A preclinical framework to epigenetically reprogram BCMA-specific CD8αβ+ memory T cells using iPSC technology to target MM.
BCMA-specific iPSC–derived hematopoietic progenitor cells having specific regulatory elements that control CD8+ T-cell lineage commitment.
Visual Abstract
A major hurdle in adoptive T-cell therapy is cell exhaustion and failure to maintain antitumor responses. Here, we introduce an induced pluripotent stem cell (iPSC) strategy for reprogramming and revitalizing precursor exhausted B-cell maturation antigen (BCMA)-specific T cells to effectively target multiple myeloma (MM). Heteroclitic BCMA72-80 (YLMFLLRKI)–specific CD8+ memory cytotoxic T lymphocytes (CTL) were epigenetically reprogrammed to a pluripotent state, developed into hematopoietic progenitor cells (CD34+ CD43+/CD14− CD235a−), differentiated into the T-cell lineage and evaluated for their polyfunctional activities against MM. The final T-cell products demonstrated (1) mature CD8αβ+ memory phenotype, (2) high expression of activation or costimulatory molecules (CD38, CD28, and 41BB), (3) no expression of immune checkpoint and senescence markers (CTLA4, PD1, LAG3, and TIM3; CD57), and (4) robust proliferation and polyfunctional immune responses to MM. The BCMA-specific iPSC–T cells possessed a single T-cell receptor clonotype with cognate BCMA peptide recognition and specificity for targeting MM. RNA sequencing analyses revealed distinct genome-wide shifts and a distinctive transcriptional profile in selected iPSC clones, which can develop CD8αβ+ memory T cells. This includes a repertoire of gene regulators promoting T-cell lineage development, memory CTL activation, and immune response regulation (LCK, IL7R, 4-1BB, TRAIL, GZMB, FOXF1, and ITGA1). This study highlights the potential application of iPSC technology to an adaptive T-cell therapy protocol and identifies specific transcriptional patterns that could serve as a biomarker for selection of suitable iPSC clones for the successful development of antigen-specific CD8αβ+ memory T cells to improve the outcome in patients with MM.
Introduction
Effective cancer immunotherapy strategies aim to boost effector T-cell development and function while abrogating mechanisms mediating immunosuppression in the tumor microenvironment. Adoptive T-cell therapy using ex vivo expanded cytotoxic T lymphocytes (CTL) against tumor-associated antigens provides an important immune defense against cancer and has achieved durable remissions in selected malignancies.1,2 Although there are clear benefits of using chimeric antigen receptor T-cell (CAR-T) or adoptive T-cell therapy, progress in the field has been impeded by exhaustion and senescence of these tumor targeting effector cells, which limits proliferation and functional activities against tumor cells.3-9 Among proposed methodologies to revitalize T cells, epigenetic reprogramming into induced pluripotent stem cell (iPSC), followed by T-cell lineage redifferentiation has shown encouraging results.10-13 However, there remains several unaddressed challenges before clinical application, including elimination of animal-derived feeder cells and identification of the iPSC clones that successfully differentiate into functional mature CD8αβ+ T cells.14-16 By overcoming these challenges, T cells differentiated from iPSC (iPSC–T cells) can provide functionally rejuvenated antigen-specific T cells that can be expanded ex vivo without repeated antigen stimulation, which is a driver of exhaustion and immune functional dysregulation.
Here, we have presented solutions to these challenges and used multiple myeloma (MM) as our model by targeting B-cell maturation antigen (BCMA), which is selectively expressed on a subset of mature B cells and long-lived plasma cells and has a favorable therapeutic index in MM.17-20 Parental BCMA-specific CTL, generated by repeated stimulation with our novel immunogenic heteroclitic BCMA72-80 (YLMFLLRKI) peptide,9 resulted in antigen-specific memory CTL with a precursor exhaustion phenotype (CD45RO+ CD83+) and polyfunctional anti-MM activities. To overcome this limitation, we applied iPSC technology to revitalize BCMA-specific CTL with a goal to enhance their functional activities against MM. The iPSC process begins with isolation of interferon gamma (IFN-γ) producing BCMA-specific memory CTL, epigenetically reprograming them into BCMA-specific iPSC, establishing hematopoietic progenitor cells (HPCs) from embryoid bodies and finally redifferentiating them into fully functional BCMA-specific CD8αβ+ memory T cells. The revitalized iPSC–T cells displayed a memory phenotype lacking expression of immune inhibitory and senescence markers (CTLA4, PD1, LAG3, and TIM3; CD57), which were abundantly expressed on the parental memory BCMA72-80 (YLMFLLRKI) peptide–specific CD8+ CTL (BCMA-CTL). In addition, our RNA sequencing (RNA-seq) analyses identified specific transcriptional profiles that could be useful to select specific iPSC clones that successfully differentiate into CD8αβ+ memory T cells. The improved iPSC process was further refined and shortened to allow for potential clinical application.
These results highlight the potential benefit of epigenetic reprogramming of precursor exhausted BCMA-specific CD8+ CTL that express immune checkpoints into iPSC, which provide a renewable source of pluripotent progenitor cells into rejuvenated, antigen-specific memory CTL under optimized feeder-free conditions. Furthermore, RNA-seq analyses identified unique whole-genome profiles and delineated specific T-cell pathways used by iPSC-derived HPCs for T-cell development. Early transcriptome identification of T-cell regulatory pathways will allow for the selection of appropriate iPSC clones in future studies for the optimal direction of cell differentiation and improve the overall process and production of exhaustion-free, antigen-specific T cells for therapeutic application. Together, these studies provide insights into improving the translation of iPSC technology into the clinic for treatment of MM or potentially other cancers.
Materials and Methods
Cells
Tumor cell lines, cells from patients with MM, cells from healthy donors, and various types of antigen-presenting cells were cultured at 37°C, 5% CO2. Detailed information is described in supplemental Methods, available on the Blood website.
Reagents
Detailed information on reagents including antibodies, cytokines, peptides, and tetramers is described in supplemental Methods.
Establishment of BCMA-specific iPSC
Heteroclitic BCMA-CTL were generated ex vivo by repeated peptide stimulation of T cells from HLA-A2+ donors as described previously.9 Upon MM stimulation, IFN-γ–producing BCMA-specific memory CTL were sorted using fluorescence-activated cell sorter (FACS) and used for iPSC establishment. The details are presented in supplemental Methods.
Development of BCMA-specific iPSC and evaluation of their pluripotency status, germ differentiation, karyotype, and Sendai virus residue
BCMA-specific iPSC (BCMA-iPSC) colonies were established, evaluated for specific phenotypes by FACS, pluripotency, and Giemsa banding patterns and karyotype and investigated on Sendai virus residue. The details are presented in supplemental Methods.
Formation of BCMA-specific embryoid body and differentiation of HPCs into antigen-specific T lymphocytes
Detailed procedures on embryoid body (EB) formation from BCMA-specific iPSC, isolation of BCMA-specific HPCs and differentiation into BCMA-specific iPSC-T lymphocytes are described in the supplemental Methods.
Characterization of iPSC-derived HPCs, BCMA iPSC–T cells, and apoptotic pathways in MM cell lines
iPSC-HPCs and iPSC-T cells were analyzed for their specific phenotype, proliferation, cytotoxic activities, CD107a degranulation, and T helper 1 (Th1) cytokine production, as appropriate. MM cells were analyzed for their proliferative- or apoptotic-related cascades upon exposure to BCMA iPSC–T cells. Details are presented in supplemental Methods.
Single-cell sequencing of TCR on BCMA-iPSC–T cells
T-cell receptor (TCR) sequence analyses were performed on single cells isolated from BCMA-iPSC–T cells using the RNase H-dependent PCR-enabled T-cell receptor sequencing (rhTCRseq) protocol.21 Details are described in the supplemental Methods.
Whole transcriptome profiling and GO enrichment analyses
FAC-sorted iPSC-derived HPCs were evaluated by RNA-seq analyses. For library preparation, SMART-Seq V4 ultralow input RNA-Seq kit (Takara Bio) was used to synthesize complementary DNA (cDNA). This involved priming the RNA's 3′ end with the coding sequence (CDS) primer, synthesizing the first strand of DNA by reverse transcription (RT), and employing the SMARTseq oligos for template switching, thus creating the second strand by RT before amplification by polymerase chain reaction. To make Illumina libraries, we sheared the cDNA to 150 bp using the Covaris M220 sonicator and then used the SMARTer Thruplex DNA-Seq kit (Takara Bio) to ligate the Illumina adapters and barcodes to the cDNA fragments. The libraries were sequenced with paired-end 50 bp reads targeting 30 million read pairs per sample on an Illumina NovaSeq 6000 platform. Sequenced reads were aligned to the UCSC hg19 reference genome assembly, and gene counts were quantified using STAR (version 2.5.1b).22 Differential gene expression testing was performed using DESeq2 (version 1.10.1)23 and normalized read counts (fragments mapped per kilobase per million [FPKM]) were calculated using cufflinks (version 2.2.1).24 RNA-seq analysis was performed using the VIPER snakemake pipeline.25 The database for annotation, visualization, and integrated discovery (DAVID 6.8 February 2021)26 was used to identify gene ontology (GO) categories, with P < .05. In addition, U266 MM cells exposed to iPSC–T cells or control U266 MM cells (no exposure) were sorted using FACS and evaluated for their transcriptional modifications by RNA-seq analyses using Partek Flow Genomic analysis software. Significant gene changes in MM cells were identified with a false discovery rate of 0.01 and a fold change of 2.
Statistical analysis
Summary results are presented as the mean ± standard deviation. Groups were compared using unpaired Student t tests. Differences were considered significant when ∗P < .05.
Results
BCMA-specific iPSC have pluripotency potential and normal karyotypes
The process for reprograming of BCMA-specific IFN-γ+ CTL, establishment of specific iPSC and EB, and differentiation into iPSC–T cells is outlined in Figure 1A. In brief, heteroclitic BCMA-CTL were generated ex vivo9, and the peptide-specific IFN-γ+ CTL were sorted using FACS and epigenetically reprogrammed into iPSC using OCT3/4, SOX2, KLF4, and c-MYC transcription factors delivered via Sendai viral vectors under iMatrix-511 feeder-free conditions. The BCMA-iPSC clones demonstrated continuous growth and development on days 8, 12, and 16 after transduction (supplemental Figure 1). Each iPSC clone on day 21 had high expression of SSEA-4 and TRA-1-60, which are stem-cell markers (N = 3; 96%-100%), as did the Epstein-barr virus (EBV)–specific iPSC clone (78%), which served as a positive control (established from EBV LMP2A426-434 peptide [CLGGLLTMV]–specific T cells; a kind gift from Kyoto University, Japan; Figure 1B). Parental BCMA-CTL served as a negative control and did not express the stem-cell markers. Another key marker of iPSC establishment was high expression (94%-100%) of the 3 representative germ layer markers, SOX-17 for endoderm, Brachyury for mesoderm, and Pax-6 for ectoderm (Figure 1C). In addition, immunohistochemical analyses revealed high expression of alkaline phosphatase, a marker of stem cell development and self-renewal potential, on BCMA-iPSC and EBV-specific iPSC, but not on primary T cells from blood (negative control; Figure 1D). Cytogenetic analyses of BCMA-iPSC chromosomes in metaphase revealed genomic stability and a normal karyotype without abnormalities (Figure 1E). Analysis of BCMA-iPSC clones (N = 3) using RT polymerase chain reaction (supplemental Figure 2) showed no residual Sendai virus, confirming viral vector clearance. These data demonstrate the pluripotency and self-renewal potential27 of the BCMA-iPSC, as demonstrated by the expression of stem-cell markers, capacity to differentiate into 3 germ layers, high alkaline phosphatase expression, and a normal karyotype.
Establishment of BCMA-iPSC from parental BCMA-specific CTL. (A) Schematic presentation of the protocol for reprograming of parental BCMA-specific IFN-γ+ CTL into iPSC and differentiation into BCMA-iPSC–T cells. (B, top) Representative flow cytometric analyses showing the pluripotency potential of BCMA-iPSC and EBV-iPSC (positive control) by expression of the stem-cell markers SSEA-4 and TRA-1-60 but not on parental BCMA-CTL (negative control). (B, bottom) Summary of 3 independent analyses (N = 3; mean ± standard deviation [SD]). (C, top) Representative flow cytometric analyses showing the expression of key germ cell layer markers, SOX-17 (endoderm), Brachyury (mesoderm), and Pax-6 (ectoderm), on BCMA-iPSC and EBV-iPSC (positive control). (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D) Representative immunohistochemistry analysis demonstrating alkaline phosphatase upregulation, a characteristic marker of stem cell development and self-renewal potential, on BCMA-iPSC and EBV-iPSC (positive control) but not on primary T cells. Photomicrographs (original magnification ×100) taken with an inverted microscope (Carl Zeiss). (E) Representative cytogenetic analysis of chromosomes for Giemsa banding patterns demonstrating genomic stability and a normal karyotype in BCMA-iPSC.
Establishment of BCMA-iPSC from parental BCMA-specific CTL. (A) Schematic presentation of the protocol for reprograming of parental BCMA-specific IFN-γ+ CTL into iPSC and differentiation into BCMA-iPSC–T cells. (B, top) Representative flow cytometric analyses showing the pluripotency potential of BCMA-iPSC and EBV-iPSC (positive control) by expression of the stem-cell markers SSEA-4 and TRA-1-60 but not on parental BCMA-CTL (negative control). (B, bottom) Summary of 3 independent analyses (N = 3; mean ± standard deviation [SD]). (C, top) Representative flow cytometric analyses showing the expression of key germ cell layer markers, SOX-17 (endoderm), Brachyury (mesoderm), and Pax-6 (ectoderm), on BCMA-iPSC and EBV-iPSC (positive control). (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D) Representative immunohistochemistry analysis demonstrating alkaline phosphatase upregulation, a characteristic marker of stem cell development and self-renewal potential, on BCMA-iPSC and EBV-iPSC (positive control) but not on primary T cells. Photomicrographs (original magnification ×100) taken with an inverted microscope (Carl Zeiss). (E) Representative cytogenetic analysis of chromosomes for Giemsa banding patterns demonstrating genomic stability and a normal karyotype in BCMA-iPSC.
BCMA-iPSC polarization results in mesoderm development during EB formation
The BCMA-iPSC underwent EB formation for efficient induction of antigen-specific HPCs. During EB formation, germ layer bias and polarization were evidenced by gradual increases (EB day 7 > EB day 4 > EB day 2 > iPSC) in the genes associated with mesoderm development, including ABCA4, ALOX15, BMP10, CDH5, CDX2, FOXF1, HAND1, HAND2, HOPX, NKX2-5, PDGFRA, PLVAP, SNAI2, and TBX328,29 (Figure 2A), which were not detected in the CD8+ T lymphocyte somatic cell controls (N = 2; Figure 2B). In summary, the genes contributing to mesoderm development were gradually upregulated (∗P < .05) during EB formation from BCMA-iPSC clones (N = 2; Figure 2C), whereas the genes contributing to self-renewal were downregulated (∗P < .05). Morphologically, the EB was fully developed by day 11 (photomicrograph original magnification ×100; supplemental Figure 3). These results provide evidence that the protocols applied for EB formation are effective in directing development of mesoderm lineage for blood cell formation.
Polarization of BCMA-iPSC into mesoderm germ layer. (A) Representative ScoreCard analysis demonstrating a gradual upregulation in genes associated with mesoderm development during EB formation on days 2, 4, and 7 from BCMA-iPSC relative to an undifferentiated reference. (B) ScoreCard analysis demonstrating no specific regulation in genes associated with germ layer development in somatic cell control CD8+ T lymphocytes (from donor 1 [D1] or donor 2 [D2]) relative to an undifferentiated reference. (C) Summary analyses of differential gene expression profiles associated with primary germ layer development in BCMA-iPSC (clone 1 and clone 2; N = 2) and EB (N = 2) (top) but not in somatic cell control CD8+ T lymphocytes (D1 and D2; N = 2) (bottom).
Polarization of BCMA-iPSC into mesoderm germ layer. (A) Representative ScoreCard analysis demonstrating a gradual upregulation in genes associated with mesoderm development during EB formation on days 2, 4, and 7 from BCMA-iPSC relative to an undifferentiated reference. (B) ScoreCard analysis demonstrating no specific regulation in genes associated with germ layer development in somatic cell control CD8+ T lymphocytes (from donor 1 [D1] or donor 2 [D2]) relative to an undifferentiated reference. (C) Summary analyses of differential gene expression profiles associated with primary germ layer development in BCMA-iPSC (clone 1 and clone 2; N = 2) and EB (N = 2) (top) but not in somatic cell control CD8+ T lymphocytes (D1 and D2; N = 2) (bottom).
BCMA iPSC–T cells differentiated from reprogrammed HPCs have high activation markers expression without immune checkpoint expression
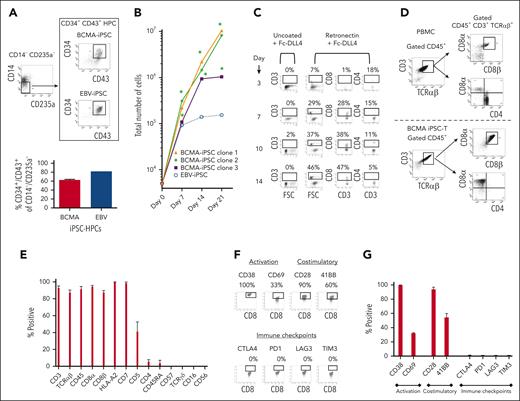
After BCMA-specific EB formation, the HPCs (CD34+ CD43+/CD14− CD235a−) were sorted using FACS (Figure 3A) and cultured in the presence of retronectin and Fc-DLL4 (chimera protein–recombinant human δ-like protein 4) to activate notch signaling for T-cell differentiation. The BCMA iPSC–derived HPCs (iPSC-HPCs; N = 3) demonstrated significant (∗P < .05) cell expansion on day 7 (total cell numbers; 183 × 103 ± 67 × 103), day 14 (440 × 103 ± 130 × 103), and day 21 (6600 × 103 ± 1047 × 103) from the number of cells initially seeded (5 × 103; Figure 3B). During the first 14 days of culture, iPSC-HPCs demonstrated a gradual cell differentiation (day 14 > 7 > 10 > 3) into CD3+ T lymphocytes with an increasing proportion of CD8+ cytotoxic T cells and decreasing proportion of CD4+ Th cells (Figure 3C). Full differentiation was completed by day 21 of culture, providing a high yield of CD3+ TCRαβ+/CD45+ and CD8αβ+ T cells and a corresponding low level (<5%) of CD4+ Th cells (Figure 3D). The final T-cell products (N = 3) exhibited a high (>85%) expression of CD3, TCRαβ, CD45, CD8α, CD8β, HLA-A2, and CD7 markers, lower CD5 (<5%), and minimum expression of CD4, CD45RA, TCRγδ, CD16, and CD56 (Figure 3E). In addition, the iPSC–T cells expressed a high level of CD38 (late activation), CD28, and 41BB (costimulatory), along with a lower level of CD69 (early activation; Figure 3F-G). Morphologically, BCMA iPSC–T cells were like normal T lymphocytes (supplemental Figure 4). Although parental BCMA-CTL expressed high levels of immune checkpoints (CTLA4, PD1, LAG3, and TIM3) and CD83, characteristic of precursor exhausted cells30,31 (supplemental Figures 5 and 6), iPSC–T cells lacked expression of such exhaustion as well as senescence (CD57) markers (Figure 3E-G). Additionally, the final iPSC–T cells had no CD4+ regulatory T cells (CD25+ FoxP3+; supplemental Figure 7), supporting their phenotypic characterization as a rejuvenated T-cell product. We further evaluated BCMA-iPSC as a renewable pool of cells for future BCMA iPSC–T–cell development. Successful T-cell differentiation was consistently demonstrated by both subcloned (supplemental Figure 8) and cryopreserved (8 or 16 months) (supplemental Figure 9) BCMA-iPSC. The final iPSC–T cells demonstrated a mature CD8αβ+ T-cell phenotype and high levels of CD3, CD45, and TCRαβ expression. These results support further subcloning and storage of antigen-specific iPSC in an as-needed basis.
Isolation and differentiation of iPSC-HPCs into BCMA iPSC–T cells with a mature CD8αβ+ cell phenotype. (A, top) Representative gating strategy for isolation of HPCs (CD34+ CD43+/CD14− CD235a−) from BCMA-specific EBs sorted using FACS. (A, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (B) Summary of the increase in total cell numbers during differentiation of BCMA-iPSC HPCs (N = 3) or EBV-specific iPSC HPCs from the initial number of cells (5 × 103; day 0) (∗P < .05). (C) Representative flow cytometric analyses demonstrating a gradual differentiation of BCMA iPSC-HPCs (day 14 > 10 > 7 > 3) into CD3+ and CD8+ T lymphocytes using retronectin-coated culture plates in the presence of Fc-DLL4. (D) Representative flow cytometric analysis comparing phenotype of primary T cells (CD3+ CD45+ TCR αβ+ / CD4+ CD8α+ CD8β+) in healthy donor’s peripheral blood mononuclear cells (top) and BCMA iPSC–T cells (CD3+ CD45+ TCR αβ+/CD4− CD8α+ CD8β+) on day 21 of T-cell differentiation (bottom). (E) Summary of overall BCMA iPSC–T-cell (N = 3; mean ± SD) phenotype on day 21 of the T-cell differentiation process. (F) Representative flow cytometric analyses of BCMA iPSC–T cells on day 21 of differentiation showing T-cell activation (CD38 and CD69) and costimulatory molecule (CD28 and 41BB) expression without immune checkpoint (CTLA-4, PD1, LAG3, and TIM3) expression. (G) Summary analyses of T-cell activation, costimulatory molecule, and immune checkpoint expression on BCMA iPSC–T cells (N = 3; mean ± SD) on day 21 of differentiation.
Isolation and differentiation of iPSC-HPCs into BCMA iPSC–T cells with a mature CD8αβ+ cell phenotype. (A, top) Representative gating strategy for isolation of HPCs (CD34+ CD43+/CD14− CD235a−) from BCMA-specific EBs sorted using FACS. (A, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (B) Summary of the increase in total cell numbers during differentiation of BCMA-iPSC HPCs (N = 3) or EBV-specific iPSC HPCs from the initial number of cells (5 × 103; day 0) (∗P < .05). (C) Representative flow cytometric analyses demonstrating a gradual differentiation of BCMA iPSC-HPCs (day 14 > 10 > 7 > 3) into CD3+ and CD8+ T lymphocytes using retronectin-coated culture plates in the presence of Fc-DLL4. (D) Representative flow cytometric analysis comparing phenotype of primary T cells (CD3+ CD45+ TCR αβ+ / CD4+ CD8α+ CD8β+) in healthy donor’s peripheral blood mononuclear cells (top) and BCMA iPSC–T cells (CD3+ CD45+ TCR αβ+/CD4− CD8α+ CD8β+) on day 21 of T-cell differentiation (bottom). (E) Summary of overall BCMA iPSC–T-cell (N = 3; mean ± SD) phenotype on day 21 of the T-cell differentiation process. (F) Representative flow cytometric analyses of BCMA iPSC–T cells on day 21 of differentiation showing T-cell activation (CD38 and CD69) and costimulatory molecule (CD28 and 41BB) expression without immune checkpoint (CTLA-4, PD1, LAG3, and TIM3) expression. (G) Summary analyses of T-cell activation, costimulatory molecule, and immune checkpoint expression on BCMA iPSC–T cells (N = 3; mean ± SD) on day 21 of differentiation.
BCMA iPSC–T cells have enhanced antitumor activities against myeloma cells through specific recognition of cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide
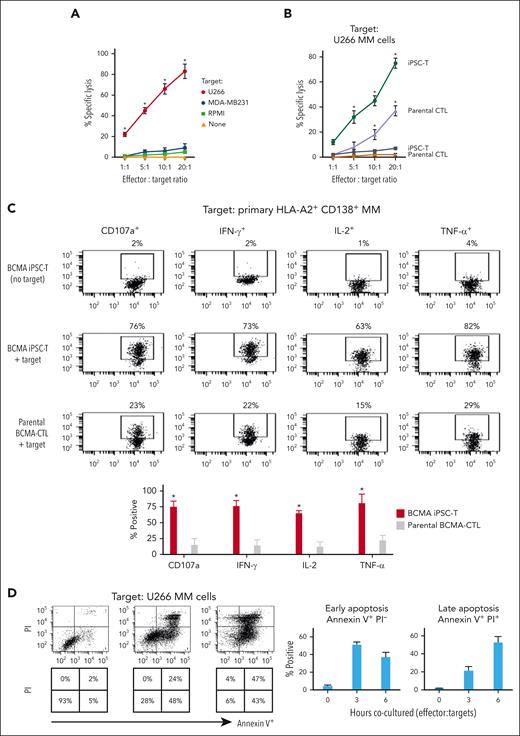
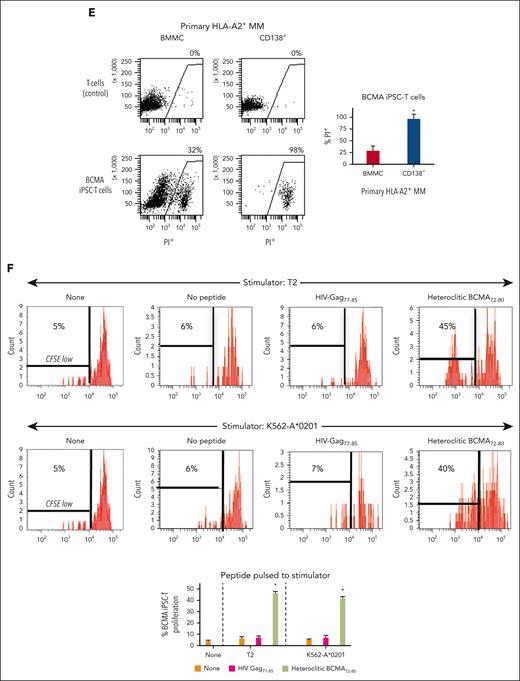
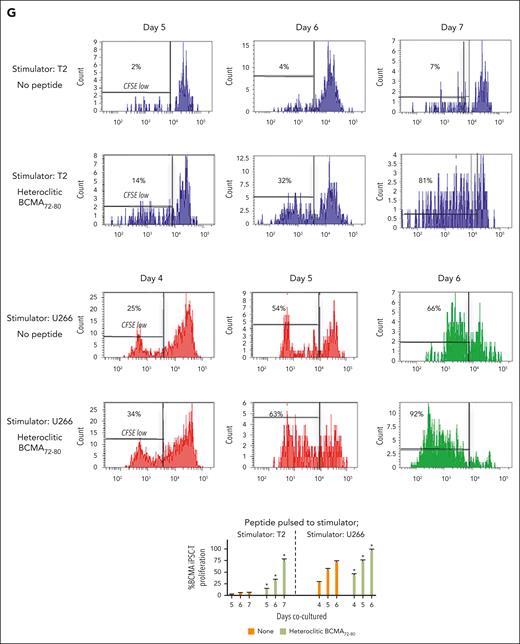
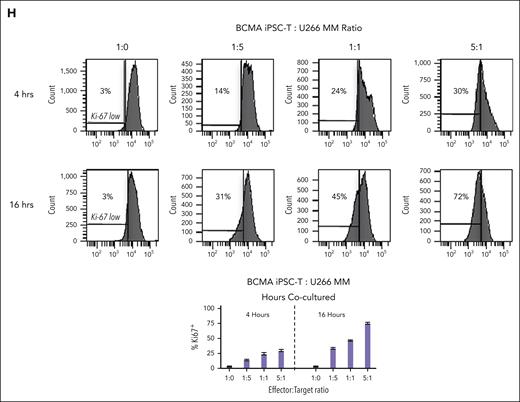
BCMA iPSC–T cells were evaluated for their functional activities and specific antitumor responses against MM. BCMA iPSC–T cells (N = 3) displayed a high (∗P < .05) level of direct cytotoxicity against HLA-A2+/BCMA+ U266 MM cells (22%-83%) at each effector-to-target cell ratio (1:1-20:1) but neither against antigen-mismatched MDA-MB231 (breast cancer; HLA-A2+/BCMA−) nor against major histocompatibility complex (MHC)–mismatched RPMI (MM; HLA-A2−/BCMA+) (Figure 4A), indicating their cytotoxic activity in an antigen-specific and HLA-A2–restricted manner. The anti-MM activities of iPSC–T cells were significantly higher (∗P < .05) than those of parental BCMA-CTL against HLA-A2+/BCMA+ U266 MM cells. Pretreatment of U266 MM cells overnight with 5 μg/mL of anti–HLA-A2–blocking antibody significantly reduced the cytotoxic activity (≤5% lysis) of both iPSC–T cells (∗P < .05) and parental BCMA-CTL as compared with their lysis of untreated U266 MM cells (Figure 4B), which further supports their HLA-A2–specific anti-MM activities. Next, polyfunctional immune responses were characterized in response to primary HLA-A2+/CD138+ cells from patients with MM or U266 MM cells. IPSC–T cells demonstrated significantly higher (∗P < .05) CD107a degranulation, a correlated measure of target cell killing,32 and Th1-type cytokine (IFN-γ, interleukin 2 [IL-2], and tumor necrosis factor α [TNF-α]) production against primary CD138+ MM target cells (Figure 4C) or U266 cells (supplemental Figure 10) than parental BCMA-CTL (N = 3) (Figure 4C). To broaden our understanding of effector cell cytotoxic activity, U266 MM target cells were evaluated for apoptosis-related cascades induced during coculture with iPSC–T cells. Viable U266 MM cells (annexin V−/propidium iodide [PI−], 0 hours) advanced through early-stage apoptosis (annexin V+/PI−, 3 hours) and late-stage apoptosis (annexin V+/PI+, 6 hours) during coculture with iPSC–T cells (Figure 4D). To determine the underlying mechanism of effector cell–induced apoptosis, caspase activation was evaluated in MM target cells using flow cytometry. Specific caspases activation in U266 cells was not detected after coculture with the iPSC–T cells (N = 3) (supplemental Figure 11), suggesting a mechanism of indirect involvement by caspase activation in the apoptosis process. To further confirm iPSC–T-cell cytotoxicity, MM target cell death was evaluated using an alternative assay with PI. The iPSC–T cells induced a significantly higher level of cell death in primary CD138+ MM cells (PI+; 98% ± 2%) compared with bulk BMMC (bone marrow mononuclear cells) (PI+; 29% ± 9%) from patients with newly diagnosed HLA-A2+ MM (∗P < .05; N = 5; Figure 4E). These results support efficient targeting of CD138+ tumor cells within BMMC by iPSC–T cells. The specific iPSC–T-cell proliferation (low in carboxyfluorescein diacetate succinimidyl ester [CFSE]) was evaluated in response to cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide presented on T2 or K562-A∗0201 cells (stimulator cells). The iPSC–T cells demonstrated specific recognition of cognate peptide with a high proliferative response (∗P < .05) as compared with irrelevant HLA-A2–specific HIV-Gag77-85 (SLYNTVATL) peptide or stimulator cells alone (Figure 4F). Significantly higher peptide-specific iPSC–T-cell proliferation (∗P < .05) occurred in a time-dependent manner in response to cognate peptide presented T2 cells (day 7 > 6 > 5) or U266 MM cells (day 6 > 5 > 4), as compared with T2 or U266 cells alone (Figure 4G). On the tumor cell side, U266 MM cells (CFSE-labeled) showed a decreased expression of Ki-67, a cellular marker of proliferation, after coculture with iPSC–T cells. The inhibited MM cell proliferation occurred in a time-dependent (16 > 4 hours) manner and iPSC–T-cell proliferation in a dose-dependent (effector-to-target cell ratio, 5:1 > 1:1 > 1:5) manner (Figure 4H). Finally, transcriptomic modifications regulating MM cell death were evaluated by comparing U266 MM cells exposed to iPSC–T cells with U266 MM cells alone (unexposed). RNA-seq analyses demonstrated that exposed U266 cells, as compared to unexposed cells, had changes in their transcriptional profiles, including upregulation of genes related to apoptosis (GNLY, DDIT4, GDF15, GOLPH3L) and a downregulation of genes (NR4A1, NR4A2) that have a pro-oncogenic role as important regulators of tumor cell growth and survival (supplemental Figure 12).33-39 Taken together, these results show that the BCMA iPSC–T cells demonstrate polyfunctional immune responses, direct antitumor killing, and/or specific proliferation in response to both primary and U266 MM cells in an antigen-specific and HLA-A2–restricted manner through their specific recognition of cognate BCMA peptide on MM cells.
Antigen-specific and HLA-A2–restricted anti-MM activities and proliferation of BCMA-specific iPSC-T cells through specific recognition of cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide. (A) Direct cytotoxic activity of BCMA iPSC–T cells (4-hour calcein-release assay) against U266 MM cells (HLA-A2+ BCMA+) occurred in an effector cell dose-dependent manner (effector-to-target cell ratios 1:1, 5:1, 10:1, and 20:1) but neither against antigen-mismatched MDA-MB231 breast cancer (HLA-A2+ BCMA−) nor against HLA-A2–mismatched RPMI MM (HLA-A2− BCMA+) cells. BCMA iPSC–T cells alone served as an effector cell control. Summary of 3 independent analyses (N = 3; mean ± SD). (B) Direct cytotoxic activity of BCMA iPSC–T cells and parental BCMA-CTLs (4-hour calcein-release assay) against HLA-A2+ BCMA+ U266 MM cells (● ▲) occurred in an effector cell dose-dependent manner (effector: target cell ratios = 1:1, 5:1, 10:1, and 20:1). There was complete inhibition of the cytotoxic activity after culturing U266 MM target cells (○ △) overnight with anti-HLA-A2 mAb (5 μg/mL), demonstrating their specific anti-tumor activities in an HLA-A2–restricted manner. Summary of 3 independent analyses (N = 3; mean ± SD). (C, top) Representative flow cytometric analyses showing higher CD107a degranulation and IFN-γ/IL-2/TNF-α cytokine production by BCMA iPSC–T cells than by parental BCMA-CTL in response to primary HLA-A2+ CD138+ MM cells (6-hour coculture). BCMA iPSC–T cells alone served as a negative control. Effector-to-target cell ratio, 1:1. (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D, top) Representative flow cytometric analyses showing early apoptosis (annexin V+ PI−; 3 hrs) continuing to late apoptosis (annexin V+ PI+; 6 hrs) in gated U266 MM target cells (CFSE-labeled) induced by the BCMA iPSC–T cells. Effector-to-target cell ratio, 1:1. (D, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (E, top) Representative flow cytometric analyses showing the cell death (PI+) of primary BMMC and CD138+ cells in BMMC from a patient with HLA-A2+ MM induced by BCMA iPSC–T cells (6-hour assay), but not in control HLA-A2+ T cells. Effector-to-target cell ratio, 1:1. (E, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (F, top) Representative flow cytometric analyses showing the specific proliferation of BCMA iPSC–T cells (CFSE-low) in response to cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide, not HLA-A2 irrelevant HIV Gag77-85 peptide–loaded stimulator cells (T2 or K562-A∗0201) on day 6 of coculture. BCMA iPSC–T cells alone or BCMA iPSC–T cells stimulated with T2 or K562-A∗0201 cells (no peptide loaded) served as controls. Responder-to-stimulator cell ratio, 1:1. (F, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (G, top) Representative flow cytometric analyses demonstrating a time-dependent increase in BCMA iPSC–T-cell proliferation (CFSE-low) in response to heteroclitic BCMA72-80 (YLMFLLRKI) peptide–loaded stimulator cells (T2, U266) on day 5, 6 or 7 of coculture. BCMA-iPSC–T cells stimulated with T2 or U266 cells (no peptide loaded) served as controls. Responder-to-stimulator cell ratio, 1:1. (G, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (H, top) Representative flow cytometric analyses showing the downregulation of Ki-67, a cellular proliferation marker, in U266 MM cells (CFSE-labeled) upon 4 or 16 hours of coculture with BCMA iPSC–T cells in a time-dependent and effector cell dose-dependent (effector-to-target cell ratios 1:5, 1:1, and 5:1) manner. (H, bottom) Summary of results (N = 3; mean ± SD).
Antigen-specific and HLA-A2–restricted anti-MM activities and proliferation of BCMA-specific iPSC-T cells through specific recognition of cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide. (A) Direct cytotoxic activity of BCMA iPSC–T cells (4-hour calcein-release assay) against U266 MM cells (HLA-A2+ BCMA+) occurred in an effector cell dose-dependent manner (effector-to-target cell ratios 1:1, 5:1, 10:1, and 20:1) but neither against antigen-mismatched MDA-MB231 breast cancer (HLA-A2+ BCMA−) nor against HLA-A2–mismatched RPMI MM (HLA-A2− BCMA+) cells. BCMA iPSC–T cells alone served as an effector cell control. Summary of 3 independent analyses (N = 3; mean ± SD). (B) Direct cytotoxic activity of BCMA iPSC–T cells and parental BCMA-CTLs (4-hour calcein-release assay) against HLA-A2+ BCMA+ U266 MM cells (● ▲) occurred in an effector cell dose-dependent manner (effector: target cell ratios = 1:1, 5:1, 10:1, and 20:1). There was complete inhibition of the cytotoxic activity after culturing U266 MM target cells (○ △) overnight with anti-HLA-A2 mAb (5 μg/mL), demonstrating their specific anti-tumor activities in an HLA-A2–restricted manner. Summary of 3 independent analyses (N = 3; mean ± SD). (C, top) Representative flow cytometric analyses showing higher CD107a degranulation and IFN-γ/IL-2/TNF-α cytokine production by BCMA iPSC–T cells than by parental BCMA-CTL in response to primary HLA-A2+ CD138+ MM cells (6-hour coculture). BCMA iPSC–T cells alone served as a negative control. Effector-to-target cell ratio, 1:1. (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D, top) Representative flow cytometric analyses showing early apoptosis (annexin V+ PI−; 3 hrs) continuing to late apoptosis (annexin V+ PI+; 6 hrs) in gated U266 MM target cells (CFSE-labeled) induced by the BCMA iPSC–T cells. Effector-to-target cell ratio, 1:1. (D, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (E, top) Representative flow cytometric analyses showing the cell death (PI+) of primary BMMC and CD138+ cells in BMMC from a patient with HLA-A2+ MM induced by BCMA iPSC–T cells (6-hour assay), but not in control HLA-A2+ T cells. Effector-to-target cell ratio, 1:1. (E, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (F, top) Representative flow cytometric analyses showing the specific proliferation of BCMA iPSC–T cells (CFSE-low) in response to cognate heteroclitic BCMA72-80 (YLMFLLRKI) peptide, not HLA-A2 irrelevant HIV Gag77-85 peptide–loaded stimulator cells (T2 or K562-A∗0201) on day 6 of coculture. BCMA iPSC–T cells alone or BCMA iPSC–T cells stimulated with T2 or K562-A∗0201 cells (no peptide loaded) served as controls. Responder-to-stimulator cell ratio, 1:1. (F, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (G, top) Representative flow cytometric analyses demonstrating a time-dependent increase in BCMA iPSC–T-cell proliferation (CFSE-low) in response to heteroclitic BCMA72-80 (YLMFLLRKI) peptide–loaded stimulator cells (T2, U266) on day 5, 6 or 7 of coculture. BCMA-iPSC–T cells stimulated with T2 or U266 cells (no peptide loaded) served as controls. Responder-to-stimulator cell ratio, 1:1. (G, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (H, top) Representative flow cytometric analyses showing the downregulation of Ki-67, a cellular proliferation marker, in U266 MM cells (CFSE-labeled) upon 4 or 16 hours of coculture with BCMA iPSC–T cells in a time-dependent and effector cell dose-dependent (effector-to-target cell ratios 1:5, 1:1, and 5:1) manner. (H, bottom) Summary of results (N = 3; mean ± SD).
BCMA iPSC memory T cells have a rejuvenated phenotype with improved antimyeloma activity and display a single specific TCR clonotype
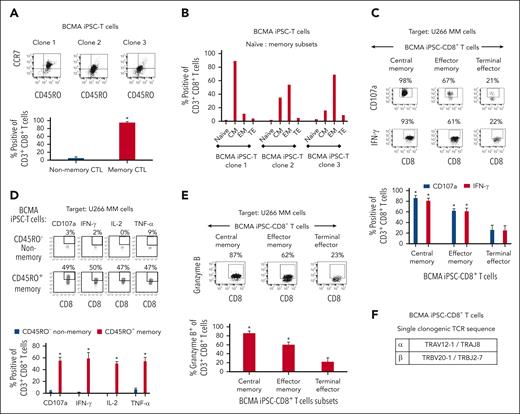
BCMA iPSC–T-cell memory subsets were evaluated for their specific roles and functional anti-MM activities. Phenotypic analyses demonstrated that BCMA iPSC–T cells (N = 3) were predominantly (∗P < .05) CD45RO+ memory CTL rather than CD45RO− nonmemory CTL (Figure 5A). Specifically, iPSC–T cells had high frequencies of central memory (CM; CCR7+/CD45RO+) and effector memory (EM; CCR7−/CD45RO+) cells (Figure 5B). Both CM and EM CD8+ T-cell subsets (N = 3) demonstrated significantly higher levels (∗P < .05) of CD107a degranulation and IFN-γ production against U266 MM cells than terminal effector CD8+ T cells with the highest activities occurring within the CM cell subset (Figure 5C). The CD45RO+ memory cell population demonstrated effective and significantly higher (∗P < .05) polyfunctional immune responses, including CD107a+ degranulation and Th1 cytokine (IFN-γ, IL-2, and TNF-α) production, than the CD45RO− nonmemory cell population in response to U266 MM cells (Figure 5D). Additionally, the CM and EM CD8+ CTL of BCMA iPSC–T cells (N = 3) demonstrated significantly higher (∗P < .05) levels of granzyme B production against U266 MM cells than the terminal effector CD8+ CTL (Figure 5E). The highest anti-MM activities were detected consistently in the CM cell subset, indicating their critical role inducing antitumor activities. Finally, the specific TCR expressed by BCMA iPSC–T cells was examined using single-cell sequencing of complementarity-determining region 3. The analyses of 88 single iPSC–T cells revealed a single clonotype (100%) of TCRα and TCRβ sequences, TRAV12-1/TRAJ8 paired with TRBV20-1/TRBJ2-7, respectively (Figure 5F).
Characterization of the high anti-MM activities within the memory subsets of BCMA iPSC–T cells. (A, top) Representative flow cytometric analyses of the naïve:memory phenotype in BCMA iPSC–CD8+ T cells differentiated from BCMA-iPSC clones 1, 2, and 3. (A, bottom) Summary of total nonmemory (CD45RO−) and memory (CD45RO+) cell distribution on gated viable CD8+ T cells of the BCMA iPSC–T cells (N = 3; mean ± SD). (B) Summary of naïve (CD45RO− CCR7+), CM (CD45RO+ CCR7+), EM (CD45RO+ CCR7−), and terminal effector (TE; CD45RO− CCR7−) cell frequencies in BCMA iPSC–CD8+ T cells differentiated from each individual BCMA-iPSC clone. (C, top) Representative flow cytometric analyses demonstrating higher levels of CD107a degranulation and IFN-γ production within the CM and EM cell subsets of BCMA iPSC–CD8+ T cells than TE cells in response to U266 MM cells (effector-to-target cell ratio, 1:1). (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D, top) Representative flow cytometric analyses demonstrating a significant induction of CD107a degranulation and Th1-type cytokine (IFN-γ/IL-2/TNF-α) production by CD45RO+ memory cells within BCMA iPSC–CD8+ T cells, not by CD45RO− nonmemory cells, in response to U266 MM cells. Effector-to-target cell ratio, 1:1. (D, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (E, top) Representative flow cytometric analyses showing higher granzyme B production by the CM and EM cell subsets within BCMA iPSC–CD8+ T cells compared with TE cells in response to U266 MM cells. Effector-to-target cell ratio, 1:1. (E, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (F) Identification of unique clonotype TCRα and TCRβ sequences based on single cell sequencing (N = 88; single cells analyses) in the complementarity-determining region 3 of BCMA iPSC–T cells.
Characterization of the high anti-MM activities within the memory subsets of BCMA iPSC–T cells. (A, top) Representative flow cytometric analyses of the naïve:memory phenotype in BCMA iPSC–CD8+ T cells differentiated from BCMA-iPSC clones 1, 2, and 3. (A, bottom) Summary of total nonmemory (CD45RO−) and memory (CD45RO+) cell distribution on gated viable CD8+ T cells of the BCMA iPSC–T cells (N = 3; mean ± SD). (B) Summary of naïve (CD45RO− CCR7+), CM (CD45RO+ CCR7+), EM (CD45RO+ CCR7−), and terminal effector (TE; CD45RO− CCR7−) cell frequencies in BCMA iPSC–CD8+ T cells differentiated from each individual BCMA-iPSC clone. (C, top) Representative flow cytometric analyses demonstrating higher levels of CD107a degranulation and IFN-γ production within the CM and EM cell subsets of BCMA iPSC–CD8+ T cells than TE cells in response to U266 MM cells (effector-to-target cell ratio, 1:1). (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D, top) Representative flow cytometric analyses demonstrating a significant induction of CD107a degranulation and Th1-type cytokine (IFN-γ/IL-2/TNF-α) production by CD45RO+ memory cells within BCMA iPSC–CD8+ T cells, not by CD45RO− nonmemory cells, in response to U266 MM cells. Effector-to-target cell ratio, 1:1. (D, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (E, top) Representative flow cytometric analyses showing higher granzyme B production by the CM and EM cell subsets within BCMA iPSC–CD8+ T cells compared with TE cells in response to U266 MM cells. Effector-to-target cell ratio, 1:1. (E, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (F) Identification of unique clonotype TCRα and TCRβ sequences based on single cell sequencing (N = 88; single cells analyses) in the complementarity-determining region 3 of BCMA iPSC–T cells.
BCMA-iPSC–derived HPCs have a distinct transcriptional profile that regulates CD8+ T-cell differentiation
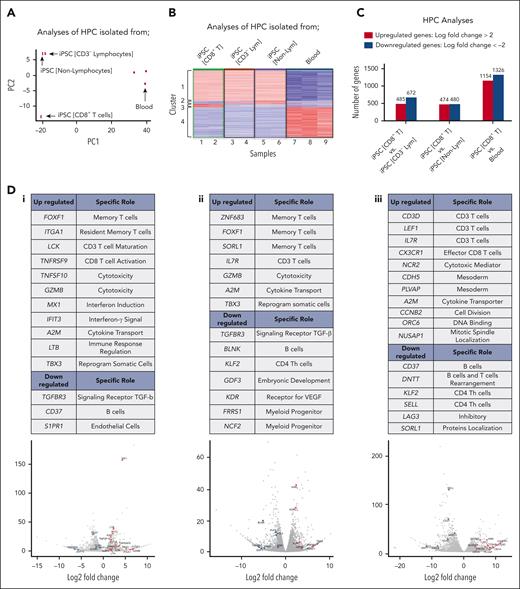
The BCMA-iPSC clones (N = 20) established in these studies differentiated into 3 distinct lineages, namely, group 1: CD8+ T cells; iPSC-HPCs (CD8+ T cells); group 2: CD3– lymphocytes (B cells and natural killer cells; iPSC-HPCs [CD3− lymphocytes]); and group 3: nonlymphocytes (monocytes and granulocytes; iPSC-HPCs [nonlymphocytes]). Based on these results, the transcriptome of HPCs enriched from each individual iPSC clone or primary blood from healthy donors were sorted and evaluated to identify specific genes regulating differentiation into the cell lineage by RNA-seq analyses. After confirmation of high-quality purified RNA by viper output analyses (supplemental Figures 13-16), principal component (PC) analyses revealed distinct clusters and a low variance in transcriptomes among the iPSC-HPCs replicates based on their normalized gene expression (Figure 6A). The PC1 axis distinguished iPSC-HPCs from primary blood HPCs (control), whereas PC2 axis distinguished iPSC-HPCs (CD8+ T cells; group 1) from iPSC-HPCs that differentiated into other cell types (groups 2 and 3). Heat map analyses of the top 1000 highly expressed genes identified 4 distinct cluster patterns based on gene expression levels across the iPSC-HPCs and primary blood HPCs. Results are shown in Figure 6B as follows: upregulated (cluster 1) or downregulated (cluster 4) genes in iPSC-HPCs compared with in the primary blood HPCs, upregulated (cluster 3), or downregulated (cluster 2) genes in iPSC-HPCs (CD8+ T cells) compared with in iPSC-HPCs (CD3− lymphocytes) and iPSC-HPCs (nonlymphocytes) (Figure 6B). Significant gene expression changes were detected in iPSC-HPCs (CD8+ T cells) as compared with in iPSC-HPC (CD3− lymphocytes) (upregulated, 485 genes; downregulated, 672 genes) or iPSC-HPC (nonlymphocytes) (upregulated, 474 genes; downregulated, 480 genes) (adjusted P value < .05; Figure 6C), indicating distinctive gene regulatory patterns controlling the commitment to T-cell development. The transcriptional profile of iPSC-HPCs (CD8+ T cells) revealed an upregulation (>1.5 log2; Padj < .05) of genes involved in pathways regulating memory T-cell differentiation (FOXF1 and ITGA1), CD3+ T-cell maturation (LCK), CD8+ T-cell activation and cytotoxicity (TNFRSF9 [4-1BB], TNFSF10 [TRAIL], and GZMB), IFN induction and signaling (MX1 and IFIT3), cytokine transport (A2M), and immune response regulation (LTB)40,41 as opposed to iPSC-HPCs (CD3− lymphocytes) (Figure 6D-1). In parallel, the iPSC-HPCs (CD8+ T cells) downregulated genes (<−1.5 log2; Padj < .05) involved in the development or function of transforming growth factor β (TGF-β) receptor (TGFBR3), B cells (CD37), and endothelial cells (S1PR1).42 Upregulation of FOXF1, GZMB, A2M, and TBX3 and downregulation of TGFBR3 were found repeatedly in the iPSC-HPC (CD8+ T cells) as opposed to in iPSC-HPC (nonlymphocytes) (Figure 6D-2). Additionally, A2M and IL7R genes were commonly upregulated, whereas CD37 and KLF2 were downregulated in the iPSC-HPCs (CD8+ T cells) compared with in iPSC-HPC committed to other cell subsets and primary blood HPCs (Figures 6Di-iii). Taken together, these results reveal a specific repertoire of genes in the “iPSC-HPCs (CD8+ T cells)”, which may serve as biomarkers to identify iPSC clones having appropriate genes controlling lineage-specific commitment into mature CD8αβ+ T cells.
Specific transcriptional profile of BCMA iPSC-HPCs that differentiate into antigen-specific CD8+ memory T cells. (A) Principal component analysis of the top 1000 highly expressed genes showing the transcriptional variance (PC1 vs PC2) within or across iPSC-derived HPCs that differentiate into different cell lineages or primary HPCs isolated from the blood. (B) Hierarchical cluster analyses demonstrating 4 distinct clusters of variably expressed genes across the data set (upregulated genes = red and downregulated genes = blue). Clusters 1 and 4: upregulated (cluster 1) or downregulated (cluster 4) genes in iPSC-HPCs compared with primary HPCs from the blood, Clusters 2 and 3: downregulated (cluster 2) or upregulated (cluster 3) genes in iPSC-HPC (CD8+ T cells) compared with iPSC-HPC (CD3− lymphocytes) and iPSC-HPC (nonlymphocytes). Nine samples evaluated; samples 1 and 2: iPSC-HPC (CD8+ T cells), samples 3 and 4: iPSC-HPC (CD3− lymphocytes), samples 5 and 6: iPSC-HPC (nonlymphocytes), and samples 7, 8, and 9: primary HPCs from the blood. (C) Summary of differential gene expression profiles reported as the total number of upregulated (log fold-change > 2) or downregulated (log fold-change <−2) genes in iPSC-HPC (CD8+ T cells) compared with in iPSC-HPC (CD3− lymphocytes), iPSC-HPC (nonlymphocytes), or primary HPCs from the blood. (D) Tables: summary of upregulated or downregulated genes in iPSC-HPC (CD8+ T cells) and their specific role in CD8+ T-cell lineage development. Volcano plots: significantly upregulated or downregulated genes in iPSC-HPC (CD8+ T cells) compared with (i) iPSC-HPC (CD3− lymphocytes), (ii) iPSC-HPC (nonlymphocytes) or (iii) primary HPCs from the blood. Lym, lymphocytes.
Specific transcriptional profile of BCMA iPSC-HPCs that differentiate into antigen-specific CD8+ memory T cells. (A) Principal component analysis of the top 1000 highly expressed genes showing the transcriptional variance (PC1 vs PC2) within or across iPSC-derived HPCs that differentiate into different cell lineages or primary HPCs isolated from the blood. (B) Hierarchical cluster analyses demonstrating 4 distinct clusters of variably expressed genes across the data set (upregulated genes = red and downregulated genes = blue). Clusters 1 and 4: upregulated (cluster 1) or downregulated (cluster 4) genes in iPSC-HPCs compared with primary HPCs from the blood, Clusters 2 and 3: downregulated (cluster 2) or upregulated (cluster 3) genes in iPSC-HPC (CD8+ T cells) compared with iPSC-HPC (CD3− lymphocytes) and iPSC-HPC (nonlymphocytes). Nine samples evaluated; samples 1 and 2: iPSC-HPC (CD8+ T cells), samples 3 and 4: iPSC-HPC (CD3− lymphocytes), samples 5 and 6: iPSC-HPC (nonlymphocytes), and samples 7, 8, and 9: primary HPCs from the blood. (C) Summary of differential gene expression profiles reported as the total number of upregulated (log fold-change > 2) or downregulated (log fold-change <−2) genes in iPSC-HPC (CD8+ T cells) compared with in iPSC-HPC (CD3− lymphocytes), iPSC-HPC (nonlymphocytes), or primary HPCs from the blood. (D) Tables: summary of upregulated or downregulated genes in iPSC-HPC (CD8+ T cells) and their specific role in CD8+ T-cell lineage development. Volcano plots: significantly upregulated or downregulated genes in iPSC-HPC (CD8+ T cells) compared with (i) iPSC-HPC (CD3− lymphocytes), (ii) iPSC-HPC (nonlymphocytes) or (iii) primary HPCs from the blood. Lym, lymphocytes.
iPSC-derived HPC use lineage-specific differentiation pathways to control commitment into mature CD8αβ+ T cells
Next, a larger transcriptional profile analyses (>10 000 genes) identified “common” or “unique” gene expressions between the following cohorts: cohort 1, iPSC-HPC (CD8+ T cells) vs iPSC-HPC (CD3− lymphocytes); cohort 2: iPSC-HPC (CD8+ T cells) vs iPSC-HPC (nonlymphocytes); and cohort 3: iPSC-HPC (CD8+ T cells) vs primary HPC from blood. A total of 57 genes were “commonly” upregulated (log2 fold-change >2; Padj < .05; Figure 7A, left Venn diagram), and 110 genes were “commonly” downregulated (log2 fold-change <−2, Padj < .05; Figure 7A, right Venn diagram) in the iPSC-HPC (CD8+ T cells) after an independent comparison of the 3 cohorts. The upregulated genes (∗P < .05) control reprogramming of somatic cells and early T-cell development (TBX3 and HOXA11), CD8+ T-cell regulation, differentiation, or activation and memory T-cell formation (IRF4, PIK3C2B, KLF15, IL-12B, and MAPK4) and immunogenic signal transduction (ITLN1/2, TRIM6, and EDA2R; supplemental Table 1).40,43,44 The downregulated genes modulate cell differentiation and cell cycle regulation (RPS6KA2, CDK3, and YPEL4); lineage-specific immune cell differentiation (BATF2, BTN3A1, USP44, CD70, and ZXDA); early embryonic and stem cell development (FGFR1, NPM2, GGN, SPAG1, and CATSPER2); regulation, organization, or development of the central nervous system (N4BP3, P2RY14, NLGN2, SHC2, GRASP, AMIGO2, TBC1D32, CACNA1A, and SLC6A9); formation and regulation of the vascular system, cytoskeletal arrangement, and angiogenesis (HEYL, NEURL, RAB39B, ANK1, PSD, LRRK1, RUNX2, and CXCL5); and promotion of inflammation, mediation of cellular stress response and homeostasis (SEMA7A, JDP2, PLA2G6, MAP3K9, PIPOX, and TNFRSF6B) (supplemental Table 2).45-47 Finally, the “commonly” upregulated genes were examined for specific functional enrichment by the means of GO annotation analysis of differentially expressed genes using DAVID Bioinformatics Resources.26 Using gene counts (≥3; cut-off ∗P < .05), several GO terms relating to cellular functions were identified, including chemotaxis, G-protein coupled receptor signaling, notch signaling, immune response, inflammatory response, cell junction, TNF–activated receptor activity, phospholipid binding, and calcium channel activity (Figure 7B). Furthermore, key GO terms related to immune and inflammatory responses were identified, which can contribute to the specific T-cell lineage commitment. Thus, these results demonstrate that iPSC-HPCs use “unique” lineage-specific transcriptional programs for their cell differentiation into specific mature CD8α+β+ T cells.
Distinct transcriptional profiles in BCMA iPSC-HPCs that commit to antigen-specific CD8+ T cells or other cell lineages (expanded analysis of >10 000 genes). (A) Venn diagrams illustrating “uniquely” or “commonly” expressed genes, either upregulated (left) or downregulated (right) in the iPSC-HPCs that differentiate into CD8+ T cells as compared with other lineages: iPSC-HPC (CD3− lymphocytes), iPSC-HPC (nonlymphocytes), or primary HPC from the blood (log2 fold change >2 : Padj < .05). (B, left) GO annotation analysis (−log 10 [P value]) identified “commonly” upregulated genes (57 genes) in iPSC-HPC (CD8+ T cells). (B, right) GO enrichment of specific categories related to immune response or inflammatory response in iPSC-HPCs that differentiate into CD8+ T cells.
Distinct transcriptional profiles in BCMA iPSC-HPCs that commit to antigen-specific CD8+ T cells or other cell lineages (expanded analysis of >10 000 genes). (A) Venn diagrams illustrating “uniquely” or “commonly” expressed genes, either upregulated (left) or downregulated (right) in the iPSC-HPCs that differentiate into CD8+ T cells as compared with other lineages: iPSC-HPC (CD3− lymphocytes), iPSC-HPC (nonlymphocytes), or primary HPC from the blood (log2 fold change >2 : Padj < .05). (B, left) GO annotation analysis (−log 10 [P value]) identified “commonly” upregulated genes (57 genes) in iPSC-HPC (CD8+ T cells). (B, right) GO enrichment of specific categories related to immune response or inflammatory response in iPSC-HPCs that differentiate into CD8+ T cells.
Discussion
Adoptive cell therapy with tumor-redirected CAR-Ts and TILs (tumor infiltrating lymphocytes) have provided significant clinical benefits, including a high response rate and durable complete remission in some patients with cancer.8,9 However, there remains significant challenges to overcome T-cell exhaustion and immune dysfunction seen with these therapies, which hampers both sustained cell expansion, tumor targeting, and overall treatment efficacy.48 Additionally, questions remain for adoptive cell therapy, including use of uncharacterized bulk T cells to generate CAR-Ts and the requirement of solid tumors to isolate TILs, further highlighting the need to develop additional therapeutic approaches. Epigenetic reprogramming of antigen-specific T cells offers an alternative strategy to provide revitalized effector cells with a high expansion potential in the absence of cognate antigen stimulation and improved functional antitumor responses. The key to iPSC technology involves epigenetic reprogramming of parental antigen-specific CD8+ CTL, which display an exhausted phenotype into a pluripotent state followed by differentiation into rejuvenated effector T cells with neither inhibitory/senescence molecules induction nor cognate antigen stimulation, which directly align with their increased polyfunctional antitumor activities. The key to current studies involves reprogramming of parental antigen-specific CD8+ CTL, which display an exhausted precursor memory phenotype (CD83+/CD45RO+),30,31 into a pluripotent state followed by differentiation into rejuvenated effector T cells with neither inhibitory/senescence molecules induction nor any antigen stimulation. Unlike CAR-Ts and TILs, iPSC–T cells provide an expandable and renewable source of phenotypically defined TAA targeting physiologically functional T cells for therapeutic application.
In this study, heteroclitic BCMA-CTL with a precursor exhausted memory phenotype30,31 were epigenetically reprogrammed to iPSC with the transcription factors OCT3/4, SOX2, KLF4, and c-MYC.9,49 Our protocol combined several key cytokines and small molecule agonists or antagonists to modulate the molecular pathways promoting mesoderm development during EB formation through a gradual polarization and activation of specific transcriptional regulators. The modulation of pathway signaling during polarization affects the balance between production of cells that resemble extraembryonic hematopoietic cells and hematopoietic progenitors.50,51 Accordingly, we found a significant induction of specific progenitor cells, which were then further induced to undergo T-cell development under feeder-free culture conditions in the presence of Fc-DLL4 signaling with retronectin. To induce effective antigen-specific responses by CD8+ CTL, the CD8αβ coreceptor binds to MHC class I molecule, stabilizes the interaction between TCR and cognate peptide–MHC-I complex, and triggers activation through intracellular interaction of the CD8α cytoplasmic tail with Src-family protein kinases Lck and LAT, which subsequently phosphorylates the TCR-CD3 complex.52 CD8+ CTL vary by several orders of magnitude in their sensitivity to peptide bound to MHC-I, which is determined by TCR affinity and CD8αβ coreceptor expression, which enhances the sensitivity to cognate antigen by 100-fold compared with CD8αα expressing T cells.53 The ultimate goal of this study was to polarize and expand highly functional mature BCMA-specific CD8αβ T cells from the antigen-specific iPSC-HPCs to efficiently target MM cells. The procedures used for T-cell differentiation was improved from previously reported studies.13-16 We achieved BCMA iPSC–T-cell maturation, producing predominantly CD45RO+ memory CTL, which correlates with higher antitumor functionality than parental BCMA-CTL. In addition, the process of differentiation into BCMA-specific CD8αβ+ T cells was accomplished in the absence of feeder cells commonly included by other studies, which avoids a safety concern and challenge of clinical translation. The procedure of T-cell differentiation was also shortened by reducing the early transitional double-positive CD4+ CD8+ developmental stage.
Previously, we and others have reported that repeated antigen stimulation to generate the antigen-specific T cells drives those effector cells into an exhausted state characterized by immune checkpoint upregulation that compromises their antitumor functionality.9,54 In this study, the differentiated memory iPSC–T cells lacked the inhibitory/senescence markers (CTLA4, PD1, LAG3, and TIM3; CD57), which were highly expressed on the parental precursor exhausted BCMA-CTL, which were used to establish iPSC. The absence of immune checkpoint expression was directly aligned with an improved proliferation capacity and increased antitumor activity as compared with the parental BCMA-CTL with their high specificities. Their effective anti-MM activities and polyfunctional immune responses were aligned with a distinct sole TCR rearrangement in both TCRα and TCRβ that was identified by single-cell sequencing of complementarity-determining region 3 analyses. This outcome supports the use of antigen-specific iPSC as an effective protocol to generate functional CD8αβ+ T cells as well as an efficient source to identify antigen-specific TCR, which can be applied to alternative therapeutic applications. We are currently evaluating these unique TCRα and TCRβ sequences identified in this study to develop a TCR-based immunotherapy strategy to efficiently target MM.
The key limitation we encountered during the T-cell differentiation was the lack of previously identified biomarkers for selecting the iPSC clones programmed for T-cell commitment, which results in an inefficient process to generate the final antigen-specific T-cell population. Mirroring differentiation by blood stem cells,55 we found that HPCs (CD34+ CD43+/CD14− CD235a−) developed from iPSC during EB formation used various cell differentiation pathways, leading to lymphoid- or myeloid-specific lineages. This study highlights the need to overcome inefficient and time-consuming procedures used to secure iPSC clones programmed for CD8+ T-cell differentiation. Overall, we established 20 different iPSC clones from parental BCMA-CTL generated from different donors and then proceeded through the entire differentiation and expansion process (53 days) before knowing the final cell lineage outcome. Without question, selection of appropriate iPSC clones destined for CD8+ T-cell lineage development would be more efficient and increase our overall success in developing an effective stem cell–based immunotherapy. Consequently, we prioritized identification of transcriptional profiles predictive of iPSC-HPCs commitment into T cells to serve as biomarkers in future studies. Our RNA-seq analyses revealed distinct genome-wide shifts and transcriptional profiles in iPSC-derived HPCs committed to CD8+ T cells development, including upregulation of genes promoting CD3+ T cells development or maturation, CD8+ CTL activation or function, memory T-cell development, IFN induction, cytokine transport, and immune response regulation.56 In parallel, specific repression genes were identified in the iPSC-HPCs, which control TGF-β receptor and B-cell development, rearrangement of immunoglobulin heavy chain, leukocyte tethering, and inhibitory receptors.57-60 Taken together, these findings identify genetic mechanisms and specific regulatory elements that play key roles in iPSC-HPCs destined for T-cell–specific commitment that could help to further design regenerative medicine protocols.
In summary, these results provide the preclinical framework to epigenetically reprogram exhausted antigen-specific CTL into iPSC capable of self-renewal and differentiation into revitalized cognate antigen-specific CD8αβ+ memory T cells through understanding of their key regulatory pathways. With the proof-of-principle platform provided here, we aim to develop an iPSC-based immunotherapy to overcome antigen-specific T-cell exhaustion/senescence and potentially promote effective and long-term antitumor immunity to improve patient outcome in MM and other cancers.
Acknowledgments
The authors acknowledge the members of our laboratories for technical advice and discussions. The authors thank Christina Usher for editing the manuscript and staffs at flow cytometry facility for their technical support.
This investigation was supported by National Institutes of Health (NIH), National Cancer Institute (NCI) grant P01 CA155258-10 and Department of Veterans Affairs grant I01 BX001584-09 (N.M.); NIH, NCI grants R01 CA207237-05, R01 CA276132-01, and R01 CA050947a, a Leukemia and Lymphoma Society Blood Cancer Discovery grant, a Paula and Rodger Riney Foundation grant, and an Adelson Program in Multiple Myeloma Research grant (K.C.A.); and NIH, NCI Specialized Programs of Research Excellence grant P50-CA100707-18 (N.C.M., K.C.A.).
Authorship
Contribution: J.B. designed the study, performed experiments, developed assays, analyzed and interpreted the data, and wrote the manuscript; S. Kitayama performed experiments, developed assays, worked on the methodology, analyzed the data, and wrote the manuscript; Z.H. carried out formal analyses; L.D. analyzed and interpreted the data and reviewed the manuscript; K.K. analyzed and interpreted the data; D.B.K. and K.L. analyzed the data; S.L. performed experiments; M.T., R.R., K.K., and M.S. analyzed and interpreted the data; N.C.M. reviewed the manuscript, performed project administration, and was responsible for funding acquisition; J.R. designed the study and reviewed the manuscript; S. Kaneko designed the study, analyzed and interpreted the data, and reviewed the manuscript; and K.C.A. designed the study, reviewed the data and the manuscript, performed project administration, and was responsible for funding acquisition.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jooeun Bae, Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, 450 Brookline Ave, Smith 602, Boston, MA 02215; email: jooeun_bae@dfci.harvard.edu.
References
Author notes
Original data are available on request from the corresponding author, Jooeun Bae (jooeun_bae@dfci.harvard.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Establishment of BCMA-iPSC from parental BCMA-specific CTL. (A) Schematic presentation of the protocol for reprograming of parental BCMA-specific IFN-γ+ CTL into iPSC and differentiation into BCMA-iPSC–T cells. (B, top) Representative flow cytometric analyses showing the pluripotency potential of BCMA-iPSC and EBV-iPSC (positive control) by expression of the stem-cell markers SSEA-4 and TRA-1-60 but not on parental BCMA-CTL (negative control). (B, bottom) Summary of 3 independent analyses (N = 3; mean ± standard deviation [SD]). (C, top) Representative flow cytometric analyses showing the expression of key germ cell layer markers, SOX-17 (endoderm), Brachyury (mesoderm), and Pax-6 (ectoderm), on BCMA-iPSC and EBV-iPSC (positive control). (C, bottom) Summary of 3 independent analyses (N = 3; mean ± SD). (D) Representative immunohistochemistry analysis demonstrating alkaline phosphatase upregulation, a characteristic marker of stem cell development and self-renewal potential, on BCMA-iPSC and EBV-iPSC (positive control) but not on primary T cells. Photomicrographs (original magnification ×100) taken with an inverted microscope (Carl Zeiss). (E) Representative cytogenetic analysis of chromosomes for Giemsa banding patterns demonstrating genomic stability and a normal karyotype in BCMA-iPSC.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/10/10.1182_blood.2023020528/3/m_blood_bld-2023-020528-gr1.jpeg?Expires=1770130682&Signature=Seh97Et30HQf1o5AvbKwTu9eGUIAOTsEFyUk-8haOC3sKyuZ6KQEFDoWlxLz1E5EbOSkFw9f3bmDM68a-6LsmTtFQZr1gWnH~KTI0n7TiCja8hnfaiQzxAJOiad81MOFCt05L6xBfUkSwQwUAQenMZ3wUhmD9iv~yPCR9giRdW8wCLjrwP7n4TbrYJl67PeevAyEyCBWdliM88Bmlo75~aQIqS4CM-g5gB6YP09zoBpaX2scvvdglkMl1KdcqRGKC6~6F5A4DwHY49FYX6BhxmpprzTXc-85b2cJg~Bx1K0arAaYBl4tkIYeJPkaB5aobnK2pN3KJcb~dV0-I8w7fA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Polarization of BCMA-iPSC into mesoderm germ layer. (A) Representative ScoreCard analysis demonstrating a gradual upregulation in genes associated with mesoderm development during EB formation on days 2, 4, and 7 from BCMA-iPSC relative to an undifferentiated reference. (B) ScoreCard analysis demonstrating no specific regulation in genes associated with germ layer development in somatic cell control CD8+ T lymphocytes (from donor 1 [D1] or donor 2 [D2]) relative to an undifferentiated reference. (C) Summary analyses of differential gene expression profiles associated with primary germ layer development in BCMA-iPSC (clone 1 and clone 2; N = 2) and EB (N = 2) (top) but not in somatic cell control CD8+ T lymphocytes (D1 and D2; N = 2) (bottom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/10/10.1182_blood.2023020528/3/m_blood_bld-2023-020528-gr2.jpeg?Expires=1770130682&Signature=MCEUT4MiawjeEui5npxA90nlyreMDiGmqk31dOPsGvCznUb2aSAZB57988bIC9eIrsK7kcRa06qcdFJRQYU1qQMevvURC1UCPIvad-cIcvuhu4T5s7Zfmd9BqOPQXwNTjlLiW~QAHcpzrdXJPwTyfy1OmSJeq6cGed87wO0WS8-1P88y0VamsvF7kTMlLYDi8iJ9l4lwAjG~gpkMuwzGrVJPjnpbQtRvBn~dOQ7JkbVb~mDUp8udqMQ-VPaZz3sOhUHeKRcofmTX9yNTUfoOfs~mKoy9mEW-M-7EoArjyFEgofkRDqp3Ayjc~sP~VvU2ghE5bwFsonLbENwJsoES7Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Distinct transcriptional profiles in BCMA iPSC-HPCs that commit to antigen-specific CD8+ T cells or other cell lineages (expanded analysis of >10 000 genes). (A) Venn diagrams illustrating “uniquely” or “commonly” expressed genes, either upregulated (left) or downregulated (right) in the iPSC-HPCs that differentiate into CD8+ T cells as compared with other lineages: iPSC-HPC (CD3− lymphocytes), iPSC-HPC (nonlymphocytes), or primary HPC from the blood (log2 fold change >2 : Padj < .05). (B, left) GO annotation analysis (−log 10 [P value]) identified “commonly” upregulated genes (57 genes) in iPSC-HPC (CD8+ T cells). (B, right) GO enrichment of specific categories related to immune response or inflammatory response in iPSC-HPCs that differentiate into CD8+ T cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/10/10.1182_blood.2023020528/3/m_blood_bld-2023-020528-gr7.jpeg?Expires=1770130682&Signature=rBXWvvrkg8lk-os4HT7vOpA6zqnqjp4h3SAZvaYRj~E0rTtmfSgjRE5jeWKga4WBGDD53FWrj0p4a6Z8JY25eV3Vz4R2i2R7FtV7U4bAs7j~7iLw32P4Y-553SEZIFdOlI7vff1JVz6Mab34mbJztlLYU81v20Mgd269RLJR6gqQxAeV-Acw3fVyVdaorDIC4wzvXEMXY6j853wa6AGVFlBMfahgiFfgulNy~z5mlW--pd-XCIiRZljzHa0eGQttsjETlRXUkAF8ie9hkWk2XCbtaI6374Grnd~XwZhlqmM8wJP0u8QrPGfk64LcgRy60i9kzxEyYyjx~1XrA4WhBA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal