUVECs in the HK•PK complex can cleave its receptor and native substrate, HK.
Effect of PK activation on the cleavage of HK. HUVECs were preincubated with 20 nmol/L 125I-HK for 1 hour at 37°C. Unbound 125I-HK was removed and either buffer (A) or 20 nmol/L PK (B) was added for 1 to 30 minutes. The cells were washed and solubilized in electrophoresis sample buffer containing 2% β-mercaptoethanol. The proteins were boiled, separated on 10% SDS-PAGE, and analyzed by autoradiography. The numbers to the left of the photographs represent molecular mass standards in kilodaltons. The numbers at the bottom of the gels refer to the time (in minutes) that the cells were incubated with PK. CN refers to 125I-HK directly added to the gel.
Effect of PK activation on the cleavage of HK. HUVECs were preincubated with 20 nmol/L 125I-HK for 1 hour at 37°C. Unbound 125I-HK was removed and either buffer (A) or 20 nmol/L PK (B) was added for 1 to 30 minutes. The cells were washed and solubilized in electrophoresis sample buffer containing 2% β-mercaptoethanol. The proteins were boiled, separated on 10% SDS-PAGE, and analyzed by autoradiography. The numbers to the left of the photographs represent molecular mass standards in kilodaltons. The numbers at the bottom of the gels refer to the time (in minutes) that the cells were incubated with PK. CN refers to 125I-HK directly added to the gel.
The effect of PK activation on two-chain urokinase and plasmin formation.
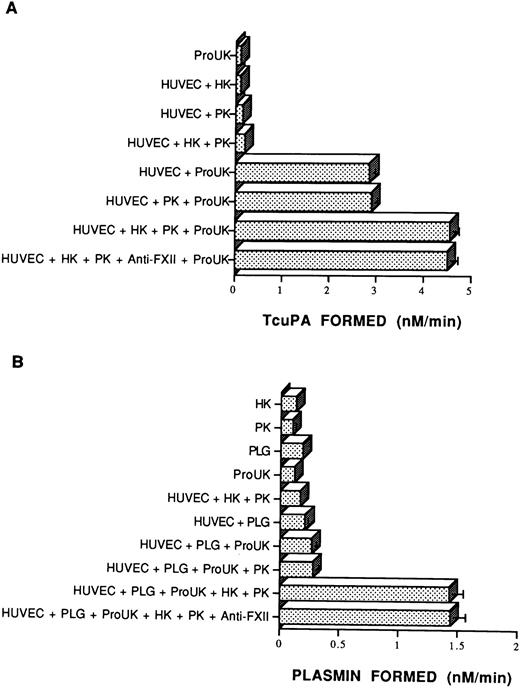
Because kallikrein is a potent activator of Pro-UK in vitro,15 the capacity of kallikrein formed on HUVECs to activate Pro-UK was examined (Fig 8A). Incubation of Pro-UK with HUVECs increased urokinase activity almost threefold over that seen with Pro-UK alone.45 The addition of HK and PK to HUVECs increased the level of urokinase activity an additional 1.6-fold (Fig8A). In these experiments, no attempt was made to inhibit the synthesis of plasminogen activator inhibitor-1. The same amount of Pro-UK activation occurred in the presence of a neutralizing concentration of an antibody to FXII (Fig 8A).
The influence of HK and PK on pro-urokinase and plasminogen activation. (A) Pro-urokinase activation. Empty microtiter plate wells or wells coated with a monolayer of endothelial cells (HUVECs) were incubated with HK (20 nmol/L) or buffer for 1 hour. Unbound HK was removed and the cells were incubated with PK (20 nmol/L) for another 1 hour and washed. Pro-UK (20 nmol/L) and 0.6 mmol/L S2444 were added to empty wells or wells coated with HUVECs and hydrolysis was monitored continuously over 75 minutes at 37°C. In one set of experiments, 0.4 mg/mL of a neutralizing antibody to FXII was added along with the PK. Formation of tcuPA was determined by comparing substrate hydrolysis on cells with known concentrations of soluble tcuPA. The data presented are the mean ± SEM of three experiments. (B) Plasminogen activation. Empty microtiter plate wells or wells coated with a monolayer of HUVECs were incubated for 1 hour with 1 μmol/L plasminogen (PLG) before 0.3 mmol/L S2251 was added either alone or in the presence of 2 nmol/L Pro-UK. In other experiments, HUVEC-coated wells were incubated for 1 hour with 20 nmol/L HK. After removal of the HK, the wells were incubated with 20 nmol/L PK for another 1 hour. After removal of the excess PK, the cells were incubated with 1 μmol/L plasminogen (PLG) for a third hour. As indicated, in one case, 0.4 mg/mL of a neutralizing antibody to FXII was added along with the PK. Hydrolysis of the substrate was measured over 210 minutes at 37°C. Plasmin formation was determined using a standard curve made by adding known amounts of purified plasmin to S2251 (see the Materials and Methods). The data shown are mean ± SEM of four independent experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually.
The influence of HK and PK on pro-urokinase and plasminogen activation. (A) Pro-urokinase activation. Empty microtiter plate wells or wells coated with a monolayer of endothelial cells (HUVECs) were incubated with HK (20 nmol/L) or buffer for 1 hour. Unbound HK was removed and the cells were incubated with PK (20 nmol/L) for another 1 hour and washed. Pro-UK (20 nmol/L) and 0.6 mmol/L S2444 were added to empty wells or wells coated with HUVECs and hydrolysis was monitored continuously over 75 minutes at 37°C. In one set of experiments, 0.4 mg/mL of a neutralizing antibody to FXII was added along with the PK. Formation of tcuPA was determined by comparing substrate hydrolysis on cells with known concentrations of soluble tcuPA. The data presented are the mean ± SEM of three experiments. (B) Plasminogen activation. Empty microtiter plate wells or wells coated with a monolayer of HUVECs were incubated for 1 hour with 1 μmol/L plasminogen (PLG) before 0.3 mmol/L S2251 was added either alone or in the presence of 2 nmol/L Pro-UK. In other experiments, HUVEC-coated wells were incubated for 1 hour with 20 nmol/L HK. After removal of the HK, the wells were incubated with 20 nmol/L PK for another 1 hour. After removal of the excess PK, the cells were incubated with 1 μmol/L plasminogen (PLG) for a third hour. As indicated, in one case, 0.4 mg/mL of a neutralizing antibody to FXII was added along with the PK. Hydrolysis of the substrate was measured over 210 minutes at 37°C. Plasmin formation was determined using a standard curve made by adding known amounts of purified plasmin to S2251 (see the Materials and Methods). The data shown are mean ± SEM of four independent experiments. The absence of standard error bars in some columns indicates that the variation was too little to portray visually.
Additional experiments were then performed to elucidate the effect of contact proteins on the kinetics of Pro-UK activation on the endothelial cell surface. The Km of Pro-UK activation (61 to 86 nmol/L) was not changed when Pro-UK was activated in the absence of either PK or HK compared with that seen when both were present (64 nmol/L; Table 2). However, the Vmax of Pro-UK activation (4 to 1.4 nmol/L/min) on HUVECs was reduced 2.5- to 7.1-fold when either PK or HK were omitted compared with cells preincubated with both HK and PK (10 nmol/L/min; P ≤ .01 v each condition). Addition of FXII had little or no effect on the kinetics of Pro-UK activation (Table 2). These data indicated that added FXII was not required for optimal Pro-UK activation by the assembled HK•PK complex on HUVECs.
Activation of Pro-Urokinase by the Contact Proteins
| Condition* . | Km (nmol/L) . | Vmax (nmol/L/min) . |
|---|---|---|
| HK + PK + FXII† | 135 ± 81 | 14.5 ± 8 |
| HK + PK | 64 ± 5 | 10 ± 0.1 |
| HK + FXII | 61 ± 12 | 1.4 ± 0.7 |
| PK + FXII | 86 ± 44 | 4 ± 1.4 |
| Condition* . | Km (nmol/L) . | Vmax (nmol/L/min) . |
|---|---|---|
| HK + PK + FXII† | 135 ± 81 | 14.5 ± 8 |
| HK + PK | 64 ± 5 | 10 ± 0.1 |
| HK + FXII | 61 ± 12 | 1.4 ± 0.7 |
| PK + FXII | 86 ± 44 | 4 ± 1.4 |
*HUVECs were preincubated with 20 nmol/L HK for 1 hour at 37°C. Unbound HK was removed, HUVECs were incubated with 20 nmol/L PK for another 1 hour, and the cells were washed. The capacity of the cell-associated PK/HK complex to activate Pro-UK was determined by adding 0.6 mmol/L S2444, 20 nmol/L factor XII (FXII) where indicated, and variable concentrations of pro-urokinase (5 to 1,000 nmol/L). Hydrolysis was measured over 75 minutes at 37°C. Cell-associated urokinase activity was determined using a standard curve made with known amounts of soluble tcuPA. The data presented are the mean ± SD of three experiments.
The reactants added included high molecular weight kininogen (HK), prekallikrein (PK), and factor XII (FXII), as indicated.
Investigations next were performed to determine if the assembly of the contact proteins and Pro-UK on HUVECs with bound plasminogen translated into enhanced plasmin formation (Fig 8B). The addition of plasminogen alone onto HUVEC monolayers was associated with little activity. Consistent with previous reports, a little more plasmin was generated when Pro-UK alone was added to plasminogen on HUVECs45 (Fig8B). However, the addition of HK and PK to Pro-UK and plasminogen was associated with a 4.3-fold increase in plasminogen activation. Furthermore, the same increase in plasmin formation occurred when a neutralizing concentration of antibody to FXII was present (Fig 8B). Moreover, the addition of FXII and βFXIIa did not potentiate plasmin formation above that seen with the HK and PK complex alone (data not shown). Furthermore, the substitution of kallikrein for PK resulted in a 1.7-fold decrease in plasmin formation (data not shown). These results indicated that optimal formation of measured plasmin on HUVECs did not require FXII or its activated forms.
DISCUSSION
These studies indicate that assembly of contact proteins, HK and PK, on cultured endothelial cells leads to the formation of kallikrein, which, in turn, cleaves HK, presumably liberating bradykinin, and promotes the activation of Pro-UK and the generation of plasmin. Kallikrein formation on HUVECs is critically dependent on the sequential binding of HK and PK and was independent of added or endogenous FXII or αFXIIa-like enzyme. Furthermore, the extent of kallikrein activity is autoregulated, because, once formed, the enzyme proteolyzes its receptor (HK) resulting in its liberation from the HUVEC surface. These results stand in stark contrast to the well-established role of FXII in the activation of PK in plasma and on artificial surfaces. These studies show for the first time contact system activation on a biologic surface in the absence of a negatively charged artificial surface. It is noteworthy to point out that the so-called elusive, physiologic negatively charged surface for contact protein zymogen activation may really be the assembly of contact proteins by binding to putative receptors on endothelial cells or other cell membranes. Recognition of a PK activating mechanism on HUVECs supports this notion. The finding that the cell surface binding region on HK's domain 5 is identical to its artificial surface binding region on domain 5 also lends credence to this hypothesis.28
The initial requirement for kallikrein formation on HUVECs is the binding of HK to HUVECs. Our data indicate clearly that HK is the predominant binding site for PK. PK binding to HUVECs is inhibited by MoAbs that block the sites on PK and on HK necessary for them to form a complex, whereas an antibody to a neighboring domain of HK has no effect. Binding of PK is also inhibited by a peptide corresponding to its binding site for PK on HK. Furthermore, binding of PK to HUVECs is optimal at concentrations of HK that saturate its cellular receptors.25 The apparent Kd of biotin-PK binding to HUVEC-bound HK is 23 nmol/L, similar to theKd of complex formation between HK and PK in solution.40 46 However, these data are an estimate of the kinetics of this interaction because PK binding to HK on HUVECs is not in apparent equilibrium. Rather, upon binding to cell-associated HK, PK is activated to kallikrein, which, in turn, proteolyzes its receptor (ie, HK), allowing for its removal from the cell membrane.
PK also binds to HUVECs in the absence of added HK. We estimate at least 17 million binding sites for PK on HUVECs in the presence of HK but only 10 million binding sites for HK.25 MoAb HKL16, which completely blocks PK binding to HUVECs preincubated with HK,35 36 inhibits only 75% of PK binding to washed HUVECs in the absence of added HK. MoAb HKL16 blocks PK binding but does not recognize bovine HK, confirming the expression of human HK on HUVEC. However, because 25% of PK binding in the absence of added HK is not blocked by HKL16, a distinct binding site(s) that is not human HK is also present. Possible candidates include bovine HK acquired from the conditioned medium or, more likely, an endothelial cell protein that forms a 116-kD SDS-stable complex with PK or kallikrein (see below). It is of interest that, in the absence of added HK, the protein(s) that participates in formation of the 116-kD stable complex accounts for about 28% of PK-kallikrein binding, whereas in the presence of exogenous HK, it accounts for only 5% of total binding. Because 95% of the specific binding of PK to HUVECs can be attributed to endogenous and added human HK and because PK is found in plasma complexed with HK, our subsequent studies were devoted to understanding the biologic consequences of forming HK•PK complexes on endothelial cells.
We first asked whether PK bound to HK on HUVECs is activated through the same mechanism that operates in plasma. To our surprise, kallikrein amidolytic activity is generated when the HK•PK complex formed on cells and kallikrein itself is evident on SDS-PAGE under reduced conditions. This finding suggests that HUVECs have the capacity to activate PK in the absence of exogenous FXII. Indeed, the addition of FXII, αFXIIa, or βFXIIa does not enhance PK activation when HK and HUVECs are present, in contrast to the requirement for FXII and its activated species for PK to be activated on artificial surfaces. Indeed, more kallikrein activity is measured when PK and HK are permitted to assemble on HUVECs in the absence of exogenous FXII, αFXIIa, or βFXIIa or kallikrein (Fig 4A). The reasons for these findings are not completely known, but it is possible that addition of these enzymes proteolyze HK directly or accelerate the activation of PK, which proteolyzes HK, leading to the loss of the complex from the cell surface at a faster rate.18 In support of this interpretation, we showed that activating PK when bound to HK on HUVECs actually cleaves the HK it is bound to. Furthermore, it has been reported that 50% of HUVEC-bound 125I-PK converted to kallikrein in the presence of αFXIIa dissociates from the cell membranes within 15 minutes.18 A second possibility for increased measured kallikrein activity from the activating HK•PK complex comes from the observation that PK forms a 116-kD SDS-stable complex on HUVECs. Kallikrein formed from cell-bound HK•PK complex may be afforded relative protection from inhibitors compared with that produced by activated FXII, αFXIIa, or βFXIIa added directly to the cells or when kallikrein is substituted for PK. The endothelial cell component of the 116-kD complex was not identified but is unlikely to be plasminogen activator inhibitor-1 or C1-inhibitor because, on immunoblot, neither antigen is detected in the 116-kD band.
The specific enzyme(s) responsible for the conversion of PK to kallikrein by HUVECs cannot be determined from our studies, but our experiments do provide some insights into its mechanism of action and cellular location. First, no serine protease inhibitor or the addition of neutralizing antibodies to FXII prevented cleavage of PK bound to HK on HUVECs. It is theoretically possible that a cell-bound form of αFXIIa that is protected from high concentrations of serine protease inhibitors and a neutralizing concentration of antibody to FXII could be responsible for this cleavage. However, the concentrations of these inhibitors exceed any possible cell-bound αFXIIa by many orders of magnitude and there is no precedent for this possibility. Second, the finding that PK activation is blocked by antipain, HgCl2, cysteine, β-mercaptoethanol, and DTT indicates that kallikrein formation is by an enzyme that is not factor XIIa-like and not due exclusively to a conformational change upon binding. Third, at least one PK activating enzyme(s) has properties consistent with it being a thiol protease. It does not appear to be calpain itself because E64 does not inhibit kallikrein formation at all. Furthermore, HK is itself a potent cysteine protease inhibitor (Ki = 0.71 nmol/L), but it clearly promoted rather than inhibited PK activation.47 However, it could be argued that the capacity of HK to function as a cysteine protease inhibitor was nullified by it being bound to endothelial cells, because its cellular binding site on domain 3 overlaps with the region that expresses cysteine protease inhibitory activity.27 Inhibition by certain metal ion chelating agents can be explained by these compounds removing Zn+2 from the complex of HK•PK, thus dissolving the protein-protein assembly necessary for the PK activating enzyme to function. Last, the PK activator(s) on HUVECs also appears to have different requirements than a Hageman factor activator, which has been found previously in homogenates of cultured rabbit endothelial cells.48
Kallikrein formed on the surface of HUVECs from the HK•PK complex is positioned to exert several potentially important functions. First, cell-bound HK was cleaved by the formed kallikrein on HUVECs. This result may be an important control step in this pathway, releasing kallikrein from its binding site. Second, the pattern of cleavage of HK is identical to that seen in plasma and on cells by kallikrein when bradykinin is liberated.44,49 Bradykinin formed in proximity to the endothelium may be especially potent, because, theoretically, it could engage its receptors before inactivation by plasma kininases.50 Moreover, bradykinin liberation can result in potent tissue plasminogen activator release from endothelial cells in vivo.51 52
Kallikrein formed from HK•PK complex also activated Pro-UK, which, in turn, caused a 4.3-fold increase in plasminogen activation on HUVECs. In that these experiments were performed without taking measures to inactivate plasminogen activator inhibitor-1, they represent minimum estimates of urokinase activity and suggest that the kallikrein-dependent mechanism described here may be more important to cell surface plasminogen-activating activity than in plasma. The data may explain why mice genetically engineered to lack plasminogen have normal concentrations of tcuPA found in their urogenital tract.53 These investigations indicate a possible mechanism whereby a small amount of Pro-UK can be converted to tcuPA independent of plasmin, fibrin, or tissue plasminogen activators. This pathway may potentiate Pro-UK activation associated with its binding to its receptor.54
Supported in part by Grant No. 93/4124-0 from Fundacão de Amparoà Pesquisa do Estado de São Paulo to G.M., Grants No. HL35553 and HL52776 to A.H.S., and Grants No. HL40387 and HL50790 to D.B.C.
Address reprint requests to Alvin H. Schmaier, MD, University of Michigan, 5301 MSRB III, 1150 W Medical Center Dr, Ann Arbor, MI 48109-0640.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal