EBV infects lymphoid, myeloid, and stem cells in patients with CAEBV, so HSCT is currently the only curative treatment for CAEBV disease.
EBV-infected HSCs have a higher rate of differentiation, which speeds up the cellular infection and alters immune response in CAEBV.
Visual Abstract
Chronic active Epstein-Barr virus (EBV) disease (CAEBV) is a lethal syndrome because of persistent EBV infection. When diagnosed as CAEBV, EBV infection was observed in multiple hematopoietic lineages, but the etiology of CAEBV is still elusive. Bone marrow and peripheral cells derived from 5 patients with CAEBV, 1 patient with EBV-associated hemophagocytic lymphohistiocytosis, and 2 healthy controls were analyzed. Multiple assays were applied to identify and characterize EBV-infected cells, including quantitative polymerase chain reaction, PrimeFlow, and single-cell RNA-sequencing (scRNA-seq). Based on scRNA-seq data, alterations in gene expression of particular cell types were analyzed between patients with CAEBV and controls, and between infected and uninfected cells. One patient with CAEBV was treated with allogeneic hematopoietic stem cell transplantation (HSCT), and the samples derived from this patient were analyzed again 6 months after HSCT. EBV infected the full spectrum of the hematopoietic system including both lymphoid and myeloid lineages, as well as the hematopoietic stem cells (HSCs) of the patients with CAEBV. EBV-infected HSCs exhibited a higher differentiation rate toward downstream lineages, and the EBV infection had an impact on both the innate and adaptive immunity, resulting in inflammatory symptoms. EBV-infected cells were thoroughly removed from the hematopoietic system after HSCT. Taken together, multiple lines of evidence presented in this study suggest that CAEBV disease originates from the infected HSCs, which might potentially lead to innovative therapy strategies for CAEBV.
Introduction
Epstein-Barr virus (EBV), a member of the gammaherpesvirus family, is a highly effective pathogen that infects >95% of the world’s population.1 Although primary EBV infection is frequently asymptomatic, EBV has been revealed to be associated with various diseases, including infectious mononucleosis,2 chronic active EBV infection (CAEBV),3 EBV-associated hemophagocytic lymphohistiocytosis (EBV-HLH),4 and a variety of cancers.1
CAEBV is a relatively rare syndrome with EBV infection, feathered by a high proportion of infected T and/or natural killer (NK) cells.5 Despite the heterogeneous clinical outcomes, CAEBV can lead to an aggressive and fatal condition with generally poor prognosis.5 However, the etiology of CAEBV is still elusive, such as the mechanism of EBV infection to T and NK cells remains unsolved. It is widely recognized that CD21 and CD35 are the receptors that facilitate EBV entry to B cells, through the interactions with the EBV envelope protein gp350.1 Although a recent study has demonstrated that CD21 is also expressed in T cells and is likely the cellular receptor for T-cell infection,6 the potential of EBV infection to progenitor lymphoid cells has also been proposed to explain the observation of EBV-infected cells of multiple lineages.3,7,8
To address possible origination of CAEBV, here we analyzed bone marrow (BM) and peripheral cells derived from patients with CAEBV and control donors with multiple molecular technologies, including quantitative polymerase chain reaction (qPCR) assays for EBV-DNA copies, PrimeFlow RNA assays, and single-cell RNA-sequencing (scRNA-seq) for EBV-encoded small RNAs (EBERs). The collective evidence points to a notion that CAEBV originates from EBV-infected hematopoietic stem cells (HSCs). This conclusion well explains that the only effective treatment of patients with CAEBV is HSC transplantation (HSCT).
Methods
Ethical approval
The study received ethical approval from the Ethics Committee of Beijing Friendship Hospital, Capital Medical University, Beijing, China. Written informed consent was obtained from all patients and healthy volunteers in accordance with the Declaration of Helsinki.
Patients
The diagnostic criteria for CAEBV were in accordance with the World Health Organization classification, including persistent infectious mononucleosis–like symptoms for >3 months, increased EBV DNA in peripheral blood (PB), histological evidence of organ disease, and EBV RNA or viral protein detected in affected tissues.9 The EBV-HLH was defined as previously described10,11: (1) meeting the HLH-2004 diagnostic criteria12; (2) present evidence of active EBV infection, such as high values for EBV-DNA copies in PB or tissues, or EBER-containing cells in PB and tissues; and (3) excluding primary HLH and EBV infection secondary to lymphoma-associated HLH. The patients with CAEBV enrolled in this study were mainly adults except for one 17-year-old adolescent, and the patient with EBV-HLH and healthy controls were all adults (refer to Table 1 for more details).
Clinical data of patients and healthy controls, and the analysis of EBV DNA in the BM aspirate and PB
| Subject no. . | Age (y)/sex . | Diagnosis . | Treatment: response . | Outcome . | EBV DNA: copies per mL . | EBV DNA: copies per million cells . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | PB . | PB cell types . | ||||||||||
| Cells . | Liquid part . | Cells . | Plasma . | CD3+CD4+ (CD4+ T) . | CD3+CD8+ (CD8+ T) . | CD3-CD19+ (B) . | CD56+ (NK) . | |||||
| 1 | 17/Male | CAEBV, EBV+LPD | None | Alive | 3 × 105 | NP | 2 × 104 | 7 × 103 | 2 × 103 | 3 × 104 | 6 × 104 | 2 × 104 |
| 2 | 21/Female | CAEBV with HLH | L-DEP: NR | Death | 2 × 106 | 5 × 105 | 4 × 106 | 2 × 105 | N | 4 × 104 | N | 1 × 106 |
| 3 | 26/Male | CAEBV, EBV+LPD | Pegaspargase-anlotinib-pembrolizumab: NR | Death | 2 × 107 | 8 × 103 | 6 × 106 | 9 × 102 | 1 × 105 | 1 × 105 | 2 × 105 | 1 × 106 |
| 4 | 25/Male | CAEBV with HLH | DEP+anti-PD-1 antibody: NR; then HSCT | Alive | 5 × 106 | 3 × 104 | 2 × 106 | 2 × 103 | 2 × 105 | 2 × 105 | 7 × 104 | 3 × 106 |
| 5 | 54/Male | CAEBV | None | Death | 6 × 105 | 7 × 104 | 2 × 104 | 2 × 104 | 2 × 103 | 1 × 103 | N | 4 × 103 |
| 6 | 56/Female | EBV-HLH | DEP+anti-PD-1 antibody: NR; then L-DEP: CR | Alive | 1 × 103 | 2 × 103 | <5 × 102 | 1 × 103 | <5 × 102 | N | <5 × 102 | 7 × 102 |
| 7 | 47/Female | Healthy control | <5 × 102 | N | <5 × 102 | N | NP | NP | NP | NP | ||
| 8 | 47/Male | Healthy control | <5 × 102 | N | <5 × 102 | <5 × 102 | NP | NP | NP | NP | ||
| Subject no. . | Age (y)/sex . | Diagnosis . | Treatment: response . | Outcome . | EBV DNA: copies per mL . | EBV DNA: copies per million cells . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | PB . | PB cell types . | ||||||||||
| Cells . | Liquid part . | Cells . | Plasma . | CD3+CD4+ (CD4+ T) . | CD3+CD8+ (CD8+ T) . | CD3-CD19+ (B) . | CD56+ (NK) . | |||||
| 1 | 17/Male | CAEBV, EBV+LPD | None | Alive | 3 × 105 | NP | 2 × 104 | 7 × 103 | 2 × 103 | 3 × 104 | 6 × 104 | 2 × 104 |
| 2 | 21/Female | CAEBV with HLH | L-DEP: NR | Death | 2 × 106 | 5 × 105 | 4 × 106 | 2 × 105 | N | 4 × 104 | N | 1 × 106 |
| 3 | 26/Male | CAEBV, EBV+LPD | Pegaspargase-anlotinib-pembrolizumab: NR | Death | 2 × 107 | 8 × 103 | 6 × 106 | 9 × 102 | 1 × 105 | 1 × 105 | 2 × 105 | 1 × 106 |
| 4 | 25/Male | CAEBV with HLH | DEP+anti-PD-1 antibody: NR; then HSCT | Alive | 5 × 106 | 3 × 104 | 2 × 106 | 2 × 103 | 2 × 105 | 2 × 105 | 7 × 104 | 3 × 106 |
| 5 | 54/Male | CAEBV | None | Death | 6 × 105 | 7 × 104 | 2 × 104 | 2 × 104 | 2 × 103 | 1 × 103 | N | 4 × 103 |
| 6 | 56/Female | EBV-HLH | DEP+anti-PD-1 antibody: NR; then L-DEP: CR | Alive | 1 × 103 | 2 × 103 | <5 × 102 | 1 × 103 | <5 × 102 | N | <5 × 102 | 7 × 102 |
| 7 | 47/Female | Healthy control | <5 × 102 | N | <5 × 102 | N | NP | NP | NP | NP | ||
| 8 | 47/Male | Healthy control | <5 × 102 | N | <5 × 102 | <5 × 102 | NP | NP | NP | NP | ||
Bold entries indicate negative detection of EBV. CR, complete response; EBV+LPD, EBV-associated lymphoproliferative diseases; DEP, doxorubicin-etoposide-methylprednisolone; L-DEP, pegaspargase (PEG)-asparaginase in combination with liposome doxorubicin, etoposide and high-dose methylprednisolone; N, negative; NP, not performed; NR, no response.
Sample collection
Fresh PB or BM aspirates were collected in EDTA from patients and healthy controls. Samples were analyzed within 24 hours of sample collection. The samples were centrifuged at 3000 rpm for 10 minutes, and the plasma or supernatant from BM aspirates was collected and stored at −80°C until further use. Cells were isolated after red blood cell removal with red blood cell lysis buffer (Beijing Solarbio Science & Technology Co, Ltd, no. R1010). Freshly prepared cells were used in subsequent assays for analysis of EBV-infected cell populations.
Assays of EBV-infected cell subpopulations
After freshly prepared cells were washed once in phosphate-buffered saline (PBS) (Beijing Solarbio Science & Technology Co, Ltd, no. P1020), all samples were sorted on the FACSMelody (BD). Cells from PB samples were stained with anti-CD3, anti-CD4, anti-CD8, anti-CD19, and anti-CD56 antibodies (supplemental Table 1, available on the Blood website) and subsequent sorting for the CD3+CD4+ T cells, CD3+CD8+ T cells, CD3−CD19+ B cells, and CD56+ NK/NKT cells. Cells from BM aspirates were isolated by staining with anti-CD34 antibody (supplemental Table 1) and subsequent sorting for the CD34+ cells. EBV-DNA copies were quantified in BM aspirates, PB, and sorted cell populations using a commercial PCR diagnostic kit for EBV (Shanghai ZJ Bio-Tech Co, Ltd, no. P20220201) according to the manufacturer’s recommendations.
PrimeFlow assays for EBERs
One million to 10 million cells per assay were stained with antibodies (supplemental Table 1) for surface markers and distinguishing live/dead cells. Then, RNA targets of EBERs, human β2-microglobulin (B2M), and Bacillus subtilisdapB were detected in cells by the PrimeFlow RNA assay kits (ThermoFisher) according to the manufacturer’s instructions. β2-B2M (AF488) and B subtilisdapB (AF488) were used as the positive and negative controls for the PrimeFlow assays, respectively. EBER target probes (AF488) were used for detecting EBV-infected cells. Cells were acquired using a DxFlex or CytoFlex (Beckman Coulter) and analyzed using the CytExpert for DXFLEX software version 2.0.2.18 (Beckman Coulter).
Single-cell suspensions preparation
Individual cells were isolated by density gradient centrifugation using Ficoll-Paque Plus medium (GE Healthcare) and washed with Ca/Mg-free PBS. To remove red blood cells, 2 mL GEXSCOPE red blood cell lysis buffer (RCLB, Singleron) was added at 25°C for 10 minutes. The solution was then centrifuged at 500g for 5 minutes and suspended in PBS. After the removal of red blood cells, the remaining cells were isolated by centrifugation at 400g for 10 minutes at 4°C. The supernatant was discarded, and the cells were resuspended in PBS to obtain single-cell suspension. Finally, the samples were stained with trypan blue, and the cell viability was evaluated microscopically.
Reverse transcription and library construction
Single-cell suspensions (2 × 105 cells per mL) with PBS (HyClone) were loaded onto microwell chip using the Singleron Matrix Single Cell Processing System. Subsequently, barcoding beads were collected from the microwell chip, followed by reverse transcription of the messenger RNA (mRNA) captured by the barcoding beads to obtain complementary DNA, and PCR amplification. The amplified complementary DNA was then fragmented and ligated with sequencing adapters. The scRNA-seq libraries were constructed according to the protocol of the FocuSCOPE Single Cell mRNA × EBV Library Kit (Singleron).13 Individual libraries were diluted to 4 nM, pooled, and sequenced on Illumina NovaSeq 6000 with 150 bp paired-end reads.
scRNA-seq data analyses
Raw reads from scRNA-seq were processed to generate gene expression matrices using CeleScope (https://github.com/singleron-RD/CeleScope) pipeline, and the downstream analyses were conducted according to the guideline.14 Additional details of the data analyses are provided in the supplemental Methods.
Statistics and repeatability
Signature score comparisons between 2 cell groups were performed using unpaired two-tailed Wilcoxon rank-sum tests. All statistical analyses and presentations were performed using R. Other statistical tests used in figures are shown in figure legends, and statistical significance was set at P < .05. Exact value of n and what n represents are shown in the figure legends.
Results
EBV infects both lymphoid and myeloid cells of patients with CAEBV
We enrolled 5 patients with CAEBV, 1 patient with EBV-HLH, and 2 healthy controls (Table 1; refer to “Patient information” in supplemental Materials for more details) for EBV infection analysis. Assays of EBV-DNA copy number based on qPCR applied to the PB and BM samples confirmed substantial EBV infection to the hematologic cells of the patients with CAEBV, which included multiple lymphoid cell populations (Table 1).
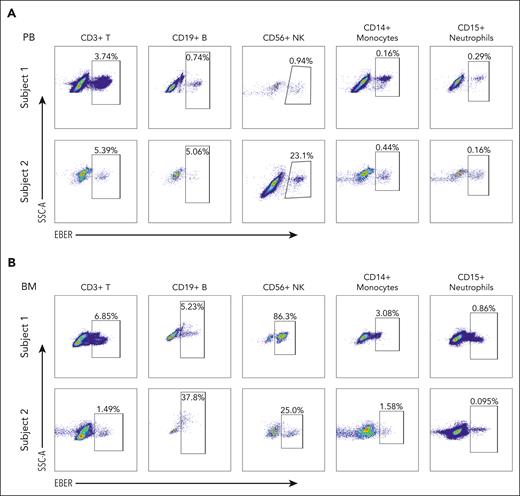
On the ground of successful benchmark of the PrimeFlow assay for detecting EBV-infected cells using the EBV-positive Raji cell line and the EBV-negative Jurkat cell line (supplemental Figure 1), we further applied this assay to different types of immune cells obtained from PB and BM samples of the patients with CAEBV (supplemental Figure 2A-B) and calculated the proportion of the EBV-infected cells (Figure 1; supplemental Figure 3A). Consistent with the literature,5 the EBV-infected cells were mainly enriched in CD56+ NK/NKT cells; however, there was also a significant portion of EBV-infected cells identified in other cell types, such as peripheral CD19+ B cells (Figure 1A, subject 1), peripheral CD14+ monocytes (Figure 1A, subject 2), and BM CD14+ monocytes (Figure 1B, subjects 1 and 2). This observation indicated that not only lymphocytes but also myeloid cells were EBV-infected to develop CAEBV. Moreover, we found that there were relatively more EBV-infected cells in the BM than in PB (BM 15.8% vs PB 1.2% on average for subject 1; BM 12.1% vs PB 6.8% on average for subject 2). Altogether, our results suggest that EBV infecting multiple hematologic lineages in the BM is a hallmark of the CAEBV disease.
Representative PrimeFlow assay data showed EBV-infected hematologic cells in CAEBV. (A) EBV-infected cells are identified in CD3+ T cells, CD19+ B cells, CD56+ NK cells, CD14+ monocytes, and CD15+ neutrophils from PB samples of the patients with CAEBV (subjects 1 and 2). (B) EBV-infected cells are identified in CD3+ T cells, CD19+ B cells, CD56+ NK cells, CD14+ monocytes, and CD15+ neutrophils from BM samples of the patients with CAEBV (subjects 1 and 2). Refer to supplemental Figure 3A for megakaryocytes and erythrocytes. In each panel, the top row shows the data from subject 1, whereas the bottom row shows the data from subject 2. Both patients were diagnosed as CAEBV, and the EBV-infected cells were found in almost all hematologic cell types. SSC-A, side scatter-area.
Representative PrimeFlow assay data showed EBV-infected hematologic cells in CAEBV. (A) EBV-infected cells are identified in CD3+ T cells, CD19+ B cells, CD56+ NK cells, CD14+ monocytes, and CD15+ neutrophils from PB samples of the patients with CAEBV (subjects 1 and 2). (B) EBV-infected cells are identified in CD3+ T cells, CD19+ B cells, CD56+ NK cells, CD14+ monocytes, and CD15+ neutrophils from BM samples of the patients with CAEBV (subjects 1 and 2). Refer to supplemental Figure 3A for megakaryocytes and erythrocytes. In each panel, the top row shows the data from subject 1, whereas the bottom row shows the data from subject 2. Both patients were diagnosed as CAEBV, and the EBV-infected cells were found in almost all hematologic cell types. SSC-A, side scatter-area.
EBV-infected HSCs are found in patients with CAEBV
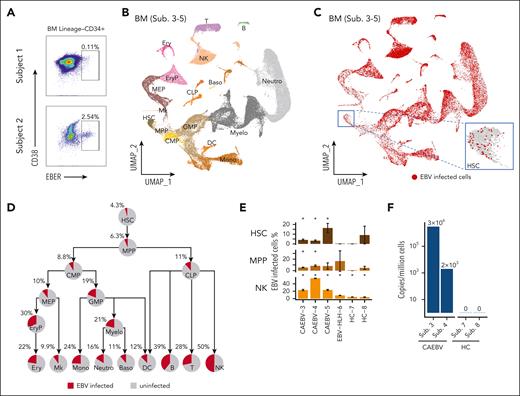
In patients with CAEBV, we found that both the myeloid and lymphoid lineages were infected by EBV, which may suggest that their common progenitors were likely to have been infected before differentiation. To verify this, we sorted Lin−CD34+ BM cells (ie, HSCs) with the other compartments (supplemental Figure 2A) for PrimeFlow assay. Indeed, EBV-infected HSCs were detected in both patients analyzed (subjects 1 and 2; Figure 2A), and the factions of EBV-infected HSCs were comparable to EBV-infected cells in the other compartments for subject 2 (Figure 2A; supplemental Figure 3B).
EBV-infected HSCs were detected in CAEBV patients with multiple assays. (A) PrimeFlow data showing EBV-infected cells were identified in BM Lineage–CD34+ HSC cells, from the patients with CAEBV (subjects 1 and 2). (B) Uniform manifold approximation and projection (UMAP) plot showing the two-dimensional (2D) projection of the BM cells derived from the 3 patients with CAEBV (subjects 3-5) based on the single-cell transcriptomics data. According to marker genes (refer to supplemental Figure 4C) identified in each cell cluster (indicated by different colors), cell types were annotated and their names were displayed in their respective locations. (C) UMAP showing a universal EBV infection in the BM hematopoietic system of the patients with CAEBV (subjects 3-5), with the inset demonstrating the infected cells in the HSC compartment. The EBV-infected cells were depicted in red dots. Notably, the red dots (ie, infected cells) were plotted on the top of the gray ones (ie, uninfected cells), so the exact infection percentages (shown in panel D) were not reflected in this plot. (D) Pie charts showing the percentages of EBV-infected cells in each hematopoietic cell population of the patients with CAEBV (subjects 3-5), and the tree-like structure shows the proportion evolution along the differentiation pathways. (E) Significant proportions of EBV-infected cells in HSC, MPP, NK cell populations of each patient with CAEBV (subjects 3-5), compared with none or nonsignificant proportions of the infected cells of the patient with EBV-HLH (subject 6) and healthy controls (HCs; subjects 7 and 8). The error bars indicate the percentage standard deviation (SD) based on bootstrapping. Please note that in some cell populations of individual samples, there were too few cells measured to make a robust estimation on the infection percentage. For example, there were only 11 measured HSCs from subject 8 (HC-8), and therefore the SD was large (9.6%). Asterisks (∗) above the bars indicate that statistically significant fractions of cells were EBV infected, whereas bars without asterisks show that none or nonsignificant fractions of cells were infected. Refer to supplemental Figure 5E-F for other cell populations from BM and cell populations from PB. (F) The EBV copy numbers quantified by RT-qPCR in the CD34+ compartment derived from patients with CAEBV (subjects 3 and 4) and HCs (subjects 7 and 8). B, B cells; Baso, basophils; CMP, common myeloid progenitors; CLP, common lymphoid progenitors; DC, dendritic cells; Ery, erythrocytes; EryP, erythroid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte/erythroid progenitors; Mk, megakaryocytes; Mono, monocytes; Myelo, myelocytes; Neutro, neutrophils; T, T cells.
EBV-infected HSCs were detected in CAEBV patients with multiple assays. (A) PrimeFlow data showing EBV-infected cells were identified in BM Lineage–CD34+ HSC cells, from the patients with CAEBV (subjects 1 and 2). (B) Uniform manifold approximation and projection (UMAP) plot showing the two-dimensional (2D) projection of the BM cells derived from the 3 patients with CAEBV (subjects 3-5) based on the single-cell transcriptomics data. According to marker genes (refer to supplemental Figure 4C) identified in each cell cluster (indicated by different colors), cell types were annotated and their names were displayed in their respective locations. (C) UMAP showing a universal EBV infection in the BM hematopoietic system of the patients with CAEBV (subjects 3-5), with the inset demonstrating the infected cells in the HSC compartment. The EBV-infected cells were depicted in red dots. Notably, the red dots (ie, infected cells) were plotted on the top of the gray ones (ie, uninfected cells), so the exact infection percentages (shown in panel D) were not reflected in this plot. (D) Pie charts showing the percentages of EBV-infected cells in each hematopoietic cell population of the patients with CAEBV (subjects 3-5), and the tree-like structure shows the proportion evolution along the differentiation pathways. (E) Significant proportions of EBV-infected cells in HSC, MPP, NK cell populations of each patient with CAEBV (subjects 3-5), compared with none or nonsignificant proportions of the infected cells of the patient with EBV-HLH (subject 6) and healthy controls (HCs; subjects 7 and 8). The error bars indicate the percentage standard deviation (SD) based on bootstrapping. Please note that in some cell populations of individual samples, there were too few cells measured to make a robust estimation on the infection percentage. For example, there were only 11 measured HSCs from subject 8 (HC-8), and therefore the SD was large (9.6%). Asterisks (∗) above the bars indicate that statistically significant fractions of cells were EBV infected, whereas bars without asterisks show that none or nonsignificant fractions of cells were infected. Refer to supplemental Figure 5E-F for other cell populations from BM and cell populations from PB. (F) The EBV copy numbers quantified by RT-qPCR in the CD34+ compartment derived from patients with CAEBV (subjects 3 and 4) and HCs (subjects 7 and 8). B, B cells; Baso, basophils; CMP, common myeloid progenitors; CLP, common lymphoid progenitors; DC, dendritic cells; Ery, erythrocytes; EryP, erythroid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte/erythroid progenitors; Mk, megakaryocytes; Mono, monocytes; Myelo, myelocytes; Neutro, neutrophils; T, T cells.
We next performed scRNA-seq for the BM and PB samples from the patients with CAEBV, and simultaneously enriched EBV transcripts EBER1, EBER2, EBNA2, EBNA1, and ZEBRA15 from the same cells for sequencing. Integrating data derived from 3 patients with CAEBV (supplemental Figure 4A; subjects 3-5), we annotated HSC and progenitor cells as well as mature cells in both the BM and PB samples (Figure 2B; supplemental Figure 4C-E). The recognition of EBV RNA expression in individual cells disclosed that the EBV-infected cells spread out all the hematologic cell populations as well as the stem and progenitor cells (Figure 2C; supplemental Figure 4F-L). On average, 4.3% HSCs derived from the patients with CAEBV were infected by EBV, and the lymphoid cell populations displayed higher infection levels, with the maximum reaching to 50% in NK cells (Figure 2D). Tracing along the stem cell differentiation tree, we discovered that all the lineage branches were EBV infected, in concordance with the PrimeFlow results, suggesting that the infection was propagated from the stem cell compartment (Figure 2D).
Because EBV ubiquitously infects humans,1 we sought to ask whether the phenomena observed above were also present in other EBV-related diseases and healthy people. To this end, we further conducted the single-cell experiments on 1 patient with EBV-HLH (subject 6) and 2 healthy donors (subjects 7 and 8). Projecting the BM and PB cells derived from the patient with EBV-HLH and the healthy controls to the uniform manifold approximation and projection formed by CAEBV cells, we detected apparently fewer infected cells in the controls (supplemental Figure 5A-B vs Figure 2C; supplemental Figure 5C-D vs supplemental Figure 4F). By quantifying the fractions of EBV-infected cells in each cell population and accounting for the quantification uncertainty, we discerned that there were significant proportions of HSCs infected in individual patients with CAEBV (subject 3 [95% confidence interval (CI)]: 4.1% [2.7-5.9]; subject 4 [95% CI]: 3.3% [2.1-4.8]; subject 5 [95% CI]: 16.4% [7.6-26.9]), whereas none or nonsignificant factions of HSCs were infected in EBV-HLH (subject 6 [95% CI]: 0 [0-0]) or healthy controls (subject 7 [95% CI]: 0 [0-0]; subject 8 [95% CI]: 9.1% [0-27.3]) (Figure 2E). This is in agreement with the reverse transcriptase qPCR (RT-qPCR) results for measuring the EBV-DNA copy numbers in the CD34+ compartment (Figure 2F). Similar patterns were observed in the multipotent progenitors (MPPs) and some other progenitor compartments, likely resulting in higher infection levels along the lineage development trajectories in the patients with CAEBV compared with the others (Figure 2E; supplemental Figure 5E-F).
EBV-infected HSCs exhibit a higher differentiation rate
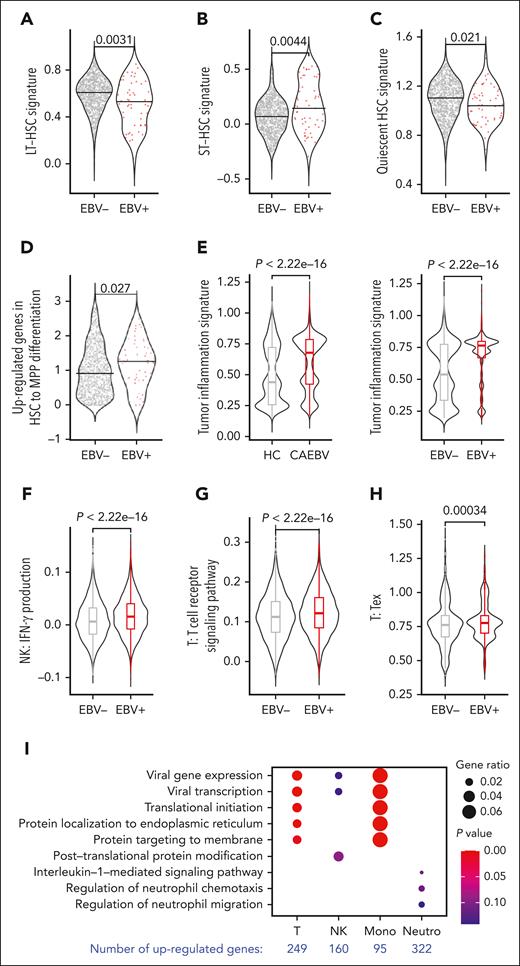
We observed an increasing percentage of cell infection along the stem cell differentiation pathway (Figure 2D), suggesting that the infected HSCs may have a higher tendency to differentiate. To verify this speculation, we characterized infected vs uninfected HSCs at the molecular level based on transcriptomic analysis. It is known that long-term HSCs (LT-HSCs) sit at the top of the hematopoietic hierarchy, possessing a quiescent signature, and thus are less active in differentiation.16,17 Based on the signature genes of LT-, short-term HSCs, and quiescent HSCs gathered from the literature,16,17 we revealed that the EBV-infected HSCs indeed displayed lower LT- and quiescent HSC signatures but a higher short-term HSC signature compared with the uninfected cells (Figure 3A-C). However, there was no obvious difference in cell cycle (supplemental Figure 6A). These results indicate that EBV infection is likely to make HSCs produce more progeny downward the differentiation hierarchy, without enlarging the HSC population size.
Molecular characteristics of the EBV-infected cells. (A) Violin plots comparing the LT-HSC signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). (B) Violin plots comparing the short-term HSC (ST-HSC) signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected HSCs exhibited a significantly lower LT-HSC signature and a significantly higher ST-HSC signature, indicating that the infected HSCs were more active in differentiating into downstream lineages. (C) Violin plots comparing the quiescent HSC signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected HSCs exhibited a significantly lower quiescent HSC signature, suggesting again that the infected HSCs were more active in differentiating. (D) Violin plots comparing the signature of upregulated genes in HSC-to-MPP differentiation between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The infected HSCs exhibited a higher signature, indicating that the infected HSCs tended to be more primed for differentiating into MPPs. (E) Violin plots and boxplots comparing the tumor inflammation signature between PB cells derived from the 3 patients with CAEBV (subjects 3-5) vs HCs (subjects 7 and 8) (left), and EBV-infected (EBV+) vs uninfected (EBV–) PB cells derived from the 3 patients with CAEBV (subjects 3-5) (right). These results indicate that the EBV infection leads to a relatively higher level of inflammation. (F) Violin plots and boxplots comparing interferon gamma production scores between EBV-infected (EBV+) vs uninfected (EBV–) peripheral NK cells derived from the 3 patients with CAEBV (subjects 3-5). A significantly higher level of interferon gamma production was found in the infected cells. (G) Violin plots and boxplots comparing the signature of T-cell receptor signaling pathway between EBV-infected (EBV+) vs uninfected (EBV–) peripheral T cells derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected T cells exhibited a significantly higher signature of T-cell receptor signaling pathway. (H) Violin plots and boxplots comparing the T-cell exhaustion score between EBV-infected (EBV+) vs uninfected (EBV–) peripheral T cells derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected T cells had a significantly higher exhaustion score. (I) Dot plot showing the Gene Ontology (GO) terms enriched in the upregulated genes identified in EBV-infected peripheral cells compared with the uninfected cells of the 3 patients with CAEBV (subjects 3-5). T cells, NK cells, monocytes (mono), and neutrophils (neutro) were examined, and the number of differentially expressed genes were indicated at the bottom. (A-D) The black horizontal lines represent the median values, and dots in the plots represent individual cells. (E-H) In the boxplots, the box represents the interquartile range (IQR), the line within the box shows the median, and the whiskers extend up to 1.5 IQR. The signature genes used in these analyses were listed in supplemental Table 2.
Molecular characteristics of the EBV-infected cells. (A) Violin plots comparing the LT-HSC signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). (B) Violin plots comparing the short-term HSC (ST-HSC) signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected HSCs exhibited a significantly lower LT-HSC signature and a significantly higher ST-HSC signature, indicating that the infected HSCs were more active in differentiating into downstream lineages. (C) Violin plots comparing the quiescent HSC signature between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected HSCs exhibited a significantly lower quiescent HSC signature, suggesting again that the infected HSCs were more active in differentiating. (D) Violin plots comparing the signature of upregulated genes in HSC-to-MPP differentiation between EBV-infected (EBV+) and uninfected (EBV−) HSCs derived from the 3 patients with CAEBV (subjects 3-5). The infected HSCs exhibited a higher signature, indicating that the infected HSCs tended to be more primed for differentiating into MPPs. (E) Violin plots and boxplots comparing the tumor inflammation signature between PB cells derived from the 3 patients with CAEBV (subjects 3-5) vs HCs (subjects 7 and 8) (left), and EBV-infected (EBV+) vs uninfected (EBV–) PB cells derived from the 3 patients with CAEBV (subjects 3-5) (right). These results indicate that the EBV infection leads to a relatively higher level of inflammation. (F) Violin plots and boxplots comparing interferon gamma production scores between EBV-infected (EBV+) vs uninfected (EBV–) peripheral NK cells derived from the 3 patients with CAEBV (subjects 3-5). A significantly higher level of interferon gamma production was found in the infected cells. (G) Violin plots and boxplots comparing the signature of T-cell receptor signaling pathway between EBV-infected (EBV+) vs uninfected (EBV–) peripheral T cells derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected T cells exhibited a significantly higher signature of T-cell receptor signaling pathway. (H) Violin plots and boxplots comparing the T-cell exhaustion score between EBV-infected (EBV+) vs uninfected (EBV–) peripheral T cells derived from the 3 patients with CAEBV (subjects 3-5). The EBV-infected T cells had a significantly higher exhaustion score. (I) Dot plot showing the Gene Ontology (GO) terms enriched in the upregulated genes identified in EBV-infected peripheral cells compared with the uninfected cells of the 3 patients with CAEBV (subjects 3-5). T cells, NK cells, monocytes (mono), and neutrophils (neutro) were examined, and the number of differentially expressed genes were indicated at the bottom. (A-D) The black horizontal lines represent the median values, and dots in the plots represent individual cells. (E-H) In the boxplots, the box represents the interquartile range (IQR), the line within the box shows the median, and the whiskers extend up to 1.5 IQR. The signature genes used in these analyses were listed in supplemental Table 2.
In addition, we compared the signature scores calculated based on upregulated genes during HSC-to-MPP differentiation. The infected HSCs exhibited significantly higher scores (Figure 3D), indicating that the infected stem cells were more prone to produce downstream progenitors. However, when comparing the common myeloid progenitor and common lymphoid progenitor differentiation signature scores in the MPP compartment, there was no obvious difference between the infected and uninfected cells (supplemental Figure 6B). Taken together, the higher differentiation rate of infected HSCs, but not the differentiation rate of infected MPPs, was likely the cause of the increasing infection levels along the hematopoietic differentiation pathways.
EBV infection affects immune response in CAEBV
The CAEBV disease has been categorized as a lymphoproliferative disorder and has the potential to develop into a malignant neoplasm.5,18 Indeed, a reduced apoptosis signature (supplemental Figure 6C) and an augmented proliferation score were witnessed in infected cells based on our scRNA-seq data (supplemental Figure 6D). Consistently, the CAEBV samples and infected cells exhibited upregulated expression in EBV infection–associated genes previously identified,19 when compared with the healthy samples and uninfected cells, respectively (supplemental Figure 6E). Together with the alteration of host gene expression, EBV genes enhanced inflammation in CAEBV (Figure 3E), partially because of the induction of proinflammatory cytokines, such as interleukin 2, and the inhibition of anti-inflammatory cytokines, such as interleukin 10, from monocytes (supplemental Figure 6F), which consequently enhanced the production of interferon gamma from NK cells (Figure 3F) and T-cell activation (Figure 3G). Owing to the lasting inflammation, the EBV infection could finally result in elevated T-cell exhaustion signature (Figure 3H), a typical phenotype of chronic inflammation and malignant tumors. Moreover, we identified lineage-specific differentially expressed genes between infected and uninfected cells, which were related to viral gene expression in T cells and monocytes and cell chemotaxis in neutrophils (Figure 3I).
HSCT clears out EBV-infected cells throughout the hematopoietic system
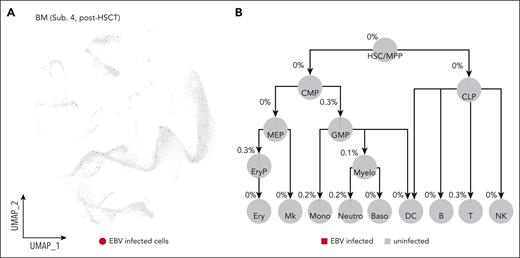
In practice, HSCT is currently the only curative therapy for CAEBV disease.5,20-22 Therefore, we performed allogeneic HSCT to 1 patient with CAEBV (subject 4), and 6 months later, we repeated the assays for EBV copy number and expression at the tissue and the single-cell levels, respectively. In BM and PB cells, the EBV-DNA copy number was below the detection limitation, and in plasma, there was completely no detectable EBV DNA. The single-cell assay confirmed that HSCT almost completely removed EBV from the hematopoietic system (Figure 4; supplemental Figure 7A-C). Overall, although it is worthy collecting further follow-up data from this patient, our data so far demonstrated that HSCT had effectively eliminated the EBV-infected hematopoietic and immune cells from this patient with CAEBV, indicating again that HSC infection is most likely the cause of CAEBV.
HSCT removes EBV-infected cells from the hematopoietic system. (A) UMAP showing that HSCT cleared out EBV infection from the hematopoietic system of the patient with CAEBV (subject 4) after HSCT. Red dots in the UMAP represent the EBV-infected cells. For comparison, the UMAP for the pre-HSCT sample of the same patient is shown in supplemental Figure 4H. (B) Pie charts showing the percentages of the EBV-infected cells in each hematopoietic cell population of the patient with CAEBV (subject 4) after HSCT. B, B cells; Baso, basophils; CMP, common myeloid progenitors; CLP, common lymphoid progenitors; DC, dendritic cells; Ery, erythrocytes; EryP, erythroid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte/erythroid progenitors; Mk, megakaryocytes; Mono, monocytes; Myelo, myelocytes; Neutro, neutrophils; T, T cells.
HSCT removes EBV-infected cells from the hematopoietic system. (A) UMAP showing that HSCT cleared out EBV infection from the hematopoietic system of the patient with CAEBV (subject 4) after HSCT. Red dots in the UMAP represent the EBV-infected cells. For comparison, the UMAP for the pre-HSCT sample of the same patient is shown in supplemental Figure 4H. (B) Pie charts showing the percentages of the EBV-infected cells in each hematopoietic cell population of the patient with CAEBV (subject 4) after HSCT. B, B cells; Baso, basophils; CMP, common myeloid progenitors; CLP, common lymphoid progenitors; DC, dendritic cells; Ery, erythrocytes; EryP, erythroid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte/erythroid progenitors; Mk, megakaryocytes; Mono, monocytes; Myelo, myelocytes; Neutro, neutrophils; T, T cells.
Discussion
CAEBV is a lethal disease caused by the neoplastic consequences of persistent EBV infection. Once the immune system or other molecular machinery is unable to control the effects of the viral infection, the infection can become acutely active.3 Tremendous efforts have been taken to understand the pathogenesis of CAEBV for better disease treatment. In this study, we have shown multiple lines of evidence that support the notion that CAEBV originates from the EBV infection of HSCs. This novel concept could therefore easily explain the observation that EBV infects both myeloid and lymphoid lineages observed by us and others,3,7,8 and the fact that currently the only curative treatment for CAEBV is HSCT.20-22 More importantly, our findings would potentially improve current CAEBV treatment strategies in conjunction with HSCT and might even enlighten novel therapeutic opportunities.
More direct evidence is still needed to consolidate our conclusions. For instance, only 5 patients with CAEBV and 3 controls were enrolled in this study, and these patients were all adults except for one 17-year-old adolescent. The inclusion of pediatric patients and a larger cohort size could further strengthen our findings. Besides, genetic tools such as linage tracing could record the progeny of infected HSCs.23 If the EBV-infected downstream progenitors and mature hematologic cells are indeed the progeny of infected stem cells, we would have more confidence to declare that the full-spectrum infection of EBV originates from HSCs. However, typical lineage tracing tools require genome engineering in human cells, which is usually infeasible. Nonetheless, spontaneous mutations accumulated in mitochondrial DNA could serve as intrinsic barcodes for lineage tracing if the mutations were detected in a highly sensitive manner.24 Moreover, EBV genome may also integrate into the human genome,25 and the nearly random integration sites in stem cells might also be useful to trace their progeny.
It is well-known that epithelia cells and B cells are the primary targets of EBV, and the receptors such as CD21 and CD35 present on B cells can be used by the virus for attachment to enter the cells.3 So far, the mechanism for EBV to infect other cell types, including HSCs, is still largely unknown. Nevertheless, in this study, we have applied RT-qPCR, PrimeFlow assays, and single-cell sequencing approaches to detect EBV+ cells. With the consistent results from all the above assays, there is no doubt that HSCs can be infected by EBV. Similarly, human cytomegalovirus DNA has been previously detected from CD34+ hematopoietic progenitors.26,27
Because the stem cells sit at the top of the differentiation tree, it is very unlikely that the infected HSCs were differentiated or specialized from other cell types after infection unless dedifferentiation takes place. However, dedifferentiation is not an unusual occurrence in mature immune cells, such as CD8 T cells28 and B cells.29 Moreover, it has been reported that HIV-1–infected T cells showed a dedifferentiated phenotype, featured by the loss of T-cell markers and gene regulation patterns similar to those of HSCs.30 Once infection has occurred, the infected HSCs could be another source of infected hematopoietic progenitors or mature periphery cells. Thus, particular cell populations such as B cells would therefore have 2 pathways for EBV infection: one would be the direct infection, and the other could be differentiated from infected HSCs. Whether there are differences in molecular characteristics and biological functions awaits further studies.
Rather than receptor-mediated EBV entry into cells, transfer of mRNA from EBV-infected cells via exosomes has been proposed as a possible alternative mechanism.31,32 However, the literature provided only limited in vitro evidence that exosomes secreted from EBV-infected cells contained both viral-encoded proteins and RNA,32 and these exosomes had a stimulatory capacity to enhance proliferation and differentiation for particular cell types.31 Therefore, the hypothesis of exosome-mediated EBV infection is yet to be verified. Recently, the coculture experiment provided another line of evidence that direct transfer of viral episomes would result in NK cell infection of EBV33; however, the in vivo data are still lacking. Altogether, receptor-, exosome-, and episome-mediated EBV infections are all plausible mechanisms to explain the ways in which the virus enters into HSCs, but further studies that can provide more details and more direct evidence are still in demand, which would certainly be helpful to dissect the pathogenesis of CAEBV.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81871633, 81901982, and 32170742), Key Scientific Project for Capital’s Health Development Research (No. 2020-1-2022), the Start Fund for Specially Appointed Professor of Jiangsu Province, and Nanjing Medical University (NMUR2020009).
Authorship
Contribution: J.W. and M.S. designed, conducted experiments, and analyzed data with input from Y.G., H.Y., and S.Y.; N.W., J.Z., and L.W. conducted human data analysis; L.W., D. Suolitiken, Y.P., D. Song, L.C., and H.L. provided technical assistance; X.W., Z.W., and J.W. wrote the manuscript with input from M.S.; and X.W. and Z.W. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zhao Wang, Department of Hematology, Beijing Friendship Hospital, Capital Medical University, 95 Yong An Rd, Xicheng District, Beijing 100050, China; email: wangzhao@ccmu.edu.cn; and Xi Wang, Prenatal Diagnosis Center, The First Affiliated Hospital, Department of Urology, The Second Affiliated Hospital, and State Key Laboratory of Reproductive Medicine and Offspring Health, Nanjing Medical University, Longmian Ave 101, Jiangning District, Nanjing 211166, China; email: xiwang@njmu.edu.cn.
References
Author notes
∗J.W. and M.S. contributed equally to this work.
The sequencing data have been deposited in the Gene Expression Omnibus database (accession number GSE231946).
All raw and processed data sets generated and used in this study are available on request from the corresponding authors, Zhao Wang (wangzhao@ccmu.edu.cn) or Xi Wang (xiwang@njmu.edu.cn).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal