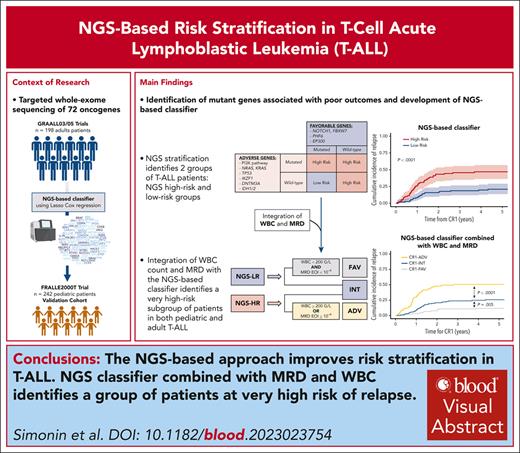
Key Points
NGS-based stratification refines risk classification in T-ALL.
NGS classifier combined with MRD and WBC identifies a group of patients with a very high risk of relapse.
Visual Abstract
We previously reported a better outcome in adult and pediatric T-cell acute lymphoblastic leukemia (T-ALL) harboring NOTCH1 and/or FBXW7 mutations without alterations of K-N-RAS and PTEN genes. Availability of high-throughput next-generation sequencing (NGS) strategies led us to refine the outcome prediction in T-ALL. Targeted whole-exome sequencing of 72 T-ALL–related oncogenes was performed in 198 adults with T-ALLs in first remission from the GRAALL-2003/2005 protocols and 242 pediatric patients with T-ALLs from the FRALLE2000T. This approach enabled the identification of, to our knowledge, the first NGS-based classifier in T-ALL, categorizing low-risk patients as those with N/F, PHF6, or EP300 mutations, excluding N-K-RAS, PI3K pathway (PTEN, PIK3CA, and PIK3R1), TP53, DNMT3A, IDH1/2, and IKZF1 alterations, with a 5-year cumulative incidence of relapse (CIR) estimated at 21%. Conversely, the remaining patients were classified as high risk, exhibiting a 5-year CIR estimated at 47%. We externally validated this stratification in the pediatric cohort. NGS-based classifier was highly prognostic independently of minimal residual disease (MRD) and white blood cell (WBC) counts, in both adult and pediatric cohorts. Integration of the NGS-based classifier into a comprehensive risk-stratification model, including WBC count at diagnosis and MRD at the end of induction, enabled the identification of an adverse-risk subgroup (25%) with a 5-year CIR estimated at 51%, and a favorable-risk group (32%) with a 5-year CIR estimated at 12%. NGS-based stratification combined with WBC and MRD sharpens the prognostic classification in T-ALL and identifies a new subgroup of patients who may benefit from innovative therapeutic approaches. The GRAALL-2003/2005 studies were registered at www.ClinicalTrials.gov as #NCT00222027 and #NCT00327678.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a class of aggressive hematological cancers arising from the transformation of thymic cell precursors arrested at specific stages of their differentiation that accounts for nearly 15% to 20% of ALL in children and adults.1,2 The outcome of pediatric and adult T-ALL has globally improved by using more poly-chemotherapy, yet T-ALL remains of poor prognosis, notably for relapsing cases.3,4 Approximately 25% of pediatric and 30% of adult patients with T-ALL ultimately relapse,5,6 with dramatically poor prognosis despite salvage chemotherapy, particularly due to secondary chemotherapy resistance acqusition.7-9 Therefore, it is urgent to improve risk stratification, which could allow for those with the highest risk of relapse to be allocated to innovative treatment strategies and/or allogeneic hematopoietic stem cell transplantation in first remission. A critical obstacle to this aim is the heterogeneous molecular nature of the disease, which is driven by a densely interconnected network of oncogenic events, making their individualization toward an optimal therapeutic stratification highly complex. To address this critical issue, the objective of this study was to identify novel molecular factors that can significantly enhance outcome prediction in both pediatric and adult T-ALL cases.
Although molecular genetic analyses and sequencing studies have led to the identification of recurrent T-ALL alterations associated with prognostic impact, allowing for more precise prediction of the risk of relapse, a significant proportion of T-ALL relapses remain unpredicted. In 2013, we provided the first T-ALL stratification based on NOTCH1, FBXW7, RAS, and PTEN (NFRP) status in adults with T-ALL.6 Thus, patients harboring NOTCH1 and/or FBXW7 mutations (N/FMut) without N/K-RAS mutation and/or PTEN (R/P) mutation and/or deletion benefited from significantly better outcomes than those with N/F wild type (N/FWT) and/or R/P altered (R/PAlt). More recently, these findings were also confirmed in the French pediatric cohort of patients with T-ALL from the FRALLE2000T trial.5 In this study, NFRP alteration profile was a robust and a minimal residual disease (MRD)–independent predictor of relapse.
Although the absence of N/FMut appears to be strongly associated with poor prognosis in T-ALL and T-cell lymphoblastic lymphoma,6,10-12 the identification of additive alterations with prognostic relevance in the oncogenetic background of N/FMut T-ALL is still unknown.13-16 Several factors explain the lack of significant success in identifying such prognostic factors; first, the size of the studies is often insufficient to reveal the impact of certain rare alterations. Secondly, the heterogeneity of the methodology applied between studies and the variability of the genes explored but also the patients (multiple chemotherapy regimen) hamper cross-study comparability. These pitfalls highlight the need for a broad and comprehensive analysis of the prognostic impact associated with specific genetic alterations in a homogeneous and large cohort of patients with T-ALL. The recent widespread adoption of next-generation sequencing (NGS) approaches has opened new possibilities for achieving this goal. Using a hybrid capture-based NGS approach to analyze a large series of genes implicated in T-ALL oncogenesis, we have developed and validated an integrative approach for creating a molecular risk-adapted classifier that can be used for both adult and pediatric patients with T-ALL.
Patients and methods
Patients and protocols
The GRAALL-2003 protocol (NCT00222027) was a multicenter phase 2 trial that enrolled adults with T-ALL between November 2003 and November 2005. The multicenter randomized GRAALL-2005 trial (NCT00327678) was the following phase 3 trial and was similar to the GRAALL-2003 trial, with the addition of a randomized evaluation of an intensified sequence of hyperfractionated cyclophosphamide during induction and late intensification (supplemental Figure 1, available on the Blood website). This study concerns the 198 patients who achieved a first complete remission (CR) after induction (CR1) and for whom diagnostic DNA and/or complementary DNA was available (supplemental Figure 2). From 2000 to 2010, a total of 405 patients with T-ALL aged 1 to 19 years were treated in 16 French pediatric FRALLE study group hematology departments according to the FRALLE2000T guidelines (as previously described) (supplemental Figure 1).5 Based on CR1 achievement and DNA availability, 242 pediatric patients of 405 were included in this study (supplemental Figure 2). No difference in clinical outcomes was observed between the included patients and the entire cohort. Informed consent was obtained from all patients at trial entry. Both adult and pediatric trials were conducted in accordance with the Declaration of Helsinki and approved by local and multicenter research ethical committees.
Gene mutation screening
A custom capture Nextera XT gene panel (Illumina, San Diego, CA) targeting all coding exons and their adjacent splice junctions of 72 genes was designed, based on available evidence in hematological neoplasms (supplemental Methods; supplemental Table 1). Genes affecting the same pathway were associated in the analysis: NOTCH1 and FBXW7, NRAS and KRAS, IDH1 and IDH2, PI3K/AKT pathway (PTEN, PIK3CA, and PIK3R1), polycomb repressive complex 2 pathway (SUZ12, EZH2, and EED), and JAK/STAT pathway (JAK1, JAK3, STAT5B, DNM2, and IL7R).
Statistical analysis
End points
The main clinical end point was the cumulative incidence of relapse (CIR), and the secondary end points were disease-free survival (DFS) and overall survival (OS) from CR1. CIR was defined from the time of CR1 achievement to relapse, considering nonrelapse mortality as a competing event. DFS was defined from CR1 until relapse or death (whichever came first), and OS was defined from CR1 until death.
Analysis plan
First, we compared results of previously published NFRP classifier with Sanger or NGS techniques in the GRAALL cohort. Then, we built, in the GRAALL cohort, a new NGS classifier integrating gene mutational status. Our objective was to stratify patients who achieved CR. Individuals with induction failure were not included. This was supported by 3 reasons: first, to exclude the impact of induction death in our analysis; but also to have a clear and noncomposite endpoint looking at CIR; and finally, because therapeutic adaptations are performed in patients after CR achievement. We performed an external validation on the pediatric FRALLE cohort. Finally, we evaluated the prognosis impact of the new NGS classifier taking into account conventional prognostic variables, white blood cell (WBC) counts and MRD at end of induction (MRD1), using previously published thresholds of ≥200 × 109/L and ≥10–4, respectively.17,18 The new NGS classifier was introduced in the global risk stratification5 and validated in the total cohort.
Statistical methods
To account for the large number of candidates genes, genes retained in the classifier were selected using a least absolute shrinkage and selection operator (LASSO) penalization in a Fine and Gray model predicting CIR.19,20 Tuning parameter lambda was selected using a fivefold cross-validation based on time-dependent area under receiver operating characteristic curves computed at 5 years.21
Additional information regarding the protocol, genetic exploration, and statistical analyses can be found in supplemental Data.
Results
NGS approach confirms and refines previous Sanger-based NFRP classifier in adult T-ALL
The NGS analysis were applied to 198 GRAALL-treated patients (Table 1). Of them, 154 had been included in the study by Trinquand et al6 and had also been previously sequenced by Sanger analysis.
Clinico-biological and outcome characteristics of adult and pediatric T-ALL (GRAALL and FRALLE protocols)
| Variable . | GRAALL03/05 (training cohort) . | FRALLE2000T (validation cohort) . | Total . | P value∗ . |
|---|---|---|---|---|
| (N = 198) . | (N = 242) . | (n = 440) . | ||
| Male | 144 (73%) | 188 (78%) | 332 (75%) | .3 |
| Age, y† | 30 (23-39) | 10 (5-13) | 15 (9-28) | <.001 |
| WBC count, G/L† | 34 (12-108) | 93 (25-191) | 62 (17-158) | <.001 |
| CNS involvement‡ | 22 (11%) | 23 (10%) | 45 (10%) | .6 |
| Immunophenotype | ||||
| ETP phenotype | 36/165 (22%) | 13/114 (11%) | 49/279 (18%) | .03 |
| Immature (IM0/d/g)§ | 48/189 (25%) | 32/198 (16%) | 80/387 (21%) | .03 |
| Cortical (IMb/pre-ab) | 100/189 (53%) | 99/198 (49%) | 199/387 (51%) | .6 |
| Mature TCRab | 22/189 (12%) | 41/198 (20%) | 63/387 (16%) | .02 |
| Mature TCRgd | 19/189 (10%) | 26/198 (13%) | 45/387 (12%) | .4 |
| Oncogenetic classification | ||||
| TLX1 | 45/183 (25%) | 9/199 (5%) | 54/382 (14%) | <.001 |
| TLX3 | 23/183 (13%) | 41/199 (21%) | 64/382 (17%) | .04 |
| SIL-TAL1 | 16/183 (9%) | 38/199 (19%) | 54/382 (14%) | .005 |
| CALM-AF10 | 9/183 (5%) | 3/199 (2%) | 12/382 (3%) | .08 |
| Treatment response | ||||
| Good prednisone response | 111/198 (56%) | 141/237 (57%) | 252/435 (58%) | .5 |
| Chemosensitivity | 118/198 (60%) | 211/237 (89%) | 329/435 (76%) | <.001 |
| MRD1 >10−4 | 36/120 (30%) | 80/212 (38%) | 116/332 (35%) | .2 |
| Allo-HSCT | 77/198 (39%) | 24/242 (10%) | 101/440 (23%) | <.001 |
| Variable . | GRAALL03/05 (training cohort) . | FRALLE2000T (validation cohort) . | Total . | P value∗ . |
|---|---|---|---|---|
| (N = 198) . | (N = 242) . | (n = 440) . | ||
| Male | 144 (73%) | 188 (78%) | 332 (75%) | .3 |
| Age, y† | 30 (23-39) | 10 (5-13) | 15 (9-28) | <.001 |
| WBC count, G/L† | 34 (12-108) | 93 (25-191) | 62 (17-158) | <.001 |
| CNS involvement‡ | 22 (11%) | 23 (10%) | 45 (10%) | .6 |
| Immunophenotype | ||||
| ETP phenotype | 36/165 (22%) | 13/114 (11%) | 49/279 (18%) | .03 |
| Immature (IM0/d/g)§ | 48/189 (25%) | 32/198 (16%) | 80/387 (21%) | .03 |
| Cortical (IMb/pre-ab) | 100/189 (53%) | 99/198 (49%) | 199/387 (51%) | .6 |
| Mature TCRab | 22/189 (12%) | 41/198 (20%) | 63/387 (16%) | .02 |
| Mature TCRgd | 19/189 (10%) | 26/198 (13%) | 45/387 (12%) | .4 |
| Oncogenetic classification | ||||
| TLX1 | 45/183 (25%) | 9/199 (5%) | 54/382 (14%) | <.001 |
| TLX3 | 23/183 (13%) | 41/199 (21%) | 64/382 (17%) | .04 |
| SIL-TAL1 | 16/183 (9%) | 38/199 (19%) | 54/382 (14%) | .005 |
| CALM-AF10 | 9/183 (5%) | 3/199 (2%) | 12/382 (3%) | .08 |
| Treatment response | ||||
| Good prednisone response | 111/198 (56%) | 141/237 (57%) | 252/435 (58%) | .5 |
| Chemosensitivity | 118/198 (60%) | 211/237 (89%) | 329/435 (76%) | <.001 |
| MRD1 >10−4 | 36/120 (30%) | 80/212 (38%) | 116/332 (35%) | .2 |
| Allo-HSCT | 77/198 (39%) | 24/242 (10%) | 101/440 (23%) | <.001 |
MRD1 corresponds to MRD evaluation after induction and was performed by allele-specific oligonucleotides polymerase chain reaction. T-cell receptor (TCR) status and molecular characterization were performed as described in supplemental Methods. P values <.05 are indicated in bold.
Allo-HSCT, allogeneic hematopoietic stem cell transplantation; CNS, central nervous system.
Statistical tests performed: Fisher exact test; Wilcoxon rank-sum test.
Statistics presented as median (Q1-Q3).
CNS involvement; CNS3 in FRALLE2000T trial; CNS2 and/or CNS3 in GRAALL2003 and GRAALL2005 trials.
T-ALL are divided into 4 subclasses as follows: (1) immature (no detectable TCRb variable diversity joining [IM0/d/g]): IM0 (TCRd and TCRg germline), IMd (TCRd rearranged but not TCRg), and IMg (both TCRd and TCRg rearranged); (2) T-ALL early-cortical IMb/Pre-ab; (3) mature sTCRab+; and (4) mature sTCRgd.1
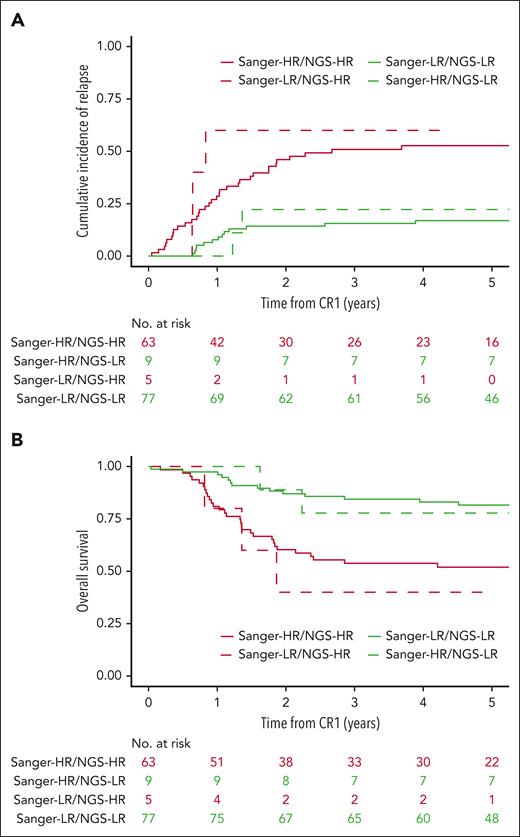
NGS approach was associated with higher sensitivity than Sanger, leading to the identification of NOTCH1 and FBXW7 mutations previously undetected (14 and 7 mutations, respectively; supplemental Figure 3). Furthermore, NGS approach allowed for pan-exon sequencing of N/K RAS and PTEN, whereas Sanger was restricted to the mutational hot spot; hence, NGS revealed 5 N/K RAS and 2 PTEN mutations previously undetected. Collectively, the NFRP classifier was conserved in 140 of 154 of the cases (91%). Conversely, in 14 of 154 cases (9%), NGS led to a classification modification: 5 patients previously classified as low risk (LR) became high risk (HR) based on NGS analysis (RAS/PTEN abnormalities), and 9 patients previously classified as HR became LR (N/F mutations; Figure 1A-B). At 5 years, area under receiver operating characteristic curves of Sanger NFRP classifier and NGS NFRP classifier were 0.670 (95% confidence interval [CI], 0.587-0.752) vs 0.720 (95% CI, 0.641-0.799) for CIR, 0.656 (95% CI, 0.574-0.738) vs 0.722 (95% CI, 0.646-0.799) for DFS, and 0.639 (95% CI, 0.553-0.725) vs 0.688 (95% CI, 0.605-0.771) for OS, respectively. Taken together, these data confirm that the NGS approach both validates and refines the NFRP classification previously based on Sanger analysis.
NGS-based approach confirms and refines the historical classifier in adult T-ALL. (A) Comparison of CIR according to NGS- or Sanger-based method to identify NFRP classifier in adult T-ALL. At 5 years, CIR was 53% (95% CI, 39-64) in HR Sanger/HR NFRP NGS, 17% (95% CI, 10-26) in LR Sanger/LR NFRP NGS, 60% (95% CI, 7-91) (at 4.29 years, no longer follow-up) in LR Sanger/HR NFRP NGS, and 22% (95% CI, 3-53) in HR Sanger/LR NFRP NGS. (B) Comparison of OS according to NGS or Sanger-based method to identify NOTCH1/FBXW7/RAS/PTEN (NFRP) classifier in adult T-ALL. At 5 years, OS was 52% (95% CI, 41-66) in HR Sanger/HR NFRP NGS, 82% (95% CI, 73-91) in LR Sanger/LR NFRP NGS, 40% (95% CI, 14-100) in LR Sanger/HR NFRP NGS, and 78% (95% CI, 55-100) in HR Sanger/LR NFRP NGS.
NGS-based approach confirms and refines the historical classifier in adult T-ALL. (A) Comparison of CIR according to NGS- or Sanger-based method to identify NFRP classifier in adult T-ALL. At 5 years, CIR was 53% (95% CI, 39-64) in HR Sanger/HR NFRP NGS, 17% (95% CI, 10-26) in LR Sanger/LR NFRP NGS, 60% (95% CI, 7-91) (at 4.29 years, no longer follow-up) in LR Sanger/HR NFRP NGS, and 22% (95% CI, 3-53) in HR Sanger/LR NFRP NGS. (B) Comparison of OS according to NGS or Sanger-based method to identify NOTCH1/FBXW7/RAS/PTEN (NFRP) classifier in adult T-ALL. At 5 years, OS was 52% (95% CI, 41-66) in HR Sanger/HR NFRP NGS, 82% (95% CI, 73-91) in LR Sanger/LR NFRP NGS, 40% (95% CI, 14-100) in LR Sanger/HR NFRP NGS, and 78% (95% CI, 55-100) in HR Sanger/LR NFRP NGS.
Univariable prognostic impact of genes alterations in adults T-ALL
We conducted an analysis of gene alterations using NGS data from our panel of 72 genes (supplemental Table 1) to investigate their incidence and prognostic significance in 198 adult patients with T-ALL in CR1 in the GRAALL03/05 trials (Table 1).
NOTCH1 mutations were the most frequent alterations observed in our cohort, affecting 77% of adults with T-ALL, followed by CDKN2A alterations (66%), PHF6 alterations (50%), BCL11B mutations (22%), DNM2 mutations (20%), and FBXW7 mutations (19%) (Figures 2A-B; supplemental Table 2).
Identification of new prognostic molecular in adult T-ALL. (A) Oncoplot depicting the genetic anomalies observed in HR-NGS or LR-NGS patients of the GRAALL03/05 trials. Genes are classified by functional groups. (B) Bar plot highlighting the frequency of gene alterations in the 198 adults with T-ALL issued from the GRAALL03/05 trial (cutoff of ≥4 mutations per gene [2%]). (C) SHRs of relapse according to genes alterations in adult T-ALL. Error bars indicate 95% CI. P values are from univariable Fine and Gray models. (D) Trace plot of Fine and Gray model with LASSO penalization. The trace plot visualizes the result of model selection process for predicting CIR in adult T-ALL using gene alterations at diagnosis. The selected model, chosen by fivefold cross-validation, is marked with a vertical dashed line on the trace plot. The variables colored and labeled were included in the chosen model with nonnull coefficients and were used to construct the NGS classifier. Red and blue colors indicate variables with an increased or a reduced risk of relapse, respectively. LASSO, least absolute shrinkage and selection operator; SE, standard error.
Identification of new prognostic molecular in adult T-ALL. (A) Oncoplot depicting the genetic anomalies observed in HR-NGS or LR-NGS patients of the GRAALL03/05 trials. Genes are classified by functional groups. (B) Bar plot highlighting the frequency of gene alterations in the 198 adults with T-ALL issued from the GRAALL03/05 trial (cutoff of ≥4 mutations per gene [2%]). (C) SHRs of relapse according to genes alterations in adult T-ALL. Error bars indicate 95% CI. P values are from univariable Fine and Gray models. (D) Trace plot of Fine and Gray model with LASSO penalization. The trace plot visualizes the result of model selection process for predicting CIR in adult T-ALL using gene alterations at diagnosis. The selected model, chosen by fivefold cross-validation, is marked with a vertical dashed line on the trace plot. The variables colored and labeled were included in the chosen model with nonnull coefficients and were used to construct the NGS classifier. Red and blue colors indicate variables with an increased or a reduced risk of relapse, respectively. LASSO, least absolute shrinkage and selection operator; SE, standard error.
In univariable analysis, only NOTCH1/FBXW7 mutations (158/198 patients [80%]) and PHF6 alterations (99/198 [50%]) were significantly associated with a lower risk of relapse, whereas alterations affecting PI3K pathway (PTENAlt, PIK3CA, and PIK3R1 mutations; 36/198 [18%]) and IDH1/2 (8/198 [4%]) conferred a significantly higher risk of relapse (Figure 2C; supplemental Table 3).
N/F, PHF6 alterations, EP300, N-K-RAS, PI3K pathway, IKZF1, TP53, IDH1/2, and DNMT3A mutational status build a new oncogenetic classifier in adult T-ALL
We used penalized regression to examine the impact of genetic mutations on the risk of relapse in a Fine and Gray model among the 39 of 72 genes from our NGS panel that affected >2% of the patients. Our analysis revealed that N/F, PHF6, and EP300 alterations were associated with favorable outcomes. Conversely, N-K-RAS, PI3K pathway, TP53, IDH1/2, IKZF1, and DNMT3A mutations were significantly associated with a higher risk of relapse (Figure 2D).
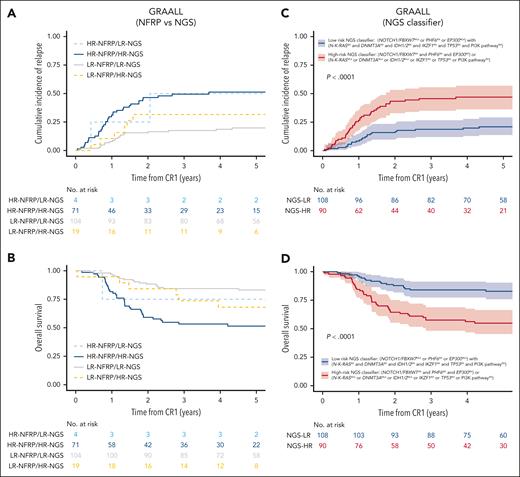
Presence of at least 1 favorable gene without adverse gene was associated with a low CIR (5-year CIR, 21%; 95% CI, 14-29). On the contrary, CIR was higher in the 3 other subgroups: presence of adverse gene with or without favorable gene was associated with a high CIR (5-year CIR, 45% [95% CI, 32-57] and 53% [95% CI, 26-74], respectively); and absence of favorable and of adverse gene was also associated with a high CIR (5-year CIR, 50% [95% CI, 19-75]; supplementary Figure 4). These observations led us to propose a T-ALL NGS–based oncogenetic classifier defining NGS low-risk (LR-NGS) patients as those with at least 1 favorable gene, either N/F mutations or EP300 mutations or PHF6 alteration, but no adverse gene, including N-K-RAS mutation, PI3K pathway alterations, IKZF1 alterations, TP53 alterations, IDH1/2 mutations, and DNMT3A mutations (here, 108/198 patients [55%]); and all other patients as NGS high-risk (HR-NGS) patients (90/198 [45%]). Importantly, 19 patients of 123 (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup based on their PI3K (other than PTEN alterations), DNMT3A, TP53, IDH1/2 or IKZF1 status, and 4 of 75 patients (5%) who would have been previously classified in HR based on their NFRP status joined the LR-NGS subgroup based on their PHF6 or EP300 status (Figure 3A-B).
Creation of a NGS-based classifier in adult T-ALL validated in pediatric T-ALL. (A-B) CIR (A) and OS (B) in 198 adult T-ALL (GRAALL03/05 protocols) by NFRP and NGS–based classifier status; 19 of 123 patients (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup based on their PI3K (other than PTEN alterations), DNMT3A, TP53, IDH1/2, or IKZF1 status, and 4 of 75 patients (5%) who would have previously been classified in HR based on their NFRP status joined the LR-NGS subgroup based on their PHF6 or EP300 status. (C) In GRAALL03/05, CIR at 5 years was 21% (95% CI, 14-29) in LR-NGS patients (n = 108), compared with 47% (95% CI, 36-57) in HR-NGS patients (n = 90; P < .0001). (D) In GRAALL03/05, OS at 5 years was 83% (95% CI, 76-90) for LR-NGS patients, compared with 55% (95% CI, 45-66) for HR-NGS patients (P < .0001). (E-F) CIR (E) and OS (F) in 242 pediatric T-ALL (FRALLE2000T protocol) by NFRP classifier and NGS-based classifier status; 21 patients of 144 (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup; and 3 patients of 98 (3%) who would have previously been classified in HR based on their NFRP status joined the LR-NGS subgroup. (G) In FRALLE2000T, CIR at 5 years was 17% (95% CI, 11-24) in LR-NGS patients (n = 126), compared with 35% (95% CI, 26-44) in HR-NGS patients (n = 116; P = .001). (H) In FRALLE2000T, OS at 5 years was 88% (95% CI, 83-94) in LR-NGS patients, compared with 69% (95% CI, 61-79) in HR-NGS patients (P = .0003).
Creation of a NGS-based classifier in adult T-ALL validated in pediatric T-ALL. (A-B) CIR (A) and OS (B) in 198 adult T-ALL (GRAALL03/05 protocols) by NFRP and NGS–based classifier status; 19 of 123 patients (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup based on their PI3K (other than PTEN alterations), DNMT3A, TP53, IDH1/2, or IKZF1 status, and 4 of 75 patients (5%) who would have previously been classified in HR based on their NFRP status joined the LR-NGS subgroup based on their PHF6 or EP300 status. (C) In GRAALL03/05, CIR at 5 years was 21% (95% CI, 14-29) in LR-NGS patients (n = 108), compared with 47% (95% CI, 36-57) in HR-NGS patients (n = 90; P < .0001). (D) In GRAALL03/05, OS at 5 years was 83% (95% CI, 76-90) for LR-NGS patients, compared with 55% (95% CI, 45-66) for HR-NGS patients (P < .0001). (E-F) CIR (E) and OS (F) in 242 pediatric T-ALL (FRALLE2000T protocol) by NFRP classifier and NGS-based classifier status; 21 patients of 144 (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup; and 3 patients of 98 (3%) who would have previously been classified in HR based on their NFRP status joined the LR-NGS subgroup. (G) In FRALLE2000T, CIR at 5 years was 17% (95% CI, 11-24) in LR-NGS patients (n = 126), compared with 35% (95% CI, 26-44) in HR-NGS patients (n = 116; P = .001). (H) In FRALLE2000T, OS at 5 years was 88% (95% CI, 83-94) in LR-NGS patients, compared with 69% (95% CI, 61-79) in HR-NGS patients (P = .0003).
Overall, in adults, CIR at 5 years was 21% (95% CI, 14-29) in LR-NGS patients (n = 108), compared with 47% (95% CI, 36-57) in HR-NGS patients (n = 90; P < .0001). The 5-year OS was 83% (95% CI, 76-90) for LR-NGS patients, compared with 55% (95% CI, 45-66) for HR-NGS classifier patients (P < .0001; Figure 3C-D).
External validation of this NGS-based classifier in pediatric patients
We then performed external validation of the NGS-based classifier in a distinct cohort of 242 pediatric patients with T-ALL treated according to the FRALLE2000T protocol (Table 1). The proportion of pediatric patients was equally distributed between the 2 NGS classifier groups: 126 of 242 (52%) in LR-NGS and 116 of 242 (48%) in HR-NGS. Of note, 21 of 144 patients (15%) who would have previously been classified in LR based on their NFRP status joined the HR-NGS subgroup based on their PI3K pathway, DNMT3A, IDH1/2, TP53, or IKZF1 status, and 3 patients of the 98 (3%) who would have previously been classified in HR based on their NFRP status joined the LR-NGS subgroup based on their PHF6 or EP300 status (Figures 3E-F).
Overall, at 5 years, CIR was 17% (95% CI, 11-24) in LR-NGS patients (n = 126), compared with 35% (95% CI, 26-44) in HR-NGS patients (n = 116; P = .001). The 5-year OS was 88% (95% CI, 83-94) in LR-NGS patients, compared with 69% (95% CI, 61-79) in HR-NGS patients (P = .0003; Figures 3G-H).
Taken together, these data demonstrate that the detection of PHF6, EP300, DNMT3A, TP53, IKZF1, IDH1/2, and PI3K pathway alterations adds significant prognostic value to the assessment of NFRP status and allows for the identification of a significant proportion (206/440 [47%]) of poor-prognosis adult and pediatric patients with T-ALL including 40 of 206 (19%) who were not identified with the previous NFRP classifier, whereas their outcomes were similar to that of HR patients. Impact of NGS classifier was also observed after censoring individuals who received an allogeneic hematopoietic stem cell transplantation in CR1 (supplemental Figure 5).
Early T-cell precursor (ETP) ALL status was available for 279 of 440 patients (63%), including 114 pediatric patients and 165 adult patients. In our study, 49 patients (36 adults and 13 pediatric) were classified as ETP ALL based on phenotype classification. Interestingly, in univariable analysis, ETP status was not associated with a poor prognosis in both adult (5-year CIR, 31% [95% CI, 23-39] for non-ETP vs 36% [95% CI, 21-52]; P = .38) and pediatric T-ALL (5-year CIR, 23% [95% CI, 16-32] for non-ETP vs 46% [95% CI, 18-71] for ETP; P = .11). Of note, among the 36 adult patients with ETP ALL, our NGS classifier was able to identify patients with poor outcomes (5-year CIR of 13% [95% CI, 2-35] for LR-NGS and 52% [95% CI, 29-72] for HR-NGS; P = .023).
NGS-based stratification combined with MRD and WBC refines stratification of patients in first remission
Because our NGS-based classifier improved the risk stratification, we sought to further enhance its prognostic value by integrating clinically significant characteristics previously associated with prognostic impact in adult and pediatric T-ALL.
First, we evaluated the impact of the genetic classifier along with high WBC and high MRD1 levels (≥200 × 109/L and ≥10–4 cutoffs, respectively). A subset of 332 of 440 patients (75%; 181 new LR-NGS and 151 HR-NGS patients; 120 in GRAALL03/05 and 212 in FRALLE2000T protocols) were evaluated for genomic immunoglobulin heavy chain/TCR MRD level at time of CR achievement after the first induction course. In both settings, NGS classifier was strongly associated with a higher CIR independently of high WBC and high MRD1 in multivariable analyses. In adults (GRAALL03/05 cohort, n = 120), HR-NGS classifier was associated with a higher CIR (subdistribution hazard ratio [SHR], 2.84 [95% CI, 1.45-5.57]; P = .002) independently of high WBC and high MRD1. HR-NGS classifier was also associated with an inferior DFS (hazard ratio, 3.36 [95% CI, 1.75-6.45]; P = .0003) and an inferior OS (hazard ratio, 4.15 [95% CI, 1.9-9.08]; P = .0004) independently of high WBC and high MRD1 (supplemental Table 4).
In the pediatric population (FRALLE2000T cohort, n = 212), HR-NGS classifier was also associated with a higher CIR (SHR, 2.29 [95% CI, 1.31-4.02]; P = .004) independently of high WBC and high MRD1. Similarly, HR-NGS classifier was associated with an inferior DFS (hazard ratio, 2.72 [95% CI, 1.56-4.73]; P = .0004) and an inferior OS (hazard ratio, 2.66 [95% CI, 1.36-5.18]; P = .004; supplemental Table 4).
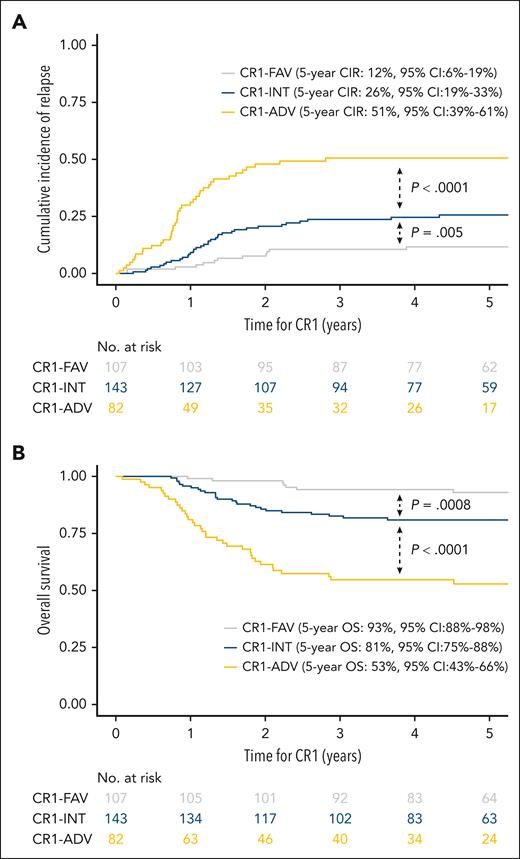
To predict prognosis after CR achievement, we clustered patients into 3 groups as described in the study of Petit et al, combining NGS classifier with WBC count and MRD1: 82 patients (25%) with a HR-NGS–based classifier with a high WBC and/or high MRD1 were classified as adverse risk (CR1-ADV); 107 patients (32%) with a LR-NGS–based classifier with a low WBC and low MRD1 were classified as favorable risk (CR1-FAV); and the remaining 143 patients (43%) were considered as intermediate risk (CR1-INT).
At 5 years, the CIR was 12% (95% CI, 6-19) in CR1-FAV patients, 26% (95% CI, 19-33) in CR1-INT patients (SHR, 2.34 [95% CI, 1.26-4.35]; P = .005, compared with CR1-FAV), and 51% (95% CI, 39-61) in CR1-ADV patients (SHR, 2.49 [95% CI, 1.59-3.92]; P < .0001, compared with CR1-INT). Similarly, the 5-year OS was 93% (95% CI, 88-98) in CR1-FAV patients, 81% (95% CI, 75-88) in CR1-INT patients (hazard ratio, 3.63 [95% CI, 1.59-8.28]; P = .0008, compared with CR1-FAV) and 53% (95% CI, 43-66) in CR1-ADV patients (hazard ratio, 2.73 [95% CI, 1.68-4.43]; P < .0001, compared with CR1-INT; Figures 4A-B; supplemental Figures 6-7).
NGS-based classifier combined with WBC and MRD identifies an adverse group of patients with T-ALL. (A) CIR in 332 patients with T-ALL by NGS-based classifier combined with WBC and MRD. The 5-year CIR in 332 patients with T-ALL was 12% (95% CI, 6-19) in CR1-FAV patients, 26% (95% CI, 19-33) in CR1-INT patients (P = .005, compared with CR1-FAV), and 51% (95% CI, 39-61) in CR1-ADV patients (P < .0001, compared with CR1-INT). (B) At 5 years, OS was 93% (95% CI, 88-98) in CR1-FAV patients, 81% (95% CI, 75-88) in CR1-INT patients (P = .0008, compared with CR1-FAV), and 53% (95% CI, 43-66) in CR1-ADV patients (P < .0001, compared with CR1-INT).
NGS-based classifier combined with WBC and MRD identifies an adverse group of patients with T-ALL. (A) CIR in 332 patients with T-ALL by NGS-based classifier combined with WBC and MRD. The 5-year CIR in 332 patients with T-ALL was 12% (95% CI, 6-19) in CR1-FAV patients, 26% (95% CI, 19-33) in CR1-INT patients (P = .005, compared with CR1-FAV), and 51% (95% CI, 39-61) in CR1-ADV patients (P < .0001, compared with CR1-INT). (B) At 5 years, OS was 93% (95% CI, 88-98) in CR1-FAV patients, 81% (95% CI, 75-88) in CR1-INT patients (P = .0008, compared with CR1-FAV), and 53% (95% CI, 43-66) in CR1-ADV patients (P < .0001, compared with CR1-INT).
Taken together, the NGS classifier improves prediction of the risk of relapse and outcome of patients. Interestingly, the oncogenetic classifier enables, to our knowledge, for the first time, the early identification of a significant subset of patients at very high risk of relapse when associating HR-NGS with WBC count and MRD.
Discussion
In this study, our objective was to enhance the accuracy of outcome predictions in adult and pediatric T-ALL by identifying novel molecular factors. To accomplish this, we performed a thorough analysis of both clinical and oncogenetic variables within a substantial and uniform cohort of treated patients with T-ALL. Our primary focus was the development of an oncogenetic-related risk classifier comprising the most relevant genes that could effectively forecast patients' outcomes. We used LASSO penalization analysis as a robust methodology for constructing this prognostic classifier. By using this method, we were able to both identify the most informative genes and avoid overfitting, thereby enhancing the robustness of our prognostic model.
By adopting this approach, we made significant strides in identifying specific alterations associated with adverse outcomes. Among these alterations, DNMT3A mutation, PI3K pathway alteration, IKZF1 alteration, TP53 alteration, and IDH1/2 mutations have been previously reported as indicators of poor prognosis. Through the integration of these factors into a combined model, we emphasize their individual significance in determining the risk of relapse. This comprehensive approach provides valuable insights into the factors influencing patient outcomes and may pave the way for more targeted and effective interventions in the future.
By incorporating newly identified alterations associated with poor outcomes, we reclassified a substantial proportion of patients (19%) previously considered low risk but ultimately had poor outcomes. We further identified new alterations associated with favorable outcomes, namely EP300 and PHF6 alterations, resulting in the reclassification of a small number of patients into the low-risk group. Importantly, NGS alterations associated with MRD (MRD1, 10–4 cutoff) and WBC at diagnosis (200 × 109/L cutoff) allowed for us to identify a subgroup of patients with very high risk, comprising nearly 25% of the cohort. These patients had an extremely poor prognosis, with a 5-year CIR of 51%. Early identification of CR1-ADV patients with T-ALL is crucial, because it allows for therapeutic intensification and/or innovative targeted therapies to be implemented in the front line. The primary goal of risk stratification should be to minimize potential overtreatment toxicity in low-risk patients while appropriately treating those at higher risk of poor outcomes. Given the excellent prognosis of the CR1-FAV subgroup, it raises the question of therapeutic de-escalation to mitigate the short- and long-term risks associated with intensive chemotherapy.
The oncogenesis of T-ALL is complex and involves a wide range of alterations. Beyond the alterations identified in this study, previous research by our group and others suggests that copy number variations, alterations in noncoding regions and epigenetic profiles may also have prognostic implications.22-25
However, our main objective was precisely to identify the minimum number of alterations present in T-ALL capable of establishing a prognostic classification feasible in routine practice based only on genomic DNA analysis. This approach deliberately focuses on a limited number of alterations, thus avoiding an exhaustive analysis that is unfeasible in clinical practice.
In addition to the potential significance of integrating a new risk-stratification approach for T-ALL into frontline therapeutic decision-making, identifying primary alterations within NGS-based classifiers could further aid in pinpointing novel actionable therapeutic targets, such as alterations of TP53, IDH1/2, DNMT3A, RA, and PI3K pathways. This is particularly pertinent for patients in the CR1-ADV subgroup, characterized by elevated rates of inherently treatment-resistant disease.
Overall, our study has identified, to our knowledge, the first NGS-based classifier for T-ALL that includes several oncogenes targetable with precision medicine. Furthermore, we have identified a subgroup of patients with a particularly poor prognosis who may benefit from innovative and/or intensified therapy.
Identifying genetic alterations with prognostic significance in T-ALL proves challenging compared with B-cell precursor–ALL. Previously, we illustrated how the NFRP classifier predicts outcomes in both pediatric and adult T-ALL cohorts.5,18 In line with this, the recent GRAALL2014/T French protocol (NCT02619630) for young adult T-ALL incorporates this classifier to identify high-risk patients who should beneficiate from nelarabine in frontline treatment. However, replication of the prognostic value associated with the NFRP classifier has been inconsistent across studies.15,26,27 Various factors, including limited patients cohort sizes, methodological variations in detecting alterations, in particular differences in NGS analysis panels, may contribute to these disparities. Additionally, the dynamic interaction between prognostic alterations and chemotherapy regimens likely plays a pivotal role.
To address these concerns, validating our NGS-based classifier within current adult and pediatric T-ALL protocols would be crucial to ensure its applicability across different treatment approaches. Furthermore, evaluating the cumulative impact of later MRD time points, such as MRD at the end of consolidation, would be critical. These efforts will enhance our understanding of our NGS classifier’s prognostic capabilities and refine its clinical usefulness in guiding treatment strategies for patients with T-ALL.
Acknowledgments
The authors thank all the participants of the GRAALL-2003 and GRAALL-2005 study groups, the Société Française des Cancers et des leucémies de L’Enfant et de l’adolescent (SFCE), and the investigators of the 16 SFCE centers involved in collection and provision of data and patient samples, and V.L. for collection of clinical data.
This work was supported by grants to Necker Laboratory from the Cancer Research for Personalized Medicine (CARPEM), Association pour la Recherche contre le Cancer (ARC, Equipe Labellisée), la Ligue contre le Cancer (Equipe Labellisée), and Institut National du Cancer PRT-K 18-071. The GRAALL was supported by grants P0200701 and P030425/AOM03081 from the Programme Hospitalier de Recherche Clinique, Ministère de l’Emploi et de la Solidarité, France, and the Swiss State Secretariat for Education, Research and Innovation, Switzerland. Samples were collected and processed by the AP-HP Direction de Recherche Clinique Tumor Bank at Necker-Enfants Malades. M.S. was supported by Action Leucémie, the Ligue contre le Cancer, and Soutien pour la formation à la recherche translationnelle en cancérologie. This work was supported by the Fondation pour la Recherche Médicale (grant number FDM202306017181; L.V.).
Authorship
Contribution: V.A. and N.B. conceived the study; E.L., L.L., A.C.-H., M.B., A.S., M.-E.D., A.T., C.G., N.G., J.-M.C., I.A., V.G., F.H., S.D., V.L., Y.C., N.I., H.D., E.M., A.P., P.R., A.B., and N.B. provided study materials or patients; M.S., L.V., J.L., and N.B. analyzed the data and performed statistical analysis; M.S., L.V., V.A., and N.B. wrote the original manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: Y.C. has received consulting fees for advisory board from Merck Sharp & Dohme, Novartis, Incyte, Bristol Myers Squibb, Pfizer, AbbVie, Roche, Jazz, Gilead, Amgen, AstraZeneca, and Servier; and travel support from Merck Sharp & Dohme, Roche, Gilead, Amgen, Incyte, AbbVie, Janssen, AstraZeneca, Jazz, and Sanofi, all via the institution. F.H. has received consulting fees for advisory boards and travel support from Novartis, Incyte, Pfizer, Amgen, and Servier. The remaining authors declare no competing financial interests.
Correspondence: Vahid Asnafi, Laboratory of Onco-Hematology, Necker-Enfants Malades Hospital, 149 rue de Sèvres, 75015 Paris, France; email: vahid.asnafi@aphp.fr; and Nicolas Boissel, Adolescent and Young Adult Hematology Unit, Saint Louis University Hospital, Assistance Publique–Hôpitaux de Paris, 1 avenue Claude Vellefaux, 75010 Paris, France; email: nicolas.boissel@aphp.fr.
References
Author notes
M.S. and L.V. contributed equally to this study.
Data are available upon request from the corresponding authors, Vahid Asnafi (vahid.asnafi@aphp.fr) and Nicolas Boissel (nicolas.boissel@aphp.fr).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Identification of new prognostic molecular in adult T-ALL. (A) Oncoplot depicting the genetic anomalies observed in HR-NGS or LR-NGS patients of the GRAALL03/05 trials. Genes are classified by functional groups. (B) Bar plot highlighting the frequency of gene alterations in the 198 adults with T-ALL issued from the GRAALL03/05 trial (cutoff of ≥4 mutations per gene [2%]). (C) SHRs of relapse according to genes alterations in adult T-ALL. Error bars indicate 95% CI. P values are from univariable Fine and Gray models. (D) Trace plot of Fine and Gray model with LASSO penalization. The trace plot visualizes the result of model selection process for predicting CIR in adult T-ALL using gene alterations at diagnosis. The selected model, chosen by fivefold cross-validation, is marked with a vertical dashed line on the trace plot. The variables colored and labeled were included in the chosen model with nonnull coefficients and were used to construct the NGS classifier. Red and blue colors indicate variables with an increased or a reduced risk of relapse, respectively. LASSO, least absolute shrinkage and selection operator; SE, standard error.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/15/10.1182_blood.2023023754/3/m_blood_bld-2023-023754-gr2.jpeg?Expires=1769081886&Signature=jjtdNd2oCl2I1AjOneAY7fZiVwfT0mYoh2ftXItc0m0K09GlThuIvSICXe79QyG44htwdXl~ezTsjXzB0~qEevRSdEcJPbqobkALSRHZYO~3on15-BldYONtMw80TxfPnOj0wQQFxogiIYmpFJttn6T~ajnKuE7uWo7IsDnymasTom3ngSjRvMre424QL9VqO0das9EpK-bZFHyWrxWy1tWwYH8ChZ8PHvmb1HxdbUt-jAXFIp1ilk6pCy7WvI~hYKGD2h4rudcLw8H4~wL51MzCJvxj11jb0huSQhZ~JFYfdbLHVirqN-Tqoa-h0JDH~BicOxLozQ5MuyBPZyRxjg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal