In this issue of Blood, Dupéré-Richer et al1 show that the histone demethylase KDM6A directly regulates the immunogenicity of multiple myeloma (MM) cells and compromises immune cell infiltration that could be restored by HDAC3 inhibition.
MM is characterized in part by structural variations occurring during the germinal center reaction. In about half of patients with MM, translocations are present that place oncogenes adjacent to the powerful enhancers of the immunoglobulin heavy chain locus.2 These initial events are quickly succeeded by further genetic changes that contribute to disease advancement and may impact patient outcomes.3 Furthermore, deregulation of epigenetic enzymes that play an important role in MM pathophysiology have also been reported.4 Among these, the histone demethylase KDM6A is associated with missense and exon deletions in around 3% of patients with MM at diagnosis.5KDM6A is located on the X chromosome and is frequently lost in MM. KDM6A helps activate enhancers by removing repressive H3K27me3 and recruiting histone acetyltransferases. EZH2, the histone methyltransferase catalyzing the H3K27 trimethylation, is overexpressed in MM and is associated with a poor outcome and resistance to treatment,6,7 suggesting that an imbalance in chromatin regulation of enhancers could play a major role in MM biology. To address these issues, Dupéré-Richer et al probed the biological function of KDM6A in MM using CRISPR-mediated gene editing, chromatin immunoprecipitation sequencing, and assay for transposase-accessible chromatin sequencing to identify the KDM6A binding sites and define the chromatin structure changes due to KDM6A deficiency. Using the CoMMpass cohort, the authors found that 52% of female patients showed monoallelic chromosome X copy number loss affecting the KDM6A locus. In female patients with MM, KDM6A copy number loss correlated with reduced progression-free survival and poorer responses to treatments, including lenalidomide. They also found that KDM6A depletion initially triggered increased cell growth in MM cell lines. However, over time, cells appeared to adjust to this change, establishing a new equilibrium in their growth patterns. In view of these results, the authors investigated other potential biological effects of KDM6A depletion.
Using chromatin immunoprecipitation sequencing, the authors show that KDM6A-bound genes were significantly enriched in cell signaling, motility, adhesion, and, importantly, immune system regulation genes. They also show that KDM6A's binding pattern closely resembled those of KMT2D and EP300, suggesting that KDM6A may functionally interact with the COMPASS-like complex within MM cells. The gene networks suppressed by KDM6A deletion were primarily associated with immune functions and cell movement. Specifically, pathways related to lymphocyte activation, immune system development, and cellular motility showed reduced activity in cells lacking KDM6A. The authors identified that the absence of KDM6A resulted in widespread changes to the chromatin landscape. At active enhancers and promoters where KDM6A normally binds, there was a decrease in H3K27ac, a finding associated with active chromatin. Conversely, the repressive H3K27me3 increased, particularly in gene bodies and in regions adjacent to enhancers and promoters usually bound by KDM6A, suggesting that KDM6A plays a crucial role in maintaining open chromatin and active gene expression states.
Dupéré-Richer et al used CRISPR-mediated gene editing to create isogenic cell lines with and without KDM6A. They demonstrate that loss of KDM6A in MM cell lines led to downregulation of genes involved in major histocompatibility complex (MHC) class I and II expression, potentially affecting immune recognition of these cells. They confirm the decrease of MHC I (HLA-A/B/C) and MHC II (HLA-DR/DQ/DP) expression by flow cytometry together with the downregulation of NLR5C and CIITA gene expression in KDM6A-depleted cells. Furthermore, they identify altered cytokine profiles in KDM6A-depleted cells, including decreased levels of T-cell chemotactic cytokines and inflammatory cytokines.
The authors then turn their attention to the fact that KDM6A-null cells showed decreased H3K27ac, suggesting lower CREBBP/EP300 activity at KDM6A binding sites. Interestingly, HDAC3 was previously found to counterbalance CREBBP/EP300-mediated histone acetylation in MM cell lines.8 They demonstrate that the treatment of KDM6A knockout MM cells with RGFP966, a selective inhibitor of HDAC3, resulted in increased total levels of acetylated H3K27; upregulation of NLRC5, CIITA, CD38, CD48, and SLAMF7 mRNAs; and restoration of MHC I and II cell surface expression. HDAC3 inhibition can partially compensate for the loss of KDM6A in MM cells, restoring the expression of genes involved in antigen presentation and immune recognition. The findings provide insight into potential therapeutic strategies for patients with MM also with KDM6A deficiency (see figure).
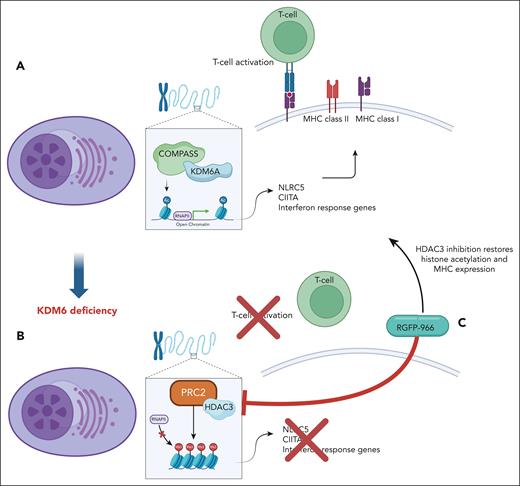
KDM6A controls genes related to immune response in MM. (A) Dupéré-Richer et al show that KDM6A regulates transcription of MHC’s master regulatory genes CIITA and NLRC5 in multiple myeloma cells by promoting acetylation of enhancers. (B) KDM6A deficiency results in decreased MHC’s expression and T-cell infiltration. (C) HDAC3 inhibitor reinstates acetylation of histones together with MHC expression in KDM6A-deficient MM cells.
KDM6A controls genes related to immune response in MM. (A) Dupéré-Richer et al show that KDM6A regulates transcription of MHC’s master regulatory genes CIITA and NLRC5 in multiple myeloma cells by promoting acetylation of enhancers. (B) KDM6A deficiency results in decreased MHC’s expression and T-cell infiltration. (C) HDAC3 inhibitor reinstates acetylation of histones together with MHC expression in KDM6A-deficient MM cells.
To determine the contribution of KDM6A loss on tumor immunogenicity in vivo, Dupéré-Richer et al created mouse embryonic fibroblasts with loxP-flanked Kdm6a. Kdm6a was disrupted using Cre recombinase and mouse embryonic fibroblasts were transformed with oncogenic KRAS (G12V mutation). There was a modest increase in cell growth and a downregulation of MHC class I genes, Nlrc5 mRNA levels, and genes involved in antigen presentation and type I interferon signaling. These effects were inhibited by RGFP-966, an HDAC3 inhibitor.
Focusing on in vivo tumor growth, the authors report that Kdm6a-null tumors grew larger than wild-type tumors in both immunocompromised and immunocompetent mice. However, the growth difference was more pronounced in the immunocompetent mice, suggesting KDM6A's tumor-suppressive functions are partially dependent on the immune system. The authors then reveal that Kdm6a-null tumors showed significant decreased infiltration of various immune cells, including infiltrating T cells, natural killer cells, granulocytes, and monocytes. Future investigations testing the therapeutic interest of HDAC3 inhibitor in vivo will be of major interest, especially to establish whether it could modulate immune cell invasion and in vivo tumor cell growth. Furthermore, HDAC3 has been reported to inhibit effector differentiation of T cells and cytotoxicity gene programs,9 suggesting that HDAC3 inhibition may have a dual advantage by promoting immune-cell infiltration and activation.
Finally, in analyzing the Cistrome TIMER2.0 database, the authors found a positive correlation between KDM6A expression and both NLRC5 and CIITA expression in several tumors. Furthermore, higher levels of KDM6A were associated with increased CD8 T-cell infiltration within tumors, suggesting that loss of KDM6A may contribute to tumor immune evasion, not only in MM, but in a wider variety of tumors. The authors conclude that KDM6A plays an important role in the immune response to tumors, potentially through its regulation of MHC expression. This suggests that loss of KDM6A may contribute to immune evasion of several tumors.
Dupéré-Richer and colleagues reveal a previously unrecognized role for the epigenetic regulator KDM6A in myeloma immune evasion and open the door for epigenome targeting to restore immune surveillance and tumor recognition in myeloma.
Conflict-of-interest disclosure: J.M. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal