Abstract
Chromosomal translocations involving the immunoglobulin heavy chain (IGH) locus at chromosome 14q32 represent a common mechanism of oncogene activation in lymphoid malignancies. In multiple myeloma (MM), the most consistent chromosomal abnormality is the 14q+ marker, which originates in one third of cases through a t(11; 14)(q13; q32) chromosomal translocation; in the remaining cases, the identity of the partner chromosomes has not been well established. We used a Southern blot approach based on the linkage analysis of the joining (J) and the constant (C) μ, α, and γ regions to detect cases bearing IGH switch-mediated chromosomal translocations. We evaluated DNA of 88 nonkaryotyped patients with MM (78 cases) or plasma cell leukemia (PCL) (10 cases) and found the presence of “illegitimate” rearranged IGH fragments (no comigration between the J and C regions) in 21 cases. To confirm this analysis, we cloned the illegitimate rearranged fragments from three samples, and the molecular and fluorescent in situ hybridization (FISH) analyses indicated the presence of chromosomal translocations juxtaposing a switch IGH region to sequences from chromosomes 11q13 (one PCL case) or 4p16.3 (two MM cases). Interestingly, the breakpoints on 4p16.3 occurred about 14 kb apart in a genomic region located approximately 50 kb centromeric to the fibroblast growth-factor receptor 3 (FGFR3) gene. Moreover, Southern blot analysis using 4p16.3 genomic probes detected a rearrangement in an additional MM tumor. FISH analysis of the MM-derived KMS-11 cell line, reported to be associated with a t(4; 14)(p16.3; q32), showed that the FGFR3 gene was translocated on 14q32. High levels of FGFR3 mRNA expression were observed in the cloned MM tumors and KMS-11 cell line, but not in the cases that were apparently negative for this lesion. Furthermore, a point mutation at codon 373 in the transmembrane domain of the FGFR3 gene resulting in an amino acid substitution (Tyr → Cys) was detected in the KMS-11 cell line. These findings indicate that the t(4; 14)(p16.3; q32) represents a novel, recurrent chromosomal translocation in MM, and suggest that the FGFR3 gene may be the target of this abnormality and thus contribute to tumorigenesis in MM.
MULTIPLE MYELOMA (MM) is a malignant proliferation of bone marrow plasma cells characterized by osteolytic lesions, monoclonal gammopathy, and a wide spectrum of clinical entities, including localized or disseminated and indolent or aggressive forms, which may occur sequentially during the clinical course of the disease.1,2 The molecular pathogenesis of MM is still largely unknown and no genetic lesions specifically associated with this neoplasm (unlike other types of B-cell neoplasms), have yet been found. We and others have reported the presence of activated RAS oncogenes (predominantly N-RAS ) in a significant proportion of cases (30%), associated with different disease stages and the partial or complete lack of response to therapy.3,4 More recently, we have reported the occurrence of inactivating mutations of the tumor suppressor gene p53 in about 15% of cases, specifically associated with advanced and aggressive forms of MM and plasma cell leukemia.5
Chromosomal translocations affecting the IGH locus on 14q32 represent the mechanism of activation of a number of proto-oncogenes in B-cell lymphoid neoplasms.6 The best examples are the t(8; 14)(q24; q32), t(8; 22)(q24; q11), and t(2; 8)(p12; q24) chromosomal translocations leading to the deregulation of the c-MYC gene in 100% of Burkitt's lymphomas7; the t(14; 18)(q32; q21) involving the BCL-2 gene in 70% to 90% of follicular lymphomas8,9; and the t(11; 14)(q13; q32) involving the BCL-1/cyclin D1 locus in 70% to 95% of mantle cell lymphomas.10,11 Although cytogenetic analyses in MM are limited and difficult, mainly because of the low proliferation rate of malignant plasma cells, a 14q+ marker has been reported in 20% to 40% of tumors.12,13 In almost 30% of cases, this is the result of a t(11; 14)(q13; q32) chromosomal translocation involving the chromosome band where the putative oncogene BCL-1/cyclin D1 is located. However, the rearrangements of the BCL-1/cyclin D1 regions that are frequently involved in mantle-cell lymphoma rarely occur in MM,14-16 although an overexpression of cyclin D1 can be found in MM cell lines carrying such a chromosomal translocation.17,18 In the remaining cases with a detectable 14q+ marker, the translocation partners have been rarely identified.12 13
In an attempt to identify novel genetic lesions involved in the molecular pathogenesis of MM, we used Southern blot to detect rearranged IGH alleles that may be putative candidates for chromosomal translocations. We reasoned that (1) the breakpoint location in chromosomal translocations involving the IGH locus in MM would occur within the switch regions, as malignant plasma cells, such as murine plasmacytoma, have generally undergone IGH isotype switching,19 and that (2) the linkage between the joining (J) and the constant (C) region, which normally occurs in a physiologically rearranged IGH (“legitimate”) allele, would not be found in switch-translocated IGH (“illegitimate”) alleles, as a result of a chromosomal translocation. This linkage can be shown in Southern blot analysis by evaluating the comigration of restriction fragments hybridizing to both J and C probes. A similar approach has been extensively used by us and others20,21 to define the breakpoint location within the IGH locus in the endemic and sporadic forms of Burkitt's lymphoma, and its general validity has been supported by the molecular cloning of several cases.20
This study was performed on tumor biopsies from 88 patients with plasma cell disorders (MM or plasma cell leukemia [PCL]) for which no karyotypic analyses were available, and illegitimate rearranged IGH alleles were identified in about 25% of the cases. Molecular cloning and the fluorescent in situ hybridization (FISH) analysis of three cases showed the presence of a t(11; 14)(q13; q32) in one case, and a novel t(4; 14)(p16.3; q32) chromosomal translocation in the others. The breakpoints on 4p16.3 clustered in a genomic region located approximately 2 Mb telomeric to the gene of Huntington's disease (HD)22 and about 50 kb centromeric to the fibroblast growth-factor receptor 3 (FGFR3) gene.23 Southern blot analysis using probes derived from this region detected a rearrangement in an additional MM tumor. Furthermore, we show that the FGFR3 gene is translocated on chromosome 14q32 and is overexpressed in cases carrying the t(4; 14)(p16.3; q32), which suggests that it may represent the target of this chromosomal translocation in MM.
MATERIALS AND METHODS
Pathological samples and MM cell lines. The bone marrow or peripheral blood samples of the 88 patients admitted at our Hematology Service were collected during standard diagnostic procedures. The diagnosis and clinical staging were made according to the criteria described by Durie and Salmon.24 Sixty-two patients were at first diagnosis (12 indolent and 50 in a symptomatic phase), 16 patients in relapse, and 10 patients were affected by PCL (seven at diagnosis and three in relapse). The U266 cell line was obtained from the American Type Culture Collection, Rockville, MD; the KMS-11 cell line was previously reported.25
Mononuclear cell suspensions of more than 95% viability were obtained from the pathological samples and prepared by Ficoll-Hypaque gradient centrifugation; in all cases, the percentage of malignant plasma cells was between 22% and 98% at morphologic and immunophenotypic analyses.
DNA preparation and Southern blot analysis. DNA was purified by proteinase K digestion, extraction with phenol-chloroform, and ethanol precipitation.26 A total of 10 μg of genomic DNA was digested with BamHI or other appropiate restriction enzymes, electrophoresed in a 0.7% agarose gel, and then denatured, neutralized, and transferred to nylon filters (Amersham International, Little Chalfont, UK). The filters were hybridized to probes that were 32P-labeled by the random priming method according to the manufacturer's specifications, washed in 0.5× SSC (NaCl/Na citrate)/1% sodium dodecyl sulfate (SDS) for 1 hour at 60°C, and then autoradiographed using an intensifying screen at −80°C.26
DNA probes. IGH gene rearrangement analysis was performed using the following probes: the 6.6-kb BamHI-HindIII fragment specific for the JH region27; the 1.3-kb EcoRI fragment for the Cμ region27; the 2.9-kb SmaI fragment containing part of the Cα1 region28; and the 7-kb HindIII-BamHI fragment containing the Cγ1 region.29 The FGFR3 cDNA clone HE8 used as a probe was kindly provided by Dr K. Alitalo (University of Helsinki, Helsinki, Finland).30
Molecular cloning. Genomic libraries from cases LB375 and LB1017 were constructed by complete digestion of genomic DNA with BamHI and the ligation of gel-purified fractions into the λEMBL3 phage vectors (Stratagene, La Jolla, CA). Rearranged BamHI fragments were isolated by screening with the respective constant IGH probes. The germ-line regions of chromosomes 4 were isolated by screening a genomic library from human placental DNA (Clontech, San Diego, CA) using probes derived from recombinant clones. Library screening and plaque isolation were performed according to established procedures.26 Inserts were analyzed by restriction enzyme mapping and subcloned into plasmid vector pGEM3 (Promega, Madison, WI) for further analysis.
RNA extraction and Northern blot analysis. Total RNA from human cell lines was prepared using the guanidium thiocyanate method. Total RNA from tumors and normal donors was prepared by using the Trizol reagent (GIBCO BRL, Gaithersburg, MD). Northern blot analysis was performed according to established procedures.26
DNA sequencing. DNA was sequenced on restriction fragments cloned into pGEM3 plasmid (Promega) by “dideoxy” chain-termination analysis using the Sequenase sequencing kit (USB, Cleveland, OH).
DNA amplification. A 164-bp fragment specific for the transmembrane (TM) domain of FGFR3, located to the middle of exon 10,31 was amplified using the following previously reported primers32: sense 5′-AGGAGCTGGTGGAGGCTGA-3′; antisense 5′-GGAGATCTTGTGCACGGTGG-3′. Amplification reactions were performed for 30 cycles under the following conditions: denaturing at 94°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 30 seconds. The amplified fragments were directly sequenced as previously described.5
cDNA amplification. One microgram of total RNA was used for the first strand cDNA synthesis in 20 μL reactions containing 10 mmol/L of each deoxynucleotide-S′-triphosphate (dNTP), 0.1 mmol/L dithiotheitol (DTT), 5× reverse transcription buffer, 1 U RNasin (Promega), 200 U Super-Script reverse transcriptase (GIBCO-BRL), and 10 pmol/L of pd(N)6 random examers (Pharmacia Biotech, Uppsala, Sweden). The reaction mixes were incubated at 42°C for 15 minutes, and polymerase chain reaction (PCR) amplifications were made by diluting 5 μL of first-strand cDNA from each individual case into a 50 μL PCR mixture. A 204-bp fragment containing the TM domain and the 3′ end of the third Ig-like loop was amplified using the antisense primer reported above and a sense primer specific for a sequence located within exon 931: 5′-TCTCATCACTCTGCGTGGC-3′ (nucleotides 1081-1099 of the published cDNA sequence, see Keegan et al33 ). Amplification reactions and direct DNA sequencing were performed as reported above.
FISH. Metaphase spreads were obtained from phytohemagglutinin (PHA)-stimulated normal peripheral blood lymphocytes and the KMS-11 cell line. Chromosome preparations were hybridized in situ with probes labeled with biotin nick translation, essentially as described,34 with minor modifications. Briefly, 200 ng of labeled probe was used for each experiment, and hybridization was performed at 37°C in 2× SSC, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, 5 μg Cot1 DNA (Boehringer, Mannheim, Germany), 3 μg of sonicated salmon sperm DNA, in a volume of 10 μL. Posthybridization washing was at 42°C in 2× SSC-50% formamide (3×), followed by three washes in 0.1× SSC at 60°C. Biotin-labelled DNA was detected using Cy3-conjugated avidin (Amersham). The chromosomes were identified by simultaneous 4′-6diamidino-2-phenylindolo (DAPI) staining, which produces a Q-banding pattern. Chromosome 4 and 14 painting probes were obtained by Alu-PCR amplification of somatic cell hybrids retaining only the human chromosome 4 or 1435; the painting generates a banding pattern corresponding to R-banding,36 which helps in the subregional identification of chromosomes. Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments, Princeton, NJ). Cy3 and Dapi fluorescence signals, detected using specific filters, were recorded separately as gray scale images. Pseudocoloring and image merging were performed using Adobe Photoshop software.
RESULTS
Identification by Southern blot analysis of IGH rearranged fragment candidates for translocation breakpoints in MM. The Southern blot approach used in our study focused on the analysis of the linkage between the various IGH regions as shown by the comigration of restriction fragments. In particular, this approach is based on the finding that normally rearranged joining-switch–constant IGH regions are generally contained on a novel BamHI restriction fragment; a translocation event involving the switch region should generate a rearranged BamHI fragment containing the 3′ constant region, but not the 5′ joining sequences. We, therefore, performed the Southern blot analysis by digesting the DNA with the BamHI restriction enzyme and subsequently hybridizing the same filter with the JH, Cμ, Cα1, and Cγ1 probes. Representative examples are shown in Fig 1. By hybridization with the Cμ probe, six cases were detected in which the rearranged Cμ fragment did not contain JH-specific sequences. The same analysis using the Cα1 constant probe allowed the identification of 11 MM tumors showing no comigration between the rearranged JH and Cα fragments. Finally, analysis with the Cγ1 constant probe detected four cases in which no apparent linkage between JH and Cγ regions could be observed in the rearranged Cγ fragments. Thus, the presence of rearranged fragments that are potential candidates for switch-mediated chromosomal translocations was detected in 21 of the 88 MM cases investigated (≈25%)
Southern blot analysis of IGH locus in MM samples. DNA was digested with the BamHI restriction enzyme and the nylon filters were subsequently hybridized to the probes indicated below. Germ-line bands are indicated in kilobases (kb) by dashes. The arrows indicate “illegitimate” IGH rearranged fragments that are candidates for chromosomal translocations and were cloned in cases LB375, LB1017, and LB411 (see text). (◃) Indicate comigrating JH and Cα rearranged fragments (“legitimate allele”) in MM cases expressing the IGH α isotype. (○) Indicate a 12-kb BamHI fragment cross-hybridizing with the JH probe in our experimental conditions. Germ-line JH and Cα BamHI fragments have approximately the same size in kb (*).
Southern blot analysis of IGH locus in MM samples. DNA was digested with the BamHI restriction enzyme and the nylon filters were subsequently hybridized to the probes indicated below. Germ-line bands are indicated in kilobases (kb) by dashes. The arrows indicate “illegitimate” IGH rearranged fragments that are candidates for chromosomal translocations and were cloned in cases LB375, LB1017, and LB411 (see text). (◃) Indicate comigrating JH and Cα rearranged fragments (“legitimate allele”) in MM cases expressing the IGH α isotype. (○) Indicate a 12-kb BamHI fragment cross-hybridizing with the JH probe in our experimental conditions. Germ-line JH and Cα BamHI fragments have approximately the same size in kb (*).
Molecular cloning of IGH breakpoints in MM identifies a novel t(4; 14)(p16.3; q32) chromosomal translocation. To confirm the presence of translocation breakpoints in the illegitimate IGH rearranged fragments, we cloned the fragments from tumors LB411, LB375, and LB1017 (see rearrangement patterns in Fig 1). Molecular cloning and FISH analysis of case LB411 showed the presence of a t(11; 14)(q13,q32) chromosomal translocation, which involved a novel region from the BCL-1/cyclin D1 locus (D. Ronchetti and A. Neri, manuscript in preparation). With regard to the other two cases (LB375 and LB1017), the genomic libraries (see Materials and Methods) were screened using the Cμ and Cα1 probes, respectively. As shown in Fig 2, the recombinant clones did not contain any IGH-specific sequences that could be shown by hybridization and nucleotide sequence analysis. In case LB375, the novel region was juxtaposed to the Sμ region and, in case LB1017, to the switch region of the Cα1 gene. FISH experiments using the recombinant clones as probes showed that the novel sequences were derived from the telomeric site of chromosome 4 (4p16.3) in both cases, thus indicating that both rearrangements were a result of a t(4; 14)(q16.3; q32) (Fig 3 and data not shown). A data base homology search of these novel sequences showed that the breakpoints occurred within a previously reported cosmid clone (L75b9) telomeric to the HD gene (approximately 2 Mb)22 and centromeric to the FGFR3 gene (approximately 50 kb).23 By screening a human placental genomic library, we next isolated two overlapping phage clones (λTwt and λAB3) spanning about 30 kb and found that the two breakpoints were located about 14 kb apart (see Fig 2) within sequencing gaps of the L75b9 clone (GenBank, accession number Z69653). Sequence analysis of regions across the 4p16.3 breakpoints showed the presence of repetitive sequences. To search for additional rearrangements in 4p16.3, we performed Southern blot analysis of BamHI and EcoRI digests from 16 MM tumors with illegitimate rearranged IGH alleles and 37 tumors without evidence of such alleles using three different probes described in Fig 2. A rearrangement was found in an MM tumor (LB278) without evidence of illegitimate alleles (Fig 4); it was detected using probe A/Bg1.3 and apparently occurred within the BamHI 5.5 kb fragment located between the breakpoint sites of cases LB375 and LB1017 (see scheme in Fig 2). While this work was in progress, Bergsagel et al37 reported the results of a similar study of MM-derived cell lines, which showed that two cell lines (KMS-11 and JIM3) and a primary tumor carried a t(4:14) translocation involving the 4p16.3. In particular, the breakpoints in the cell line KMS-1125 and in a tumor were reported to be localized within the L75b9 cosmid clone, but no restriction map of the breakpoint locations was provided.37
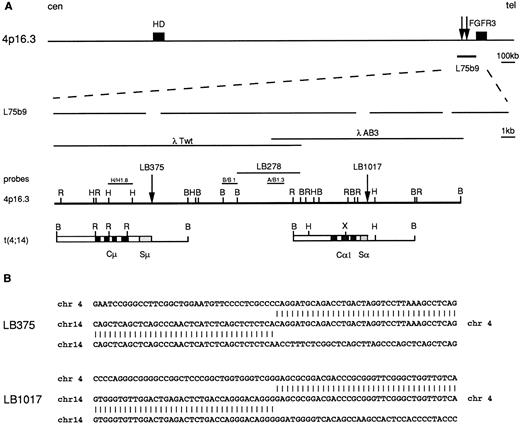
Molecular cloning of the chromosomal breakpoints from cases LB375 and LB1017. (A) Schematic representation of the cloned breakpoints and their respective germ-line counterparts (chr 14q32 and chr 4p16.3). From the top: a diagram of the 2 Mb cosmid and P1 conting region22 where the HD gene, the cosmid clone L75b9, and the FGFR3 gene23 are located; gaps in the expanded line representing the L75b9 clone indicate the sequencing gaps in the reported sequence22; the germ-line 4p16.3 restriction map, derived from phage clones λTwT and λAB3, and the probes used for Southern and Northern blot analyses (H/H1.8, B/B1, A/B1.3) are shown. The vertical arrows indicate the positions of the breakpoints; for case LB278, the BamHI fragment where the breakpoint is thought to occur is also indicated. Chromosome 14 sequences are indicated by open boxes with black or stippled boxes representing different IGH regions. Chromosome 4 regions are shown as solid lines. Restriction enzyme symbols: B, BamHI; R, EcoRI; H, HindIII; X, XhoI. (B) Nucleotide sequence analysis of the breakpoint regions in cases LB375 and LB1017 and their alignment with the corresponding 4p16.3 and 14q32 germ-line sequences.
Molecular cloning of the chromosomal breakpoints from cases LB375 and LB1017. (A) Schematic representation of the cloned breakpoints and their respective germ-line counterparts (chr 14q32 and chr 4p16.3). From the top: a diagram of the 2 Mb cosmid and P1 conting region22 where the HD gene, the cosmid clone L75b9, and the FGFR3 gene23 are located; gaps in the expanded line representing the L75b9 clone indicate the sequencing gaps in the reported sequence22; the germ-line 4p16.3 restriction map, derived from phage clones λTwT and λAB3, and the probes used for Southern and Northern blot analyses (H/H1.8, B/B1, A/B1.3) are shown. The vertical arrows indicate the positions of the breakpoints; for case LB278, the BamHI fragment where the breakpoint is thought to occur is also indicated. Chromosome 14 sequences are indicated by open boxes with black or stippled boxes representing different IGH regions. Chromosome 4 regions are shown as solid lines. Restriction enzyme symbols: B, BamHI; R, EcoRI; H, HindIII; X, XhoI. (B) Nucleotide sequence analysis of the breakpoint regions in cases LB375 and LB1017 and their alignment with the corresponding 4p16.3 and 14q32 germ-line sequences.
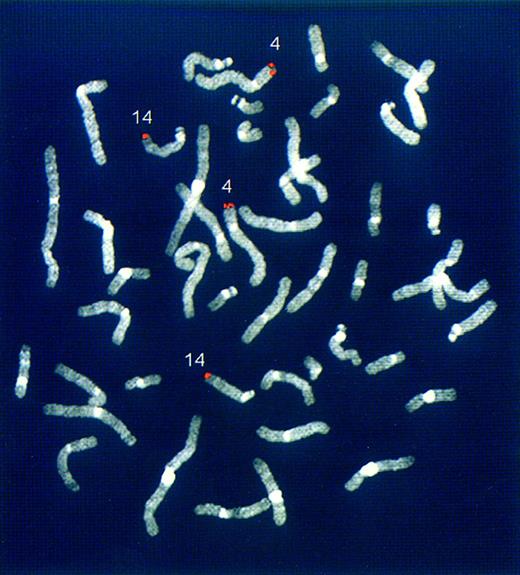
Chromosomal mapping of the novel genomic regions rearranged with the IGH locus in case LB375. FISH experiments were performed on normal human metaphases using the rearranged LB375 clone (see Fig 2) as probe. The hybridization signals (red) are located at 14q32 and 4p16.3 chromosomes.
Chromosomal mapping of the novel genomic regions rearranged with the IGH locus in case LB375. FISH experiments were performed on normal human metaphases using the rearranged LB375 clone (see Fig 2) as probe. The hybridization signals (red) are located at 14q32 and 4p16.3 chromosomes.
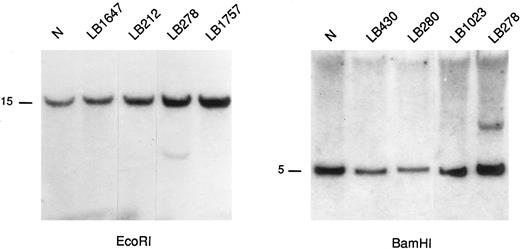
Southern blot analysis of representative MM samples using 4p16.3 probes. The DNAs were digested with the indicated restriction enzymes and hybridized with the A/Bg 1.3 probe (see Fig 2). A rearranged fragment is observed in case LB278 (see text). Germ-line bands are indicated in kilobases (kb).
Southern blot analysis of representative MM samples using 4p16.3 probes. The DNAs were digested with the indicated restriction enzymes and hybridized with the A/Bg 1.3 probe (see Fig 2). A rearranged fragment is observed in case LB278 (see text). Germ-line bands are indicated in kilobases (kb).
The FGFR3 gene is translocated on chromosome 14q32 as a result of a t(4; 14). The relatively short distance of the FGFR3 gene from the L75b9 clone22,23 prompted us to investigate its involvement in cases carrying a t(4; 14) (p16.3; q32) translocation. Unfortunately, viable cells from tumors LB375 and LB1017 were not available; therefore, we performed FISH analysis only on KMS-11 cell line. As illustrated in Fig 5A, the hybridization of KMS-11 metaphases with the FGFR3 cDNA clone30 clearly showed that the gene is translocated from one of the two chromosome 4 homologs to chromosome 14q32, thus confirming the presence of a t(4; 14)(p16.3; q32) translocation.37 Interestingly, the rearrangement of KMS-11 on 4p16.3 occurred within the region represented by the phage clone AB3 (see scheme in Fig 2), as the hybridization signals of this probe were found at the der(4) and der(14) putative chromosomes (Fig 5B). The nature of these chromosomes was confirmed by hybridization with painting probes representative of chromosomes 4 and 14 (data not shown).
FISH analyses of KMS-11 cell line. (A) Partial metaphase hybridized with a FGFR3 cDNA clone HE8.31 The signals (red) are evident at the 4p16.3 telomeric region of a shorter chromosome 4 (4s) and at the telomeric region of chromosome 14q, der(14), while absent in the normal chromosome 4 homolog, der(4). (B) Partial karyotype from a metaphase hybridized with phage λAB3 (see Fig 2). Signals (red) were found on the shorter chromosome 4 (4s), on der(4), and on der(14), indicating that the 4p16.3 breakpoint occurred within the genomic region represented by this clone.
FISH analyses of KMS-11 cell line. (A) Partial metaphase hybridized with a FGFR3 cDNA clone HE8.31 The signals (red) are evident at the 4p16.3 telomeric region of a shorter chromosome 4 (4s) and at the telomeric region of chromosome 14q, der(14), while absent in the normal chromosome 4 homolog, der(4). (B) Partial karyotype from a metaphase hybridized with phage λAB3 (see Fig 2). Signals (red) were found on the shorter chromosome 4 (4s), on der(4), and on der(14), indicating that the 4p16.3 breakpoint occurred within the genomic region represented by this clone.
The FGFR3 gene is aberrantly expressed in cases carrying the t(4; 14) and mutated in the KMS-11 cell line. Northern blot analysis of panels of total or poly(A)+ RNA from normal human tissue and tumor cell lines using 4p16.3 probes described in Fig 2 failed to detect any transcription unit in the region surrounding the breakpoints (data not shown). In addition, computer-assisted analysis failed to identify potential exons in the genomic region represented by the L75b9 cosmid clone. Because of the relative proximity of the FGFR3 locus to the breakpoint region, we looked for the aberrant expression of this gene in the MM cell line KMS-11 and in cases LB375 and LB1017 from which RNA was available. As shown in Fig 6A, Northern blot analysis indicated a higher level of FGFR3 mRNA expression in the KMS-11 than in the K562 cell line from which the FGFR3 gene was originally cloned.33 There was apparently no expression in the MM cell line U266 carrying a t(11; 14)(q13; q32) chromosomal translocation37 (and our unpublished results), in the Burkitt's lymphoma Raji, or in the leukemic T-cell line Molt-4. FGFR3 expression in tumor biopsies LB375, LB1017, and LB411, carrying a t(11; 14), was investigated by reverse transcriptase-polymerase chain reaction (RT-PCR) using a set of primers amplifying a 204-bp fragment of the FGFR3 cDNA including the 3′ end of the third Ig-like loop and the entire TM domain (see Materials and Methods). Amplification was detected in the mRNA samples from both tumors carrying the 4p16.3 breakpoint (Fig 6B), but not in bone marrow samples from two normal donors, three MM patients in clinico-hematologic remission, the LB411 tumor, and in the cell line U266 (Fig 6B and data not shown).
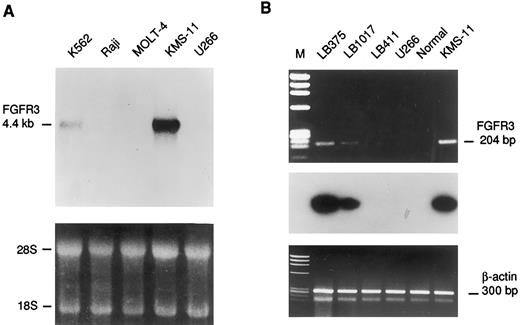
Expression analysis of the FGFR3 gene. (A) Northern blot analysis of human leukemic cell lines. The length of the FGFR3 transcript is indicated in kb. Ethidium bromide staining is shown below for loading quantification. (B) RT-PCR analysis of FGFR3 expression in MM biopsies (LB375,LB1017, LB411), cell lines (U266, KMS-11), and bone marrow from a normal donor (Normal). The length of the FGFR3 amplified fragment is shown in base pairs. The hybridization with an internal fragment obtained by nested PCR is shown below. The amplification of specific sequences of the β-actin gene is also shown as template control.
Expression analysis of the FGFR3 gene. (A) Northern blot analysis of human leukemic cell lines. The length of the FGFR3 transcript is indicated in kb. Ethidium bromide staining is shown below for loading quantification. (B) RT-PCR analysis of FGFR3 expression in MM biopsies (LB375,LB1017, LB411), cell lines (U266, KMS-11), and bone marrow from a normal donor (Normal). The length of the FGFR3 amplified fragment is shown in base pairs. The hybridization with an internal fragment obtained by nested PCR is shown below. The amplification of specific sequences of the β-actin gene is also shown as template control.
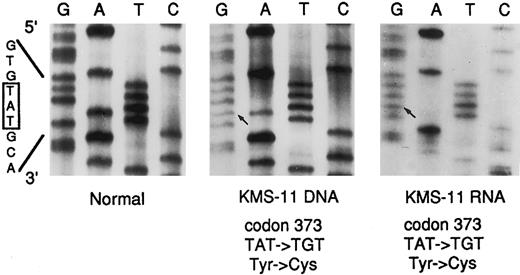
Because the most common form of dwarfism, achondroplasia,32 is associated with a specific point mutation (codon 380) within the TM domain of the FGFR3 gene, we also investigated the presence of mutations in this region by direct sequencing of a PCR-amplified 164-bp fragment (see Shiang et al32 and Materials and Methods) from DNAs of the MM samples carrying the 4p16.3 breakpoints. Interestingly, a point mutation at codon 373 (TAT → TGT) leading to the substitution of the normal tyrosine with cysteine, was detected in KMS-11 cell line, but not in the other tested cases, including the tumors LB375, LB1017, and LB278 and the cell lines K562 and U266 (see Fig 7 and data not shown). Furthermore, direct sequencing of the 204-bp amplified fragment obtained by RT-PCR from KMS-11 mRNA (see above and Fig 6B) showed the presence of the same point mutation (Fig 7). Interestingly, only the expression of the mutated FGFR3 allele was observed.
Direct DNA sequencing of the FGFR3 mutation in the KMS-11 cell line. The 3′ primer used for the amplification of the TM-containing fragments of the FGFR3 gene was used as sequencing primer for both the DNA (164 bp) and cDNA (204 bp) templates (see Text). The normal sequence across codon 373 is reported. The base pair mutation is indicated by an arrow. DNA from normal peripheral blood leukocytes was used as a control (Normal).
Direct DNA sequencing of the FGFR3 mutation in the KMS-11 cell line. The 3′ primer used for the amplification of the TM-containing fragments of the FGFR3 gene was used as sequencing primer for both the DNA (164 bp) and cDNA (204 bp) templates (see Text). The normal sequence across codon 373 is reported. The base pair mutation is indicated by an arrow. DNA from normal peripheral blood leukocytes was used as a control (Normal).
DISCUSSION
The aim of this study was to identify novel genetic lesions associated with the molecular pathogenesis of MM. We used Southern blotting to investigate a large panel of nonkaryotyped MM tumors for the presence of rearranged IGH alleles that might be candidates for chromosomal translocations. Using this approach, we identified a novel, recurrent t(4; 14)(p16.3; q32) chromosomal translocation leading to the deregulation of the FGFR3 gene. These data may provide new insights into the molecular mechanisms involved in the tumorigenesis of MM.
In our study, illegitimate rearranged IGH alleles (a fragment containing a constant region not linked to the joining region) were identified in 21 of 88 cases (≈25%). This frequency can be considered an underestimate on the basis of the following considerations. First, Southern blot analysis was only performed with the BamHI restriction enzyme, and so it is likely rearrangements could not be detected in some tumor DNA (eg, restriction fragments that were too large to be efficiently transferred). Second, the Cγ1 fragment used as a probe cross-hybridizes with the other Cγ regions (γ3,γ2,γ4), and so rearranged Cγ alleles may have comigrated with germline fragments under our experimental conditions; this is particularly important insofar as the majority of the tumor biopsies analyzed had more than 50% of normal cells. Third, but less likely, breakpoints may occur at 5′ or within the JH region, or may involve the Ig light chain loci. On the other hand, it should be considered that some of the “illegitimate” rearranged IGH alleles may represent mutations, deletions, or internal rearrangements involving the IGH locus.38 However, in our study the molecular cloning of illegitimate IGH rearranged alleles from three primary tumors confirmed the presence of translocation breakpoints. This result further supports the previously reported finding in MM-derived cell lines37 (see below) that translocations involving the IGH switch regions may represent the most frequent genetic abnormality in MM.
An important finding of our study was the identification of a novel chromosomal translocation t(4; 14)(p16.3; q32) in two primary tumors, LB375 and LB1017. Molecular analysis of the breakpoints at 4p16.3 showed that they occurred about 14 kb apart in a previous identified genomic region (cosmid clone L75b9) located approximately 50 kb centromeric to the FGFR3 locus.22,23 An additional MM tumor was found to be rearranged by Southern blot analysis using probes derived from the breakpoint regions. Chromosomal abnormalities involving the telomeric region of 4p have not generally been detected by cytogenetic analysis in MM or PCL, or in other types of lymphoid malignancies. As far as we know, a t(3; 4)(q13.3; p16) chromosomal translocation has been reported in one case of hairy cell leukemia39 and, more recently, a t(4; 7)(p11; p11) involving a more centromeric portion of 4p, has been identified by FISH in B-cell chronic lymphocytic leukemia.40 While this work was in progress, a similar analysis of a panel of MM-derived cell lines using specific IGH switch probes was reported by Bergsagel et al,37 who found the presence of illegitimate switch recombination fragments in 15 of 21 cell lines, including those without a cytogenetically detectable 14q32. The molecular analysis of several of these fragments showed the presence of translocation breakpoints involving a wide array of translocation partners, including 11q13, 6, 16q23, 8q24, 21q22, and 4p16.3. In particular, these investigators found the presence of a t(4; 14)(p16.3; q32) in three MM tumors. It is worth noting that the breakpoints in these cases involved the cosmid clone L75b9 (KMS-11 cell line and a primary tumor) or the contiguous centromeric clone L184d6 (JIM3 cell line). Our data and those reported by Bergsagel et al strongly suggest that this novel chromosomal translocation may represent a recurrent, nonrandom genetic lesion in MM. The general absence of a t(4; 14)(p16.3; q32) in cytogenetic reports could be consistent with the involvement of the most telomeric regions of the two partner chromosomes, as has also been found in the case of other translocation partners (such as 16q23 and 21q22), in cell lines carrying IGH switch-mediated 14q32 translocations.37 However, the exact frequency of this lesion in MM remains to be established. Our data, and those recently reported by others,37 suggest that breakpoints on 4p16.3 may be dispersed over a relatively large region; in addition, the high number of repetitive sequences in the breakpoint region on 4p16.3 did not allow us to make an exhaustive Southern blot analysis. The use of other techniques such as pulsed-field gel electrophoresis or FISH would therefore be helpful in better assessing the frequency of this genetic abnormality in MM.
Given the apparent absence of a transcription unit in the genomic regions surrounding the breakpoints, we investigated whether known genes located in the proximity of the breakpoints at 4p16.3 may be deregulated by the translocation. In many types of lymphoid neoplasias, such as the c-MYC in endemic Burkitt's lymphoma,7,20BCL-2 in follicular lymphoma,8,9BCL-1 in mantle cell-lymphoma11,15,17 and the PAX-5 gene in lymphoplasmacytoid lymphoma,41 the breakpoints can occur in a position that is relatively distant from the gene. Interestingly, the FGFR3 gene, a member of the fibroblast growth-factor receptor family,42 has been mapped in the proximity of the L75b9 cosmid clone.22,23 The FGFRs (four members have so far been identified) are tyrosine kinase receptors that are capable of binding a repertoire of nine related mitogenic FGFs.43 Ligand binding induces receptor homo- and heterodimerization, leading to the activation of complex signaling pathways that regulate cell proliferation, differentiation, and migration in many different tissues.42,43 FGFR3 has a relatively limited pattern of expression: in developing mouse and human, high levels of FGFR3 are observed in the cartilage growth plates, skin, and central nervous system; a lower level in lung, intestine, and kidney; and almost no detectable expression in spleen, liver, and thymus. In adult tissues, it is expressed in brain, kidney, testes, and in resting (but not hypertrophic) cartilage.30,44 This means that FGFR3 expression is apparently absent in the hematopoietic system, although, it should be mentioned that the gene has been cloned from the erythroleukemic cell line K562,30,33 and a permissive role of FGFs in primitive hematopoietic cell colony formation in culture has been suggested.45 Interestingly, missense point mutations in distinct domains of the FGFR3 gene are associated with autosomal skeletal diseases, such as hypochondroplasia, achondroplasia, and thanatophoric dysplasia type I and II32,46 (see Muenke and Schell47 for review). Recent reports indicate that point mutations associated with these distinct forms of dwarfism produce constitutively activated FGFR3, which shows autophosphorylation in the absence of ligand and are no longer regulated by FGF binding.48-50
Our data show that the FGFR3 gene is translocated to chromosome 14q32 in the KMS-11 cell line carrying the t(4; 14)(p16.3; q32) and is highly expressed. In addition, the expression of this gene was detected in the two primary tumors in which we identified such a translocation, whereas no expression was found in the MM tumors apparently negative for 4p16.3 abnormalities, or in the other types of leukemic cell lines investigated. These findings are similar to those recently reported by Bergsagel et al51 and suggest that the chromosomal translocation may contribute to FGFR3 deregulation. As is the case of other types of chromosomal translocations involving the IGH locus, it is conceivable that at the distance of the FGFR3 gene from the breakpoint (50 to 60 kb), IGH regulatory elements such as IGH transcriptional enhancers and locus control regions52 may contribute to gene deregulation. Moreover, the identification of a missense mutation at codon 373 (Tyr-Cys) within the TM domain of the gene in the KMS-11 cell line suggests that other mechanisms, alone or in combination with chromosomal translocations, may deregulate the FGFR3 in MM tumors. There are significant examples among lymphoid neoplasms in which mutations frequently occur, even independently of translocations to IG loci, such as the c-MYC gene in Burkitt's lymphoma53 and the BCL-6 gene in diffuse large-cell lymphoma.54 Thus, specific studies are needed to investigate the functional properties of this novel tumor-associated mutant form of FGFR3, as well as the possible role of FGFR3 gene mutation in MM. Furthermore, the biological role of FGFR3 deregulation in MM remains to be elucidated; an important step in this direction could be the investigation of the effects of constitutive expression of normal or tumor-associated mutant forms of FGFR3 in Epstein-Barr Virus–immortalized B lymphoblastoid cells, which can be used as targets for testing the biological activity of oncogenes.55 56
NOTE ADDED IN PROOF
After acceptance of this manuscript, Chesi et al reported similar data on the involvement of the FGFR3 gene in chromosomal translocations in multiple myeloma.57
ACKNOWLEDGMENT
We are grateful to G. Ciceri and S. Cerri for technical assistance.
Supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) (to A.N. and M.R.), and the Ministero della Sanità to Ospedale Maggiore IRCCS “Ricerca Corrente 1994.”
R.R. and D.R. contributed equally to this work.
Address reprint requests to Antonino Neri, MD, Servizio Ematologia, Centro “G.Marcora,” Ospedale Maggiore di Milano, IRCCS, Via Francesco Sforza 35, Milano, 20122, Italy.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal