Key Points
KDM6A binds NLRC5 and CIITA loci and regulates their transcription in myeloma cells.
Decreased expression of NLRC5 and CIITA in KDM6A-deficient cell lines can be rescued by HDAC3 inhibition
Visual Abstract
The histone H3 at lysine 27 (H3K27) demethylase lysine demethylase 6A (KDM6A) is a tumor suppressor in multiple cancers, including multiple myeloma (MM). We created isogenic MM cells disrupted for KDM6A and tagged the endogenous protein to facilitate genome-wide studies. KDM6A binds genes associated with immune recognition and cytokine signaling. Most importantly, KDM6A binds and activates NLRC5 and CIITA, which encode regulators of major histocompatibility complex genes. Patient data indicate that NLRC5 and CIITA are downregulated in MM with low KDM6A expression. Chromatin analysis shows that KDM6A binds poised and active enhancers and KDM6A loss led to decreased H3K27ac at enhancers, increased H3K27me3 levels in body of genes bound by KDM6A, and decreased gene expression. Reestablishing histone acetylation with an HDAC3 inhibitor leads to upregulation of major histocompatibility complex expression, offering a strategy to restore immunogenicity of KDM6A-deficient tumors. Loss of Kdm6a in Kirsten rat sarcoma virus (K-RAS)-transformed murine fibroblasts led to increased growth in vivo associated with decreased T-cell infiltration.
Introduction
Multiple myeloma (MM), the second most common hematologic cancer, is a malignant proliferation of immunoglobulin-secreting plasma cells. Despite significant advances in treatment strategies, MM remains an incurable disease and accounts for 20% of all deaths from hematologic cancers. Molecular genetics studies of MM1-6 indicated that deregulation of transcription factors (musculoaponeurotic fibrosarcoma oncogene homolog [MAF], interferon regulatory factor 4 [IRF4], and myelocytomatosis oncogene [MYC]) and cofactors (nuclear receptor binding SET domain protein 2 [NSD2]) is common in MM and often due to rearrangement with the immunoglobulin enhancers. More recently, next generation sequencing surveys indicated the presence of mutations affecting a range of epigenetic regulator genes in up to 25% of patients with MM.4 Among the affected genes are those encoding the histone demethylase KDM6A, the histone methyltransferases lysine methyltranferase 2C and 2D (KMT2C and KMT2D), the histone acetyltransferase (HAT) CREB binding protein (CREBBP), and the chromatin remodeling subunit AT-rich interaction domain 1A (ARIDIA). These proteins interact together to regulate enhancers,7 emphasizing the importance of enhancer deregulation in MM.
Examination of the cBioportal8 reveals that KDM6A undergoes inactivating mutations and deletions across many tumor types with an incidence of 4% (2075/51 380) and is associated with a >15% decrease in median survival time (59 months vs 69 months) and 28% decrease in disease-free survival 104 months vs 144 months).8,KDM6A is present on the X chromosome and is expressed from both X alleles, whereas in men the homologous UTY (ubiquitously transcribed tetratricopeptide repeat containing, Y-linked) gene is expressed alongside with KDM6A from a single gene. Loss of the X chromosome is common in MM with a reported incidence of 18% to 47% predominantly in women.9,10,11,KDM6A missense mutations and exonic deletions occur in ∼3% of MM cases at diagnosis and are associated with poorer survival.12 Notably, more than one-third of all MM cell lines have KDM6A anomalies, and these cells were generally derived from advanced cases of MM, suggesting that KDM6A loss may be a progression factor in MM. In agreement with this idea, we showed that KDM6A acts as a tumor suppressor in MM by modulating cell growth and adhesion.13
The major KDM6A isoform is 1400 amino acids long and contains tetratricoid repeats involved in protein-protein interactions14 and a C-terminal Jumonji C (JmjC) demethylase domain that removes methyl groups from histone H3 at lysine 27 (H3K27) in a Fe2+-dependent reaction, requiring a-ketoglutarate as a cofactor. KDM6A interacts and works in concert with the COMPASS-like complex, which includes the H3K4me1 methyltransferases KMT2C or KMT2D. KDM6A helps activate enhancers by removing the repressive H3K27me3 chromatin modification and recruiting P300 or CREBBP HAT to acetylate H3K27 at active enhancers.15 The enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) methyltransferase component of the enhancer of zeste 2 polycomb repressive complex 2 subunit (PRC2) complex, which catalyzes the H3K27 trimethylation reaction, is often overexpressed in MM, and also correlates with poor prognosis.16,17 Chromatin profiling studies in primary MM18 also support the concept that an imbalance in epigenetic regulation at enhancers may contribute to the biology of MM. We found that PRC2/EZH2 inhibition can compensate for the loss of KDM6A, suggesting that the catalytic activity of KDM6A plays a role in tumor suppression. In contrast, KDM6A tumor suppressive functions have been linked to its ability to interact with transcription factors and other epigenetic regulators. Indeed, mutation in the TPR domains or deletion of the tetratricopeptide repeat (TPR) of KDM6A compromises interaction with the KMT2C/D complex of proteins associated with set1 (COMPASS)-like complex19 and abolishes growth suppression by KDM6A.20 Furthermore, the recruitment of HAT can be achieved by KDM6A mutant devoid of histone demethylase activity.21
Aiming to understand the function of KDM6A in MM, we used CRISPR engineered isogenic cell lines and chromatin immunoprecipitation sequencing (ChIP-seq) to identify KDM6A binding sites in the myeloma genome. We further explored how KDM6A deficiency affects chromatin structure at these loci by mapping genome-wide histone modifications. We found KDM6A bound at enhancers in proximity to genes regulating immune function, notably those encoding the master regulators of class I and II major histocompatibility complex (MHC), NOD-like receptor family CARD domain containing 5 (NLRC5), and class II major histocompatibility complex transactivator (CIITA). Furthermore, depletion of KDM6A leads to a decrease in H3K27ac at KDM6A-associated enhancers whereas H3K27me3 is decreased in the body of genes in which the promoter or the enhancer is bound by KDM6A. We found a correlation between expression of KDM6A and CIITA and NLRC5 in patients with MM. Modeling KDM6A loss in vivo in a murine system revealed impaired T-cell infiltration in tumors in which Kdm6a is depleted, suggesting that KDM6A regulates tumor immunogenicity.
Methods
Cell lines
The MM cell line ARP-1 RRID:CVCL_D523 was established at the University of Arkansas for Medical Sciences.22 The RPMI-8226 RRID:CVCL_0014, L363 RRID:CVCL_L363, EJM RRID:CVCL_2030, KMS12 RRID:CVCL_1334, AMO-1 RRID:CVCL_1806, and Karpas-620 RRID:CVCL_1823 cell lines were gifts of the late Michael Kuehl, National Cancer Institute. Cells were maintained in advanced RPMI-1640 medium (Invitrogen, Frederick, MD), supplemented with 5% fetal bovine serum (FBS) or 20% FBS (Karpas-620) or IMDM containing 10% FBS (EJM). All growth medium were supplemented with 1% penicillin (100 U/mL) and 1% streptomycin (100 μg/mL). Cell line authenticity was confirmed by short tandem repeat profiling (Labcorp Genetica, Burlington, NC).
CRISPR-Cas9 gene editing
Annealed guide RNA (gRNA)/tracer RNA ribonucleotides were mixed with Cas9 protein (Integrated DNA Technologies, Coralville, IA) at a ratio of 1:1 (0.8 μg of each) at room temperature for 5 minutes. The complex or Cas9 only (control wild-type [WT] cells) was mixed with a nonspecific oligonucleotide (IDT) and electroporated into 5 × 105 ARP-1 cells using a NEON (ThermoFisher Scientific, Waltham, MA) apparatus at a setting of a 20 milliseconds pulse at 1600 volts. The gRNA recognition sites are the following: KDM6A exon 1: TGCGTTTCCATGAAATCCTG; KDM6A exon 4 (clone E4): CAGCATTATCTGCATACCAG; and KDM6A exon 6 (clone E6): AGCTTTTGTCGAGCCAAGGA. Two days after electroporation, clones were isolated by limiting dilution. Identification of KDM6A mutants was performed by Sanger sequencing of the amplified targeted region. Primers for generation of amplicons were as follows: KDM6A exon 1 (forward: 5'-CATGAAATCCTGCGGAGTGT-3' and reverse: 5'-TACTCGTTAACGCTCAGGGA-3'); KDM6A exon 4 (forward: 5'-TGTGGTGGGAAT CTTGTTACC-3' and reverse: 5'-GCACAAACATAAATACTCTCAACCC-3'); and KDM6A exon 6 (forward: 5’-GTTTCAATGTACTACCAAGCAAGAA-3’ and reverse: 5’-ACCCAACAACCTACCTTTAAACT-3’). Deep sequencing of the amplicons was performed at the Center for Computational and Integrative Biology at Massachusetts General Hospital (Boston).
Results
Loss of KDM6A confers a transient growth advantage to MM cells
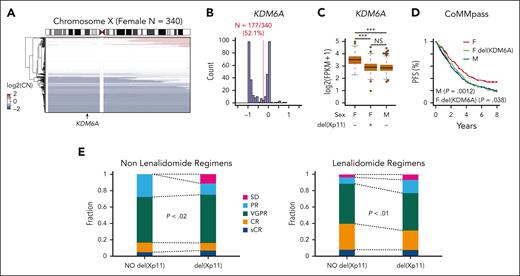
We previously demonstrated the tumor suppressive function of KDM6A in MM cell lines in vitro and in vivo using an add-back system.13 To further corroborate our previous findings, we examined KDM6A in patients with newly diagnosed MM from the clinical outcomes in MM to personal assessment of genetic profile (CoMMpass) trial. Consistent with previous results, we observed monoallelic chromosome X copy number loss, which encompasses the KDM6A locus in 52% of 340 female patients (Figure 1A-B). Copy number loss of KDM6A in females resulted in reduced KDM6A expression to levels equivalent to that of male samples (Figure 1C). KDM6A copy number loss also corresponded with reduced progression-free survival in female patients (Figure 1D). Responses to both lenalidomide and other regimens were inferior in female X chromosome with fewer complete responses in lenalidomide-treated patients and more cases of stable disease in patients treated with nonlenalidomide regimens (Figure 1E). By contrast, KDM6A expression was not associated with outcome in males (supplemental Figure 1, available on the Blood website). This suggests that KDM6A is a sex-specific tumor suppressor in MM; therefore, we continued experiments in female cell lines. Low expression of KDM6A was also found accompanying other negative lesions (eg, gain of chromosome 1q, myc translocation) and in better prognosis hyperdiploid MM, suggesting that KDM6A loss is one of many genetic factors that could play a role in MM prognosis (supplemental Table 1).
KDM6A monoallelic loss in female patients with myeloma corresponds with worse outcome. (A) Heat map of somatic copy number (CN) on the X chromosome for female patients with MM from CoMMpass. (B) Histogram of somatic CN for female patients. The red line denotes the threshold used for CN number loss and the number of female patients. (C) Expression of KDM6A in 703 patients with newly diagnosed MM from the CoMMpass trial with CN and RNA-sequencing data. P values determined by linear regression. (D) Progression-free survival for CoMMpass patients grouped into the categories shown in panel C. P values determined by Cox proportional hazards regression relative to the female (F; red) group. (E) Response of female patients with or without loss of the X chromosome, treated with lenalidomide or other regimens. SD, stable disease; PR partial response; VGPR, very good partial response; CR, complete response; sCR, stringent complete response (X2 test). FPKM, fragment per kilobase per million mapped fragment; M, male.
KDM6A monoallelic loss in female patients with myeloma corresponds with worse outcome. (A) Heat map of somatic copy number (CN) on the X chromosome for female patients with MM from CoMMpass. (B) Histogram of somatic CN for female patients. The red line denotes the threshold used for CN number loss and the number of female patients. (C) Expression of KDM6A in 703 patients with newly diagnosed MM from the CoMMpass trial with CN and RNA-sequencing data. P values determined by linear regression. (D) Progression-free survival for CoMMpass patients grouped into the categories shown in panel C. P values determined by Cox proportional hazards regression relative to the female (F; red) group. (E) Response of female patients with or without loss of the X chromosome, treated with lenalidomide or other regimens. SD, stable disease; PR partial response; VGPR, very good partial response; CR, complete response; sCR, stringent complete response (X2 test). FPKM, fragment per kilobase per million mapped fragment; M, male.
To confirm the consequences of KDM6A loss in MM, we transiently transfected female myeloma cell lines Karpas-620, AMO-1, and MM.1S with a ribonucleoprotein complex of CAS9 and KDM6A specific gRNAs. Gene editing of exon 4 was efficient with 40% to 60% of cells displaying predominantly 1, 2, and 5 bp deletions and 1 bp insertions (data not shown) within 2 days of transfection. By 8 days of growth, this number increased to 80% implying an initial growth advantage to cells devoid of KDM6A (supplemental Figure 2A). Gene editing with guides targeting exons 1, 4, or 6 yielded cell pools with dramatically reduced or absent levels of KDM6A protein in the absence of selection, suggesting a growth advantage to cells that acutely lose KDM6A expression (supplemental Figure 2B). However, after a few weeks in culture, KDM6A-depleted cell pools did not exhibit a proliferation advantage over a WT cell pool that was electroporated with a nontargeting gRNA as shown by a cell surface dye dilution assay (supplemental Figure 2C) nor a change in cell cycle distribution or rates of apoptosis (data not shown). This suggests that KDM6A may have growth suppressive function and its acute loss may stimulate cell growth, but that cells can adapt and reach a new homeostatic state. This led to us to consider other potential tumor suppressive mechanism of KDM6A.
KDM6A binds genes that regulates the immune system
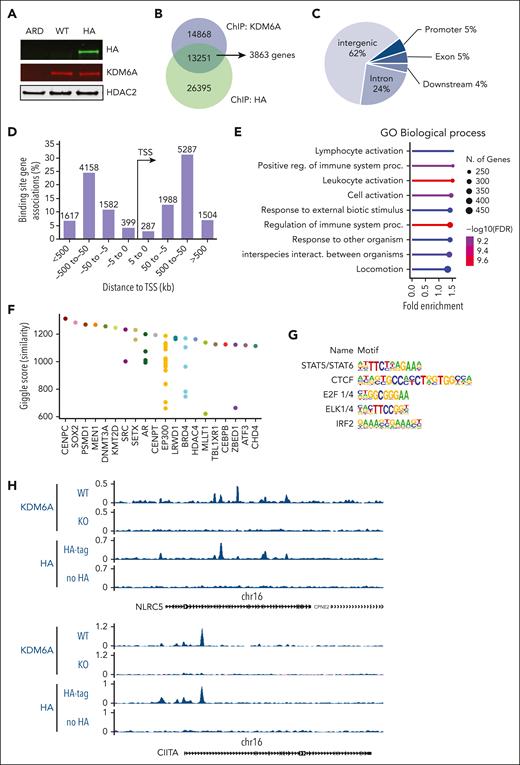
To determine direct targets of KDM6A in MM, we used ChIPmentation23 in 2 female myeloma cell lines, ARP-1 and Karpas-620, which express WT KDM6A.24 In ARP-1 cells using a polyclonal anti-KDM6A antibody, we identified 28 119 peaks that were absent in KDM6A-null ARP-1 E4 cells. To validate KDM6A binding sites and the quality of the KDM6A antibody, we inserted a dual hemagglutinin (HA) tag sequence into the C-terminal of KDM6A by CRISPR-mediated gene editing of ARP-1 cells (Figure 2A). Using the HA antibody, we detected 13 251 binding sites overlapping with the those detected with an antibody against KDM6A (Figure 2B; supplemental Table 2). These binding sites are mainly found in intergenic regions (62%) and introns (24%) (Figure 2C), consistent with the localization of KDM6A to enhancers. With the use of Genomic Regions Enrichment of Annotations Tool (version 4.0.4),25 we linked these peaks to 3863 genes at distances <100 kb. Most of the peaks (8371) were between 50 kb to 500 kb away from any gene transcription start sites (Figure 2D). To obtain a functional understanding of KDM6A regulated network in myeloma, we performed pathway enrichment analysis of the genes found less than 100 kb away from KDM6A binding sites using ShinyGo v0.8,26 using a background reference of all genes expressed in the ARP-1 cells (fragments per kilobase per million mapped fragments [FPKM] >1). Consistent with our previous findings, we observed that many bound genes are associated with cell signaling, motility, and adhesion. Interestingly however, most of the KDM6A-bound genes are related to the regulation of immune system (Figure 2E). Comparing the KDM6A binding pattern with previously reported ChIP-seq experiments reported in the Cistrome database,27 we found similarity of KDM6A binding to that of KMT2D and EP300, supporting a potential functional interaction of KDM6A with the COMPASS-like complex in myeloma cells (Figure 2F). Motif enrichment analysis of the genomic region bound to KDM6A using the hypergeometric optimization of motif enrichment (HOMER) package28 revealed significant enrichment of binding sites for CCCTC-binding factor (CTCF), E2F1/4, and the erythroiblast transformaton specific (ETS) related ELK1/4 as well as interferon regulatory factor 2 (IRF2) and signal transducer and activator of transcription (STAT5), the latter two effectors of immune signaling (Figure 2G; supplemental Table 3). Interestingly, among immune-related genes bound by KDM6A, we find genes coding for the transcriptional regulators of class I MHC (MHC I), NLRC5, and of class II (MHC II), CIITA (Figure 2H), suggesting a direct role for KDM6A in the regulation of gene transcription. In addition, KDM6A bound to the genes encoding the immunotherapeutic targets CD38 and SLAMF7 and the natural killer (NK) cell modulator CD48 (supplemental Figure 3A). To corroborate these findings, ChIP-seq analysis of Karpas-620 cells with KDM6A antibody revealed >60 000 peaks, 6163 overlapping with the highly validated peaks found by both HA and KDM6A antibodies in ARP-1 cells (supplemental Figure 3B). These binding sites were found to be within 100 kb of 2268 genes involved in the immune system, response to stimulus, and cell motility (supplemental Figure 3C). Motif enrichment analysis identified the same transcription factor recognition sites as those found in the ARP-1 cell line, ie, CTCF, STAT5, E2F1, and IRF2 (supplemental Figure 3D; supplemental Table 3).
KDM6A binding sites in myeloma cell lines. (A) Immunoblot showing the detection of KDM6A by an HA antibody in ARP-1 cells in which both alleles of endogenous KDM6A were HA tagged using CRISPR-Cas9 gene editing. ARD is a KDM6A-negative control cell line. (B) Overlap of KDM6A binding sites detected by chromatin precipitation of an HA-tagged ARP-1 cell line with anti-HA or anti-KDM6A antibodies. (C) Distribution of KDM6A binding sites in the annotated regions of the genome in the ARP-1 cell line. (D) Distance and orientation between KDM6A binding regions and their closest genes. (E) Dot plot visualization of gene ontology (GO) biological process enrichment analyses of KDM6A-bound genes using the GoShiny v0.77 tool. The size of the dots reflects the number of genes, the length of the line indicates fold enrichment, and color scale indicates false discovery rate. (F) Similarity of KDM6A binding pattern with those of transcription factors found in the Cistrome database (top 20). Each dot represents a different ChIP experiment. (G) Hypergeometric optimization of motif enrichment (HOMER)–identified enriched transcription factor binding motif within regions bound by KDM6A as identified both by KDM6A and HA antibody in ARP-1 cells expressing HA-tagged KDM6A. (H) Genome browser view of the CIITA and NLRC5 locus in ARP-1 cell line replete or knocked out or KO for KDM6A. FDR, false discovery rate; TSS, transcriptional start site.
KDM6A binding sites in myeloma cell lines. (A) Immunoblot showing the detection of KDM6A by an HA antibody in ARP-1 cells in which both alleles of endogenous KDM6A were HA tagged using CRISPR-Cas9 gene editing. ARD is a KDM6A-negative control cell line. (B) Overlap of KDM6A binding sites detected by chromatin precipitation of an HA-tagged ARP-1 cell line with anti-HA or anti-KDM6A antibodies. (C) Distribution of KDM6A binding sites in the annotated regions of the genome in the ARP-1 cell line. (D) Distance and orientation between KDM6A binding regions and their closest genes. (E) Dot plot visualization of gene ontology (GO) biological process enrichment analyses of KDM6A-bound genes using the GoShiny v0.77 tool. The size of the dots reflects the number of genes, the length of the line indicates fold enrichment, and color scale indicates false discovery rate. (F) Similarity of KDM6A binding pattern with those of transcription factors found in the Cistrome database (top 20). Each dot represents a different ChIP experiment. (G) Hypergeometric optimization of motif enrichment (HOMER)–identified enriched transcription factor binding motif within regions bound by KDM6A as identified both by KDM6A and HA antibody in ARP-1 cells expressing HA-tagged KDM6A. (H) Genome browser view of the CIITA and NLRC5 locus in ARP-1 cell line replete or knocked out or KO for KDM6A. FDR, false discovery rate; TSS, transcriptional start site.
KDM6A is found at enhancers and regulates H3K27me3 in gene bodies
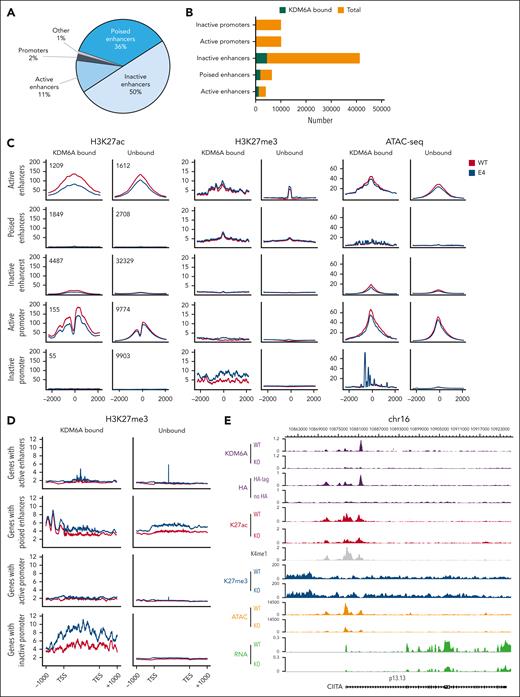
To understand how KDM6A regulates chromatin to induce transcriptional changes in MM, we mapped H3K27ac, H3K27me3, and H3K4me1 in ARP-1 clonal cell lines using ChIPmentation and assessed DNA accessibility by assay for transposase-accessible chromatin with sequencing (ATAC-seq). We defined enhancers as all H3K4me1 peaks greater than 2 kb from transcriptional start site whereas active enhancer had both H3K27ac and H3K4me1. Poised enhancers harbored H3K27me3 and H3K4me1 and inactive enhancers were marked only by H3K4me1 without H3K27ac or H3K27me3. KDM6A was found to be associated mainly with inactive enhancers (50% of binding sites) followed by poised enhancers (36%), active enhancers (11%), and very few promoters (2%; Figure 3A). Active gene networks affected by KDM6A loss were involved in lymphocyte activation, immune system development, and cell motility, and they were downregulated in KDM6A-deleted cells, reflecting an activating function of KDM6A (supplemental Figure 4A-B).
KDM6A loss impact on chromatin structure. (A) Distribution of KDM6A binding sites annotated regions of the genome in ARP-1 cell lines. (B) Box plot showing the proportion of total enhancers and promoters bound by KDM6A. (C) H3K27ac-seq, H3k27me3-seq, and ATAC-seq (assay for transposase-accessible chromatin with sequencing) signals in ARP-1 isogenic clonal cell line WT or KO for KDM6A centered on H3K4me1 peaks for enhancers and centered on transcription start sites for promoters. (D) H3K27me3 metagene analysis at locus bound by KDM6A in ARP-1 isogenic clonal cell line WT or KO for KDM6A. (E) Genome browser view of the CIITA locus in ARP-1 cell line WT or KO for KDM6A. TES, transcription end site; TSS, transcription start site.
KDM6A loss impact on chromatin structure. (A) Distribution of KDM6A binding sites annotated regions of the genome in ARP-1 cell lines. (B) Box plot showing the proportion of total enhancers and promoters bound by KDM6A. (C) H3K27ac-seq, H3k27me3-seq, and ATAC-seq (assay for transposase-accessible chromatin with sequencing) signals in ARP-1 isogenic clonal cell line WT or KO for KDM6A centered on H3K4me1 peaks for enhancers and centered on transcription start sites for promoters. (D) H3K27me3 metagene analysis at locus bound by KDM6A in ARP-1 isogenic clonal cell line WT or KO for KDM6A. (E) Genome browser view of the CIITA locus in ARP-1 cell line WT or KO for KDM6A. TES, transcription end site; TSS, transcription start site.
KDM6A deficiency caused a genome-wide decrease in DNA accessibility independent of KDM6A localization (Figure 3C; supplemental Figure 4C) and a reduction in H3K27ac at active enhancers and promoters that it bound. In addition, KDM6A loss led to increased H3K27me3 in wide genomic areas covering the body of genes and found in close proximity to enhancers and promoters bound by KDM6A (Figure 3C-D). All these changes were evident at the NLRC5, CIITA, CD38, CD48, and SLAMF7 loci, in which KDM6A was bound at multiple sites in intronic enhancers and promoters (Figure 3E; supplemental Figure 5).
Using a CRISPR knock in strategy, we created an ARP-1 cell line harboring a mutation within the enzymatic JmjC domain in which 2 amino acids essential for KDM6A enzymatic activity, H1146 and E1148,29,30 were replaced with alanine residues. ChIP-seq indicated a very modest decrease of H3K27ac at active enhancers (supplemental Figure 6A). By contrast, there was an increase in H3K27me3 in the body of genes with inactive promoters in JmjC-dead KDM6A expressing cell similar to that of KDM6A-null cells (supplemental Figure 6B).
Loss of KDM6A is associated with decreased expression of genes regulating immunity
By CRISPR-mediated gene editing, we generated additional isogenic clonal cell lines with and without KDM6A and analyzed the downstream impact on the transcriptome. In EJM cells we used a gRNA directed to exon 6 of KDM6A to generate a homozygous knockout (KO). In the AMO-1 creation of a heterozygous mutant clonal cell line was achieved using a gRNA directed against exon 1 (Figure 4A). There was no consistent pattern of gene upregulated with KDM6A loss across the cell lines (supplemental Figure 6A, right panel). There was a more consistent overlap of genes downregulated by KDM6A KO in the 3 cell lines including MHC II genes. More specifically, we found HLA-DRB1, HLA-DPA1, HLA-DR, HLA-DPB1, and the gene encoding the master regulator of MHC II gene expression, CIITA (supplemental Figure 7A; supplemental Table 4). A heat map representation of RNA-sequencing data (displaying z scores) showed decreased expression of most MHC II genes in AMO-1 and EJM clonal cell lines depleted for KDM6A, and MHC I genes were downregulated in KDM6A-null ARP-1 cells (Figure 4B). Flow cytometry analysis confirmed significant downregulation of MHC II and HLA-DR/DQ/DP expression at the cell surface of AMO-1 and EJM KDM6A KO cells (Figure 4C) whereas ARP-1 cells do not express detectable MHC II. Similarly, MHC I (HLA-A/B/C) expression decreased when KDM6A was eliminated in ARP-1 (Figure 4C; supplemental Figure 7B) and in AMO-1 cell lines (Figure 4C). We confirmed that NLRC5 and CIITA were significantly downregulated at the messenger RNA (mRNA) level in KDM6A-depleted clonal cell lines (Figure 4E). In addition, CD38 and CD48 protein expression at the cell surface was also severely decreased in ARP-1 and AMO-1 cell lines upon depletion of KDM6A (supplemental Figure 7C).
KDM6A controls MHC I and II gene expression. (A) Immunoblot showing depletion of KDM6A protein in CRISPR edited clonal cell lines. (B) Heat map of z scores for all expressed MHC genes in KDM6A WT and KO cell lines. (C) Flow cytometry analysis of HLA-A/B/C and HLA-DM/DQ/DR in clonal isogenic cell lines ARP-1, AMO-1, and EJM. Bottom panels of each histograms represent the mean fluorescence intensity (MFI) quantification (3 to 6 biological replicates; ± standard deviation [SD] Wilcoxon t test). (D) mRNA analysis by quantitative polymerase chain reaction (qPCR) normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 3-5 biological replicates; ± SD. Wilcoxon t test). (E) Normalized expression values in transcript per million (TPM) of NLRC5 and CIITA in myeloma patient tumors within the top or bottom decile of KDM6A expression (38 females and 54 males in each group, unpaired t test). (F) Multiplex enzyme-linked immunosorbent assay analysis of cytokine in ARP-1 WT and KO clonal cells line (Mann-Whitney t test). max, maximum; min, minimum; ns, not significant.
KDM6A controls MHC I and II gene expression. (A) Immunoblot showing depletion of KDM6A protein in CRISPR edited clonal cell lines. (B) Heat map of z scores for all expressed MHC genes in KDM6A WT and KO cell lines. (C) Flow cytometry analysis of HLA-A/B/C and HLA-DM/DQ/DR in clonal isogenic cell lines ARP-1, AMO-1, and EJM. Bottom panels of each histograms represent the mean fluorescence intensity (MFI) quantification (3 to 6 biological replicates; ± standard deviation [SD] Wilcoxon t test). (D) mRNA analysis by quantitative polymerase chain reaction (qPCR) normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 3-5 biological replicates; ± SD. Wilcoxon t test). (E) Normalized expression values in transcript per million (TPM) of NLRC5 and CIITA in myeloma patient tumors within the top or bottom decile of KDM6A expression (38 females and 54 males in each group, unpaired t test). (F) Multiplex enzyme-linked immunosorbent assay analysis of cytokine in ARP-1 WT and KO clonal cells line (Mann-Whitney t test). max, maximum; min, minimum; ns, not significant.
We reanalyzed the data of the Multiple Myeloma Research Foundation CoMMpass comprising 694 male and 460 female patients in which KDM6A expression levels were available. Using the top and bottom 10th percentile as a cutoff for high and low expression of KDM6A, respectively we observed that NLRC5 and CIITA were significantly decreased in both male and female KDM6A low-expressing tumors (Figure 4E). Furthermore, genes included in the antigen presentation pathway of the gene ontology were downregulated in the low KDM6A group in both male and female patients (supplemental Figure 8A). RNA expression profiles of MM cells included in the DepMap database31 show that those cell lines that express very low levels of KDM6A display lower levels of NLRC5 and CIITA and tended to have lower expression of MHC I and II genes (supplemental Figure 8B-C). Multiplex enzyme-linked immunosorbent assays showed that several important cytokines were express at lower levels in the supernatant of KDM6A-depleted cells, including the chemotactic cytokines for T cells, CC motif chemokine ligand 5 (CCL5) and 7 (CCL7), the inflammatory CCL3, interferon alfa-2, and the growth factor Fms-related receptor tyrosine kinase 3 ligand (Figure 4F). Conversely, the mitogenic factor CX3C motif chemokine receptor 1 and the platelet-derived growth factor subunit A were elevated in the media of KDM6A-depleted cells. Furthermore, transcriptional changes induced by IFNα stimulation in ARP-1 cells were attenuated when KDM6A was depleted (supplemental Figure 9A). Notably, although IFNα upregulated MHC I and II mRNA (supplemental Figure 9B) and MHC II protein at the surface of ARP-1 cells, this induction is blunted in KDM6A-depleted cells (supplemental Figure 9C).
Re-expression of KDM6A was able to induce cell surface expression of MHC II in some KDM6A-null cell lines but did not affect MHC I expression (supplemental Figure 10). The inconsistent rescue of MHC expression with re-expression of KDM6A suggests that a lasting reprogramming of the epigenome occurs in cells after KDM6A loss.
HDAC3 inhibition restores MHC expression in KDM6A-depleted cells
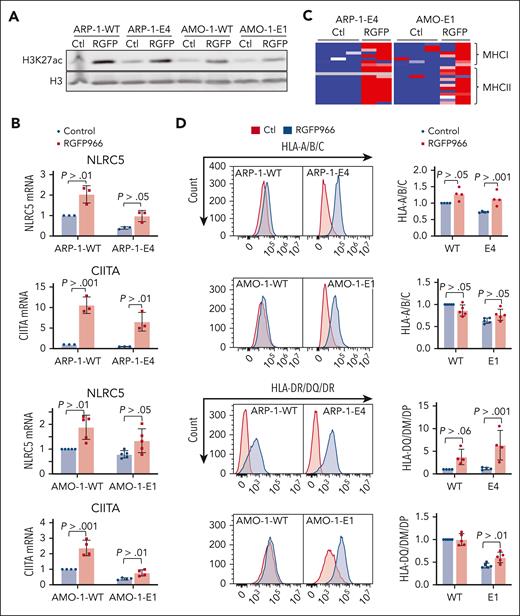
Decreased H3K27ac in KDM6A-null cells suggests that lower activity of CREBBP/EP300 at KDM6A binding sites may lead to decreased expression of KDM6A-associated genes. We previously found that histone deacetylase 3 (HDAC) was the enzyme responsible for counterbalancing CREBBP/P300–mediated acetylation of histones in myeloma cell lines.32 Accordingly, treatment of KDM6A KO ARP-1 and AMO-1 MM cells with the HDAC3-selective inhibitor RGFP966 increased total level of acetylated H3K27 (Figure 5A) and upregulated the mRNA for NLRC5 and CIITA (Figure 5B) and their downstream targets, genes encoding MHC I and II molecules (Figure 5C). In addition, HDAC3 inhibition was able to upregulate CD38, CD48, and SLAMF7 mRNA levels in KDM6A KO cell lines, AMO-1 and ARP-1 (supplemental Figure 11). Furthermore, RGFP966 restored MHC I and MHC II expression at the cell surface of KDM6A KO cells (Figure 5D).
HDAC3 inhibition restores MHC expression in MM cell lines. (A) Immunoblot of H3K27ac 24 hours after treatment with 10 μM RGFP966. (B) Heat map of z scores for all expressed MHC genes in KDM6A clonal isogenic cell lines. (C) mRNA analysis by qPCR normalized to GAPDH 24 hours after treatment with 10 μM RGFP966 (3-5 biological replicates; ± SD. Wilcoxon t test). (D) Flow cytometry analysis of HLA-A/B/C and HLA-DM/DQ/DR in myeloma isogenic cell lines treated or not with 10 μM RGFP966 for 24 hours or 72 hours. Panel on the right shows the histograms MFI quantification of HLA-A/B/C and HLA-DM/DQ/DR (4 or 5 biological replicates; ± SD, Wilcoxon t test). Ctl, control.
HDAC3 inhibition restores MHC expression in MM cell lines. (A) Immunoblot of H3K27ac 24 hours after treatment with 10 μM RGFP966. (B) Heat map of z scores for all expressed MHC genes in KDM6A clonal isogenic cell lines. (C) mRNA analysis by qPCR normalized to GAPDH 24 hours after treatment with 10 μM RGFP966 (3-5 biological replicates; ± SD. Wilcoxon t test). (D) Flow cytometry analysis of HLA-A/B/C and HLA-DM/DQ/DR in myeloma isogenic cell lines treated or not with 10 μM RGFP966 for 24 hours or 72 hours. Panel on the right shows the histograms MFI quantification of HLA-A/B/C and HLA-DM/DQ/DR (4 or 5 biological replicates; ± SD, Wilcoxon t test). Ctl, control.
KDM6A depletion decreases tumor immunogenicity in vivo
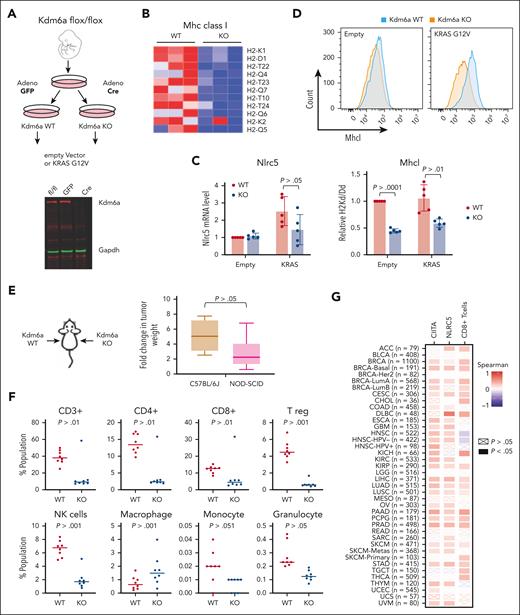
To test whether decreased MHC expression caused by KDM6A loss may affect tumor immunogenicity in vivo, we developed a mouse embryonic fibroblast (MEF) cell line by 3T3 immortalization protocol33 from C57JBL/6J mice that possess LoxP sites flanking exon 3 of Kdm6a.34 These fibroblasts were infected with an adenovirus expressing Cre recombinase to disrupt Kdm6a or a control adenovirus harboring green fluorescent protein (Figure 6A). Because activating mutations of the Ras/MAPK pathway are found in about 50% of MM,35we transformed WT and Kdm6a mutant cells by transducing a retrovirus expressing K-RAS harboring the G12V mutation. There was a modestly increased growth in Kdm6a KO MEF (supplemental Figure 12A). Growth was increased in K-RAS expressing cells but did not differ between Kdm6a-null and Kdm6a-replete cells (supplemental Figure 8A). RNA sequencing showed many MHC I genes expressed in WT fibroblasts harboring oncogenic Ras were downregulated in the KO cells (Figure 6B) along with Nlrc5 (Figure 6C) and genes involved in antigen presentation and type I interferon signaling (supplemental Figure 12B). Flow cytometry confirmed downregulation of MHC I on the cell surface of Kdm6a-null cells (Figure 6D). As in the case of the myeloma cell lines, loss of Kdm6a was associated with decreased in H3K27 acetylation at the Nlrc5 locus (supplemental Figure 12C). HDAC3 inhibition by RGFP-966 increased H3K27 acetylation (supplemental Figure 12D) and Nlrc5 mRNA levels (supplemental Figure 12E) and partially restored the expression of MHC I in Kdm6a-null MEFs (supplemental Figure 12F).
KDM6A depletion decreases MHC I expression in MEFs and decreases tumor immunogenicity in vivo. (A) Schematic of the protocol used to develop isogenic Kdm6a KO MEF from a C57BL/6J mouse homozygous for an allele of Kdm6a in which exon 3 was flanked with LoxP sites. (B) Heat map of z score for all expressed MHC I genes in Kdm6a WT and KO MEF. (C) Nlrc5 mRNA analysis by qPCR normalized to GAPDH (5 biological replicates; ± SD. Mann-Whitney t test). (D) Flow cytometry analysis of H-2kb in Kdm6a WT or KO MEFs (upper panel). MFI quantification of H-2kb surface expression from upper panel (5 biological replicates; ± SD, Mann-Whitney t test) (lower panel). (E) K-Ras-transformed WT and Kdm6a KO MEFs were injected into left and right flanks respectively of C57BL/6J and NOD-SCID mice and tumors excised and weighed after 3 weeks. The ratio of tumor weights from animal injected with Kdm6a-replete or Kdm6a-deficient K-Ras-transformed fibroblasts was calculated in immunocompetent C57BL/6J and immunodeficient NOD-SCID mice (8 biological replicates; ± SD, Mann-Whitney t test). (F) Flow cytometry quantifications of T cell, NK cells, and macrophage populations in the tumors isolated in panel E. (G) Spearman correlation between KDM6A and NLRC5 or CIITA gene expression and CD8+ T-cell infiltration in tumors isolated from various human cancers and correlation between KDM6A expression and T cell infiltrates as determined using Cistrome TIMER 2.0.36
KDM6A depletion decreases MHC I expression in MEFs and decreases tumor immunogenicity in vivo. (A) Schematic of the protocol used to develop isogenic Kdm6a KO MEF from a C57BL/6J mouse homozygous for an allele of Kdm6a in which exon 3 was flanked with LoxP sites. (B) Heat map of z score for all expressed MHC I genes in Kdm6a WT and KO MEF. (C) Nlrc5 mRNA analysis by qPCR normalized to GAPDH (5 biological replicates; ± SD. Mann-Whitney t test). (D) Flow cytometry analysis of H-2kb in Kdm6a WT or KO MEFs (upper panel). MFI quantification of H-2kb surface expression from upper panel (5 biological replicates; ± SD, Mann-Whitney t test) (lower panel). (E) K-Ras-transformed WT and Kdm6a KO MEFs were injected into left and right flanks respectively of C57BL/6J and NOD-SCID mice and tumors excised and weighed after 3 weeks. The ratio of tumor weights from animal injected with Kdm6a-replete or Kdm6a-deficient K-Ras-transformed fibroblasts was calculated in immunocompetent C57BL/6J and immunodeficient NOD-SCID mice (8 biological replicates; ± SD, Mann-Whitney t test). (F) Flow cytometry quantifications of T cell, NK cells, and macrophage populations in the tumors isolated in panel E. (G) Spearman correlation between KDM6A and NLRC5 or CIITA gene expression and CD8+ T-cell infiltration in tumors isolated from various human cancers and correlation between KDM6A expression and T cell infiltrates as determined using Cistrome TIMER 2.0.36
KRAS-transformed Kdm6a-replete or Kdm6a-deleted fibroblasts were injected into each flank of immunocompromised NOD-SCID mice and immunocompetent C57BL/6 mice. Kdm6a-null tumors were twice the size of Kdm6a WT masses in immunocompromised mice but grew 5 times bigger in immunocompetent mice (Figure 6E) suggesting that tumor suppressive functions of Kdm6a are partially dependent on an intact immune system. Flow cytometry analysis of immune cell infiltrates of Kdm6a-replete tumors grown in C57JBL/6J mice indicates an active immune environment that was compromised in Kdm6a KO tumors as exhibited by a marked decrease in infiltrating T cells (CD4+, CD8+, and Treg), NK cells, granulocytes, and monocytes (Figure 6F). Furthermore, using the Cistrome TIMER 2.0 database,36 we found a positive correlation between KDM6A and both NLRC5 and CIITA expression in a variety of tumor types, as well as a positive correlation between KDM6A expression and CD8 T-cell infiltration within the tumor (Figure 6G). Collectively, these data support an important role of KDM6A in the immune response of a variety of tumors that may be mediated partially through its ability to regulates MHC expression.
Discussion
KDM6A is a regulator of enhancer activity and a tumor suppressor in MM. Here, we found that KDM6A binds to enhancers associated with genes regulating immune recognition pathways, notably, the master regulators of MHC I and MHC II, NLRC5, and CIITA. KDM6A loss was associated with a marked decreased in H3K27ac at active enhancers, and restoration of histone acetylation using a HDAC3-selective inhibitor upregulates NLRC5, CIITA, and MHC expression in KDM6A-deficient MM cells. We further found that loss of Kdm6a compromised the immune response to a K-Ras driven malignancy, relevant to MM given that nearly 50% of patients with MM have activating mutations of the Ras/MAPK pathway.37,38
KDM6A loss led to decreased MHC I and MHC II gene expression in MM cell lines and murine fibroblasts. Loss of MHC I expression or loss of the capacity to induce upregulation of MHC I cell surface expression commonly occurs in malignant cells and serves as a strategy to escape T-cell recognition.39 MHC II is typically associated with professional antigen-presenting cells but increasing evidence shows that the MHC II antigen-presenting complex is expressed in tumors of various tissue including MM. Moreover, MHC II expression in tumors is associated with better outcomes in cancer patients and with tumor rejection in murine models (reviewed in40). Thus, therapeutic MHC I and MHC II upregulation is considered as a possible treatment maneuver in cancer and our data support considering this strategy in MM.
Studies support the idea that mutations or deregulation of epigenetic modifiers is associated with immune escape of tumors,41 and more specifically some research have suggested that KDM6A supports immunogenicity of tumor cells in bladder cancer42-44 and in medulloblastoma.45 Notably, a recent in vivo screen comparing murine tumor cell growth in immunocompetent and immunodeficient mice identifies many tumor suppressor genes encoding chromatin modifiers, including KDM6A, in the presence of an adaptive immune system only.46 Of note, PRC2, which opposes the action of KDM6A to promote gene silencing, is elevated and represses expression of MHC I antigen-presenting pathway.47-49 We extended these findings by showing that KDM6A directly binds enhancers for CIITA and NLRC5 activating their transcription, which in turn stimulate expression of MHC I and MHC II gene, respectively.
We found KDM6A bound to active and poised enhancers regulating mostly genes associated with immunity functions. Consistent with this, we observed diminished upregulation of IFNα target genes upon stimulation by IFNα in KDM6A-depleted cells. Analysis of chromatin at KDM6A-bound enhancers revealed its ability to modulate histone acetylation at H3K27 but, surprisingly, not H3K27 methylation. This is consistent with data from Wang et al,50 in which H3K27me3 levels at enhancers bound by KDM6A are not affected by its depletion. The decreased accessibility and acetylation at immune response gene enhancers support the idea that KDM6A is essential for the integrity of the COMPASS-like complex as others have suggested.50 Supporting this idea, disrupting KDM6A enzymatic activity did not reduce H3K27ac levels to that observed in KDM6A-null cells, suggesting a structural or scaffolding function of KDM6A to facilitate the recruitment or activity of CREBBP or EP300 and the COMPASS complex at enhancers. Moreover, we find that KDM6A localization across the genome in MM cells closely overlaps with previously defined patterns of EP300 and CREBBP, consistent with the ability to interact with these histone acetyl transferases (reviewed in51). We further show that enhancing histone acetylation using HDAC3 inhibitors helps to restore the expression of NLRC5, CIITA, and their respective MHC targets in both KDM6A WT and KDM6A-depleted cells, suggesting this agent might have a beneficial effect in multiple situations in which MHC expression is depressed in cancer cells. This is in line with research showing that activation of p300/CREBBP potentiates cancer chemoimmunotherapy through induction of MHC I antigen presentation.52 We also validated results very recently reported by Liu et al,53 demonstrating that KDM6A regulates the expression of CD38 and CD48 in myeloma cell lines. We extend these findings by showing direct binding of KDM6A to the CD38 and CD48 loci and an associated decreased in histone K27 acetylation at enhancers of these genes upon KDM6A loss. Moreover, we show that both H3K27ac and expression of CD38 and CD48 can be restored with HDAC3 inhibitor. This suggests that as Lui et al demonstrated with EZH2 inhibitors, HDAC3 inhibitors may provide a strategy to restore or boost the response to daratumumab and the ability to activate NK cells in KDM6A-depleted cells such as in patients with deletion of the X chromosome.
We found that methylation of H3 at lysine K27 increased in KDM6A-null cells, specifically in the body of genes in close proximity to enhancers and promoters normally bound by KDM6A. This contrasts with observations from other studies of KDM6A that focused on enhancers specific effects or relatively short genomic areas.54 Whether increased H3K27me3 is a cause or an effect of decreased gene expression is unknown. Lack of transcription was previously shown to trigger H3K27me3 accumulation in gene bodies.55 Thus, KDM6A loss may cause decreased gene expression through a decrease in H3K27ac, which in turn leads to H3K27me3 accrual.
KDM6A binding sites are enriched for motifs of IRF, STAT, and E2F transcription factors some of which were involved in regulating transcription of immunity genes. For instance, although generally recognized as a transcriptional repressor, IRF2 was found to act as a transcriptional activator for many key components of the MHC I pathway and is frequently downregulated in cancer.56
KDM6A loss and its ability to help cells escape immune surveillance may apply to other cancers. Inactivating mutations of KDM6A and KMT2C, KMT2D, and SWI/SNF components of its complex are among the most common found in all human cancers.57 In addition, the loss of X chromosome and X-linked gene expression occurs in 22% of all cancers. As such, pharmacological interventions to restore MHC and other immune function in tumor cells that have lost the activity of the enhancer activity complexes should be considered as part of the treatment approach to KDM6A mutant MM and other malignancies.
Acknowledgments
The authors thank Jonathan J. Keats and Sagar Lonial for CoMMpass genomic and clinical data, respectively.
This study was supported by the National Institutes of Health (NIH), National Cancer Institute grant number R01CA180475, the Leukemia and Lymphoma Society Specialized Center of Excellence, Samuel Waxman Cancer Research Foundation (J.D.L.), the Leukemia and Lymphoma Society Special Fellow Award (D.D.-R. and J.L.), the NIH/NCI (award numbers K22CA266739 [B.G.B.], CA260239 [W.Z.], and CA269661 [W.Z.)], the Paula and Roger Riney Foundation (L.H.B. and B.G.B.), the Myeloma Solutions Fund (J.D.L., L.H.B., and B.G.B.), the Rally Foundation for Childhood Cancer Research and Bear Necessities Pediatric Cancer Foundation (J.D.L.), and Investigador AECC (Asociacion Espanola Contra el Cancer) award from the Fundación AECC (INVES19059EZPO [T.E.]). University of Florida Health Cancer Center, supported by appropriations provided in Florida Statute 381.915 and NCI P30CA247796.
Authorship
Contribution: D.D.-R. designed and performed experiments, analyzed and interpreted data and wrote the manuscript; J.D.L. designed experiments, interpreted the data and wrote and revised the manuscript; B.G.B. analyzed the data and revised the manuscript; L.H.B., J.I.M.-S., C.S.M., R.L.B., and G.T. interpreted the data and revised the manuscript; W.Z. designed experiments and revised the manuscript; A.R. performed all bioinformatic analysis; U.D., J.L., G.Q., T.E., S.M., J.D.L., M.K., A.S. and H.C.R. performed experiments and analyzed data; and C.P. assisted with mouse work.
Conflict-of-interest disclosure: J.D.L. reports research support from Epizyme/Ipsen; and a consultancy role at AstraZeneca. L.H.B. reports a consultancy role at AstraZeneca; C.S.M. serves on the scientific advisory board of Adicet Bio; reports consultant/honoraria from Genentech, Nerviano, Secura Bio, and Oncopeptides; and research funding from EMD Serono, Karyopharm, Sanofi, Nurix, Bristol Myers Squibb, H3 Biomedicine/Eisai, Springworks, Abcuro, Novartis, and Opna. The remaning authors declare no competing financial interests.
Correspondence: Jonathan D. Licht, University of Florida Health Cancer Center, Cancer/Genetics Complex, 2033 Mowry Rd, Suite 145, Gainesville, FL 32610; email: jdlicht@ufl.edu.
References
Author notes
Next generation sequencing based data (RNA sequencing, assay for transposase-accessible chromatin with sequencing, and chromatin immunoprecipitation sequencing) are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession numbers, GSE244509, GSE244510, GSE244511, and GSE244512).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![KDM6A controls MHC I and II gene expression. (A) Immunoblot showing depletion of KDM6A protein in CRISPR edited clonal cell lines. (B) Heat map of z scores for all expressed MHC genes in KDM6A WT and KO cell lines. (C) Flow cytometry analysis of HLA-A/B/C and HLA-DM/DQ/DR in clonal isogenic cell lines ARP-1, AMO-1, and EJM. Bottom panels of each histograms represent the mean fluorescence intensity (MFI) quantification (3 to 6 biological replicates; ± standard deviation [SD] Wilcoxon t test). (D) mRNA analysis by quantitative polymerase chain reaction (qPCR) normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 3-5 biological replicates; ± SD. Wilcoxon t test). (E) Normalized expression values in transcript per million (TPM) of NLRC5 and CIITA in myeloma patient tumors within the top or bottom decile of KDM6A expression (38 females and 54 males in each group, unpaired t test). (F) Multiplex enzyme-linked immunosorbent assay analysis of cytokine in ARP-1 WT and KO clonal cells line (Mann-Whitney t test). max, maximum; min, minimum; ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/14/10.1182_blood.2024024518/2/m_blood_bld-2024-024518-gr4.jpeg?Expires=1763505047&Signature=T4FFaAtFmUtFtIlig2OnHnvZkSdeM6Ydvd3kxIxQw3~SExvSs2mZopKdn7eyTUus7nb5yeGSc2xZ3mQdBH9iipewNSD-zUoTNA1vwooFxCztcSC0uVKgJgpGYFapoBu1QNkRmgjtpOuPYWGEuco4eOq5ItCmP0P7fzKW-hesn1C2jfZHzoPRIEDUJhTFM6PVJGw1m5ekwuNekFuoWgMbCU1oVujDePII6vDrms3SWE6M2V1-Mf5xAvOsSOzuvZBBxzkCnxetW2KmrW6es1OB5pJ4G66m-OLESCLozgUpOAdM77KN86NCFhrpj9gIyLc4UVqVzGwWy~b~VexYPNEM8g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal