A B-cell–like epigenetic and transcriptomic signature is enriched in patients with t(11;14) primary MM.
A loss of “B-cell–like” epigenetic signature with gain of canonical plasma cell transcription factors is observed at the time of resistance.
Visual Abstract
The translocation t(11;14) occurs in 20% of patients with multiple myeloma (MM) and results in the upregulation of CCND1. Nearly two-thirds of t(11;14) MM cells are BCL2 primed and highly responsive to the oral BCL2 inhibitor venetoclax. Although it is evident that this unique sensitivity to venetoclax depends on the Bcl-2 homology domain 3– proapoptotic protein priming of BCL2, the biology underlying t(11;14) MM dependency on BCL2 is poorly defined. Importantly, the epigenetic regulation of t(11;14) transcriptomes and its impact on gene regulation and clinical response to venetoclax remain elusive. In this study, by integrating assay for transposase-accessible chromatin by sequencing (ATAC-seq) and RNA-seq at the single-cell level in primary MM samples, we have defined the epigenetic regulome and transcriptome associated with t(11;14) MM. A B-cell–like epigenetic signature was enriched in t(11;14) MM, confirming its phylogeny link to B-cell rather than plasma cell biology. Of note, a loss of a B-cell–like epigenetic signature with a gain of canonical plasma cell transcription factors was observed at the time of resistance to venetoclax. In addition, MCL1 and BCL2L1 copy number gains and structural rearrangements were linked to venetoclax resistance in patients with t(11;14) MM. To date, this is the first study in which both single-cell (sc) ATAC-seq and scRNA-seq analysis are integrated into primary MM cells to obtain a deeper resolution of the epigenetic regulome and transcriptome associated with t(11;14) MM biology and venetoclax resistance.
Introduction
Multiple myeloma (MM) is a genetically complex and heterogeneous neoplasia characterized by numerous recurrent translocations and mutations that lead to disease progression and drug resistance.1,2 Primary genetic aberrations often involve the immunoglobulin heavy chain on chromosome 14, where translocations juxtapose the IgH enhancers with CCND1 in t(11;14) seen in 20% of patients with MM, FGFR3/MMSET in t(4;14) reported in 10%, and less commonly MAF in t(14;16), MAFB in t(14;20), and CCND3 in t(6;14).3-5 MM cells with t(11;14) have distinct clinical features and unique biology, including lymphoplasmacytic morphology, upregulation of CCND1, and, in most cases, MS4A1 coding for the antigen CD20.6,7 This translocation is associated with overexpression of the anti-apoptotic protein B-cell leukemia/lymphoma 2 (BCL2), indicating susceptibility to BCL2 inhibition.5 The members of the BCL2 family play a crucial role in regulating the intrinsic apoptotic pathway.8 They are classified into 3 groups according to their structural and functional characteristics. They are either proapoptotic (Bcl-2 interacting mediator of cell death [BIM], BH3 interacting-domain death agonist [BID], Bcl-2 associated agonist of cell death [BAD], PUMA, and PMAIP1 [phorbol-12-myristate-13-acetate-induced protein 1] [NOXA]), antiapoptotic (BCL2, MCL1, and B-cell lymphoma–extra-large [BCLXL]), or pore-forming effectors (Bcl-2-like protein 4 [BAX] and Bcl-2 homologous antagonist/killer [BAK]).9
Venetoclax is the first clinically available drug specifically targeting the antiapoptotic BCL2 protein, displacing the proapoptotic proteins from their Bcl-2 homology domain 3 (BH3) groove and leading to cell death.10 It has demonstrated significant antitumor activity in several BCL2-dependent hematologic malignancies, including chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), and follicular lymphoma (FL).11,12 In MM, it has recently emerged as an effective therapeutic option for patients with relapsed and refractory MM harboring t(11;14), both as a single agent and in combination with other drugs.13-17 This sensitivity has been correlated with high BCL2/BCL2L1 and BCL2/MCL1 messenger RNA (mRNA) ratios,14,18 suggesting that the expression of BCL2 members could be a potential predictive biomarker of response to venetoclax. Furthermore, Gupta et al recently demonstrated by using a flow-based assay that in MM, venetoclax sensitivity corresponds to a signature enriched for B-cell–associated genes with increased BCL2 dependency,19 pointing to novel biomarkers of venetoclax sensitivity independent of t(11;14).
However, the expression of BCL2 family members in MM is variable and often associated with the upregulation of either MCL1 or BCL2L1, which can lead to venetoclax resistance. In addition, although functional tests such as BH3 profiling or in vitro BH3 mimetic drug testing have demonstrated that the unique sensitivity to venetoclax is dependent on the BH3 proapoptotic proteins BIM and BAX/BAK priming of BCL2,20 the biology that underlies t(11;14) MM dependency on BCL2 is poorly defined. Importantly, the epigenetic regulation of t(11;14) transcriptomes and its impact on gene regulation and clinical response to venetoclax remain elusive.
In this study, we have compared and integrated the chromatin accessibility (single-cell ATAC sequencing [scATAC-seq]) and transcriptome (scRNA-seq) of primary MM samples with and without t(11;14) at a single-cell level. We have analyzed the chromatin-accessibility profiles of t(11;14) patients before and after venetoclax treatment to define the epigenetic changes concurrent with venetoclax resistance. To date, this is the first study in which both scATAC-seq and scRNA-seq analysis are integrated in primary MM cells to define the epigenetic regulome and transcriptome associated with t(11;14) MM biology and obtain a deeper resolution of the changes associated with venetoclax resistance.
Methods
Processing of BM samples
Bone marrow (BM) aspirates were collected from 15 patients with MM. Written informed consent was obtained according to the Declaration of Helsinki and the Medical Center institutional review board. The clinical data of each patient included in the study are reported in Table 1.
Clinical characteristics of the patients with MM included in the study
| Clinical data . | scATAC- and scRNA-seq . | scDNA-seq . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID . | Sex . | Age (y) . | MM subtype . | t(11;14) status . | t(4;14) status . | t(14;16) status . | del13q status . | del17p status . | Stage (R-ISS) . | Number of prior therapies . | Best response to venetoclax . | Duration of response to venetoclax (mo) . | Prevenetoclax Sample . | Postvenetoclax Sample . | Prevenetoclax Sample . | Postvenetoclax Sample . |
| PM01 | M | 44 | IgGκ | Positive (17% of the cells) | Negative | Negative | Negative | Negative | II | 4 | VGPR | 8 | P1393 | P1555 | P1426 | P1555 |
| PM02 | M | 74 | IgGκ | Negative | Negative | Negative | Negative | Negative | I | 7 | NA | NA | P1723 | NA | NA | NA |
| PM03 | F | 66 | κ light chain | Negative | Negative | Negative | Negative | Negative | III | 4 | NA | NA | P1608 | P1675 | NA | NA |
| PM04 | F | 82 | IgGκ | Negative | Negative | Negative | Negative | Negative | II | 3 | NA | NA | P1299 | P1345 | NA | NA |
| PM05 | M | 78 | Λ light chain | Positive (97% of the cells) | Negative | Negative | Negative | Positive (99% of cells) | II | 8 | VGPR | 21 | P1643 | NA | NA | NA |
| PM06 | M | 65 | IgGκ | Negative | Negative | Negative | Negative | Negative | III | 4 | NA | NA | P1752 | NA | NA | NA |
| PM07 | M | 69 | IgGλ | Positive (88% of the cells) | Negative | Negative | Negative | Positive (66% of cells) | I | 4 | PR | 4 | P1389 | P1479 | P1389 | P1479 |
| PM08 | M | 60 | IgAκ | Negative | Positive (17% of the cells) | Negative | Negative | Negative | II | 6 | NA | NA | P1680 | NA | NA | NA |
| PM09 | M | 74 | IgGλ | Positive (77% of the cells) | Negative | Negative | Negative | Negative | I | 4 | VGPR | 13 | P1498 | P1698 | P1498 | P1698 |
| PM10 | F | 61 | IgGλ | Positive (94% of the cells) | Negative | Negative | Negative | Negative | II | 10 | VGPR | 11 | P1217 | P1438 | P1217 | P1438 |
| PM11 | F | 84 | IgAλ | Positive (49% of the cells) | Negative | Negative | Negative | Negative | III | 3 | VGPR | 38 | P1388 | NA | NA | NA |
| PM12 | M | 66 | IgGλ | Negative | Negative | Negative | Negative | Negative | I | 4 | NA | NA | P1751 | P1809 | NA | NA |
| PM13 | M | 81 | IgAκ | Negative | Negative | Negative | Negative | Negative | III | 1 | NA | NA | P1396 | P1461 | NA | NA |
| PM14 | F | 70 | IgAλ and IgGλ | Negative | Negative | Negative | Negative | Negative | II | 2 | NA | NA | P1395 | P1606 | NA | NA |
| PM15 | M | 60 | IgAκ | Negative | Negative | Negative | Negative | Negative | II | 2 | NA | NA | P1722 | NA | NA | NA |
| Clinical data . | scATAC- and scRNA-seq . | scDNA-seq . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID . | Sex . | Age (y) . | MM subtype . | t(11;14) status . | t(4;14) status . | t(14;16) status . | del13q status . | del17p status . | Stage (R-ISS) . | Number of prior therapies . | Best response to venetoclax . | Duration of response to venetoclax (mo) . | Prevenetoclax Sample . | Postvenetoclax Sample . | Prevenetoclax Sample . | Postvenetoclax Sample . |
| PM01 | M | 44 | IgGκ | Positive (17% of the cells) | Negative | Negative | Negative | Negative | II | 4 | VGPR | 8 | P1393 | P1555 | P1426 | P1555 |
| PM02 | M | 74 | IgGκ | Negative | Negative | Negative | Negative | Negative | I | 7 | NA | NA | P1723 | NA | NA | NA |
| PM03 | F | 66 | κ light chain | Negative | Negative | Negative | Negative | Negative | III | 4 | NA | NA | P1608 | P1675 | NA | NA |
| PM04 | F | 82 | IgGκ | Negative | Negative | Negative | Negative | Negative | II | 3 | NA | NA | P1299 | P1345 | NA | NA |
| PM05 | M | 78 | Λ light chain | Positive (97% of the cells) | Negative | Negative | Negative | Positive (99% of cells) | II | 8 | VGPR | 21 | P1643 | NA | NA | NA |
| PM06 | M | 65 | IgGκ | Negative | Negative | Negative | Negative | Negative | III | 4 | NA | NA | P1752 | NA | NA | NA |
| PM07 | M | 69 | IgGλ | Positive (88% of the cells) | Negative | Negative | Negative | Positive (66% of cells) | I | 4 | PR | 4 | P1389 | P1479 | P1389 | P1479 |
| PM08 | M | 60 | IgAκ | Negative | Positive (17% of the cells) | Negative | Negative | Negative | II | 6 | NA | NA | P1680 | NA | NA | NA |
| PM09 | M | 74 | IgGλ | Positive (77% of the cells) | Negative | Negative | Negative | Negative | I | 4 | VGPR | 13 | P1498 | P1698 | P1498 | P1698 |
| PM10 | F | 61 | IgGλ | Positive (94% of the cells) | Negative | Negative | Negative | Negative | II | 10 | VGPR | 11 | P1217 | P1438 | P1217 | P1438 |
| PM11 | F | 84 | IgAλ | Positive (49% of the cells) | Negative | Negative | Negative | Negative | III | 3 | VGPR | 38 | P1388 | NA | NA | NA |
| PM12 | M | 66 | IgGλ | Negative | Negative | Negative | Negative | Negative | I | 4 | NA | NA | P1751 | P1809 | NA | NA |
| PM13 | M | 81 | IgAκ | Negative | Negative | Negative | Negative | Negative | III | 1 | NA | NA | P1396 | P1461 | NA | NA |
| PM14 | F | 70 | IgAλ and IgGλ | Negative | Negative | Negative | Negative | Negative | II | 2 | NA | NA | P1395 | P1606 | NA | NA |
| PM15 | M | 60 | IgAκ | Negative | Negative | Negative | Negative | Negative | II | 2 | NA | NA | P1722 | NA | NA | NA |
Cytogenetic abnormalities identified based on fluorescence in situ hybridization in this patient cohort; R-ISS (revised international staging system); VGPR (very good partial response); PR (partial response); SD (stable disease); MR (minimal response); and duration of response to venetoclax.
F, female; ID, identification number; Ig, immunoglobulin; M, male; NA, not available.
After collection, the BM aspirates were diluted in phosphate-buffer saline (PBS), and the mononuclear cell separation was performed by density centrifugation (Ficoll-Paque, Sigma) for 30 minutes at 2000 rpm without a brake. Cells were carefully aspirated and washed with phosphate-buffered saline (centrifugation for 5 minutes at 2000 rpm). After cell counting, cells were separated by magnetic sorting using anti-CD138 microbeads (Miltenyi) according to the manufacturer’s protocol. Subsequently, CD138+ cells were frozen in 90% fetal bovine serum (Gibco) supplemented with 10% dimethyl sulfoxide (Sigma) and stored at −80°C until further use.
All the details related to the scATAC-seq and scRNA-seq libraries preparation, sequencing, and analysis are described in the supplemental Methods, available on the Blood website.
Data processing
Genomic sequence reads were treated with the CellRanger suite (cellranger-atac v1.1.0 and cellranger v3.0.2 for scATAC-seq and scRNA-seq, respectively) against the human reference genome GRCh38 with default parameters. scATAC-seq analysis was performed using R version 4.1.3, ArchR version 1.0.2, 14 threads, and a seed of 1. scRNA-seq analysis was performed using R version 4.1.3, Seurat version 4.1.0, and dplyr version 1.0.8.
Additional information, including quality control steps, peak calling, gene set enrichment analysis (GSEA), scRNA-seq integration, and trajectory analysis, are described in the supplemental Methods. The specifics of all the analyses performed on the different data sets are delineated in the supplemental Methods. The details of the methodologies used in the functional studies are also provided in the supplemental Methods.
Results
Epigenetic and transcriptomic characteristics of t(11;14) MM
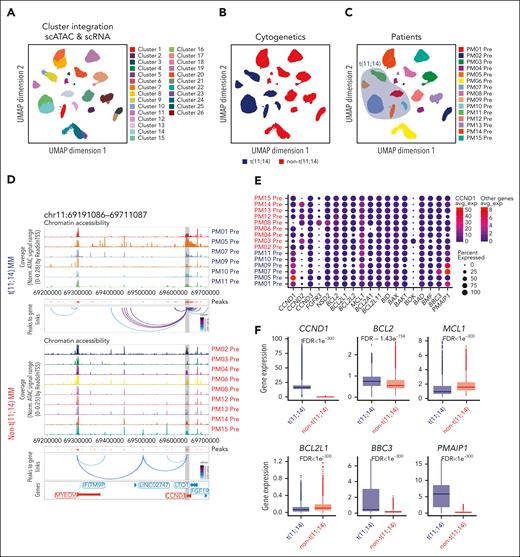
To comprehensively profile primary MM cells, we performed parallel scATAC-seq and scRNA-seq on the same samples obtained from 15 patients with relapsed and refractory MM: 6 patients with t(11;14) and 9 without t(11;14). Samples were collected before starting therapy and at the time of relapse. Patients with t(11;14) MM were treated with venetoclax (Table 1). After performing quality control steps, we first explored the epigenetic and transcriptomic landscape of MM cells in the pretreatment samples to identify the regulatory elements that define the biological states of the plasma cells (PCs) among the 2 groups. As shown in the uniform manifold approximation and projection (UMAP) (Figure 1A), after the integration of scATAC- and scRNA-seq data sets, 26 clusters were identified. The individual clustering UMAP are shown in supplemental Figure 1. Eight clusters were composed of t(11;14) cells, which were more closely related to each other than the other 18 clusters of non-t(11;14) cells (Figure 1B). Of interest, each distinct cluster corresponded to a unique sample, and some samples regrouped in distinct subclusters (Figure 1C), confirming the interpatient and intrapatient heterogeneity of MM. As expected, patients with t(11;14) have high and unique chromatin accessibility at the CCND1 locus compared with those without t(11;14) with unique cis-regulatory elements of CCND1, as shown in Figure 1D. The chromatin accessibilities observed in t(11;14) MM samples were associated with an increase in CCND1 transcription that was significantly different between the 2 subsets (Figure 1E; supplemental Figure 2A) and groups (Figure 1F).
Epigenetic and transcriptomic characteristics of t(11;14) MM. (A-C) Uniform manifold approximation and projection (UMAP) showing the distribution of CD138+ cells (n = 46 237 cells) collected from 15 patients with MM using the scATAC-seq and scRNA-seq data in the Latent Semantic Indexing method (LSI) space. Each point represents 1 cell. The cells are marked by a color code based on the different clusters (A), fluorescence in situ hybridization, cytogenetic (B), and patient identification (C). (D) Genome accessibility track visualization of CCND1 and MYEOV with peak-to-gene link identified by ArchR for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. The color of the arcs indicates the correlation strength between the peak and the gene expression (a darker color indicates a stronger correlation). The genes are indicated in red when the gene is on the positive strand (transcription start site [TSS] on the left) and blue when on the negative strand (TSS on the right). The gray boxes highlight the enhancer or promoter regions of the gene of interest. (E) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in each MM sample. The t(11;14) MM samples are highlighted in blue, whereas the non-t(11;14) MM samples are highlighted in red. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each patient. The dark blue indicates a lower gene expression count, and the light red indicates a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (F) Boxplots comparing the integrated expression of CCND1, BCL2, MCL1, BCL2L1, BBC3, and PMAIP1 in the t(11;14) vs non-t(11;14) MM groups. The false discovery rate (FDR) values associated with each gene are shown at the top of each boxplot comparison. (G) Genome accessibility track visualization of BCL2 with peak-to-gene links for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. For each group, the peaks panel highlights the different significant peaks identified by ArchR. (H-J) Boxplots indicating the BCL2/BCL2L1 (H), BCL2/MCL1 (I), and MCL1/PMAIP1 gene expression ratios (J) for each MM prevenetoclax sample (left). The boxes highlight the patients with t(11;14) MM. The comparisons between t(11;14) and non-t(11;14) MM groups are shown (right), with t(11;14) in blue and non-t(11:14) in red. The FDR values associated with each gene are shown at the top of each boxplot comparison.
Epigenetic and transcriptomic characteristics of t(11;14) MM. (A-C) Uniform manifold approximation and projection (UMAP) showing the distribution of CD138+ cells (n = 46 237 cells) collected from 15 patients with MM using the scATAC-seq and scRNA-seq data in the Latent Semantic Indexing method (LSI) space. Each point represents 1 cell. The cells are marked by a color code based on the different clusters (A), fluorescence in situ hybridization, cytogenetic (B), and patient identification (C). (D) Genome accessibility track visualization of CCND1 and MYEOV with peak-to-gene link identified by ArchR for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. The color of the arcs indicates the correlation strength between the peak and the gene expression (a darker color indicates a stronger correlation). The genes are indicated in red when the gene is on the positive strand (transcription start site [TSS] on the left) and blue when on the negative strand (TSS on the right). The gray boxes highlight the enhancer or promoter regions of the gene of interest. (E) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in each MM sample. The t(11;14) MM samples are highlighted in blue, whereas the non-t(11;14) MM samples are highlighted in red. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each patient. The dark blue indicates a lower gene expression count, and the light red indicates a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (F) Boxplots comparing the integrated expression of CCND1, BCL2, MCL1, BCL2L1, BBC3, and PMAIP1 in the t(11;14) vs non-t(11;14) MM groups. The false discovery rate (FDR) values associated with each gene are shown at the top of each boxplot comparison. (G) Genome accessibility track visualization of BCL2 with peak-to-gene links for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. For each group, the peaks panel highlights the different significant peaks identified by ArchR. (H-J) Boxplots indicating the BCL2/BCL2L1 (H), BCL2/MCL1 (I), and MCL1/PMAIP1 gene expression ratios (J) for each MM prevenetoclax sample (left). The boxes highlight the patients with t(11;14) MM. The comparisons between t(11;14) and non-t(11;14) MM groups are shown (right), with t(11;14) in blue and non-t(11:14) in red. The FDR values associated with each gene are shown at the top of each boxplot comparison.
Next, we examined whether the expression of BCL2 family genes was higher in t(11;14) MM. Although BCL2 expression trended higher in t(11;14) MM (Figure 1E-F), there was a clear overlap with patients without t(11;14) MM (supplemental Figure 2B). The chromatin-accessible regions at the BCL2 locus did not differ between the 2 groups, consistent with the mRNA data (Figure 1G). Looking at other antiapoptotic BCL2 genes, including MCL1 and BCL2L1, we also observed an overlap between the 2 groups (Figure 1F; supplemental Figure 2C-D). However, the ratio of BCL2/BCL2L1 and BCL2/MCL1 was significantly higher in patients with t(11;14) (Figure 1H-I), consistent with their BCL2 dependency. Interestingly, we observed higher expression of the BH3 sensitizers BBC3 (PUMA) and PMAIP1 (NOXA) in patients with t(11;14) (Figure 1F). Although the high expression of BBC3 in t(11;14) was driven by a high expression of the BBC3 transcript in PM07 (Figure 1E; supplemental Figure 2E), PMAIP1 was found overexpressed in all the patients with t(11;14) (supplemental Figure 2F). In addition, the chromatin-accessible regions at the PMAIP1 locus differed between patients with t(11;14) and without t(11;14), consistent with the mRNA data (supplemental Figure 3A-B). Furthermore, the ratio of MCL1/PMAIP1 was significantly lower in patients with t(11;14) MM than in those without t(11;14) (Figure 1J). NOXA is known to bind MCL1 and mediate its degradation via HUWE1 (MULE) ubiquitin E3 ligase.21 Therefore, its upregulation may suppress the anti-apoptotic effect of MCL1 and promote BCL2 priming and venetoclax sensitivity in t(11;14) MM.
t(11;14) MM cells are characterized by a B-cell–like epigenetic and transcriptomic signature
To determine the epigenetic regulation of the BCL2 dependency, we compared the genomic mapping of accessible chromatin peaks in patients with t(11;14) vs those without t(11;14). We identified 169 889 total peaks with similar numbers in both groups, and 64% overlapped (supplemental Figure 4A-B). Next, we performed an enrichment analysis of the peaks using the bulk ATAC-seq data provided by ArchR. As shown in supplemental Figure 4C, t(11;14) MM cells were characterized by a B-cell–like epigenetic signature, including memory B (MB) cells and naïve B cells. This data was confirmed in an independent bulk ATAC-seq data set of in vitro differentiated B cells and PCs.19,22 The enrichment analysis showed a PC epigenetic signature in the non-t(11;14) cells in contrast with the B-cell–like signature observed in t(11;14) cells (Figure 2A). To confirm these findings, we computed the overlaps of the differentially accessible regions (DARs) in the 2 data sets and found that the regions more accessible in the t(11;14) cells had significant overlaps with the regions more accessible in MB cells, as depicted by a positive odds ratio (Figure 2B). Similarly, the regions that were more accessible in t(11;14) cells were also depleted in regions more accessible in PCs when compared with those in MB cells. In contrast, in non-t(11;14) cells, the regions more accessible had a significant overlap with regions more accessible in PCs (Figure 2B). Additionally, GSEA performed on the integrated scRNA-seq data confirmed the B-cell–like transcriptomic signature in t(11;14) MM (Figure 2C). Next, we performed a motif enrichment analysis on the DARs using the Catalog of Inferred Sequence Binding Preferences database23 to identify transcription factors (TFs) involved in these signatures. We identified 160 TF motifs more accessible in t(11;14) and 184 in non-t(11;14) (Figure 2D-E). Among those, several TFs (>50) correlate in both groups with either the predicted scATAC-seq data or the integrated scRNA-seq data. We identified known TFs involved in the development/transition of B cells to PCs, such as the interferon regulator factor, Runt-related (RUNX), PR/SET domain zinc finger (PRDM), and the activator protein-1 (AP-1) TFs. Using the DARs, we then looked at the footprint of these positive TFs. Of interest, in the t(11;14) MM, we found increased accessibility at the motif sites in several genes involved in B-cell biology, such as the repressor of PRDM1 SPIB,24SPI1 (PU.1), and PAX5. In contrast, in non-t(11;14) MM, we found increased accessibility at the binding site of PC genes, such as PRDM1, RUNX1, and IRF7 (Figure 2F). Lastly, looking at the differentially expressed genes (DEG) based on the integrated scRNA-seq data (supplemental Figure 4D), we observed an upregulation of B-cell markers, such as PTPRC (CD45), PIK3AP1, MS4A1 (CD20), CD79A, CCR7, IRF8, CIITA, CXCR4, BEND5, and VPREB3 in t(11;14) MM. In contrast, PC markers such as CD28, ELL2, ATF6, and IL6R, were upregulated in the non-t(11;14) MM cells (Figure 2G). Together, these data suggest that a B-cell–like epigenetic and transcriptomic signature is enriched in t(11;14) MM cells, confirming its linkage to B-cell ontogeny rather than to PC biology.
t(11;14) MM cells are characterized by a B-cell–like epigenetic signature. (A) Enrichment heat maps of the peaks identified in each group (FDR ≤ 0.01 and Log2FC ≥ 1) using an independent bulk ATAC-seq data specific for MB cells and PCs. (B) Odds ratios indicate the overlap of the DARs identified in the t(11;14) MM cells (blue) and in the non-t(11;14) (red) when compared with those present in the MB cells and PCs. The error bars denote the 95% confidence interval calculated using a two-sided Fisher exact test. (C) Violin plots of the normalized enrichment score (NES) obtained in the t(11;14) vs non-t(11;14) MM groups. These data were generated on the integrated scRNA-seq counts using the Molecular Signatures databases (msigdb) for BM B cells (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL) and PCs (C8, HAY_BONE_MARROW_PLASMA_CELL). (D-E) Plots showing the top 30 most accessible motifs observed in t(11;14) (D) MM cell DARs and non-t(11;14) (E). The color scale represents the mlog10Padj with a light gray color indicating a low adjusted P value and a dark color a high adjusted P value. (F) Representation of the Tn5 bias–adjusted TF footprints for the B-cell motifs SPIB, SPI1, and PAX5; and the PC motifs PRDM1, RUNX1, and IRF7. The lines are colored in blue for the t(11;14) and red for non-t(11;14) MM cells. (G) Boxplots showing the scRNA expression of selected B-cell and PC genes identified by the DEG analysis and overexpressed in t(11:14) (blue) or non-t(11:14) (red) MM cells. The FDR values associated with each gene are shown at the top of each boxplot comparison.
t(11;14) MM cells are characterized by a B-cell–like epigenetic signature. (A) Enrichment heat maps of the peaks identified in each group (FDR ≤ 0.01 and Log2FC ≥ 1) using an independent bulk ATAC-seq data specific for MB cells and PCs. (B) Odds ratios indicate the overlap of the DARs identified in the t(11;14) MM cells (blue) and in the non-t(11;14) (red) when compared with those present in the MB cells and PCs. The error bars denote the 95% confidence interval calculated using a two-sided Fisher exact test. (C) Violin plots of the normalized enrichment score (NES) obtained in the t(11;14) vs non-t(11;14) MM groups. These data were generated on the integrated scRNA-seq counts using the Molecular Signatures databases (msigdb) for BM B cells (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL) and PCs (C8, HAY_BONE_MARROW_PLASMA_CELL). (D-E) Plots showing the top 30 most accessible motifs observed in t(11;14) (D) MM cell DARs and non-t(11;14) (E). The color scale represents the mlog10Padj with a light gray color indicating a low adjusted P value and a dark color a high adjusted P value. (F) Representation of the Tn5 bias–adjusted TF footprints for the B-cell motifs SPIB, SPI1, and PAX5; and the PC motifs PRDM1, RUNX1, and IRF7. The lines are colored in blue for the t(11;14) and red for non-t(11;14) MM cells. (G) Boxplots showing the scRNA expression of selected B-cell and PC genes identified by the DEG analysis and overexpressed in t(11:14) (blue) or non-t(11:14) (red) MM cells. The FDR values associated with each gene are shown at the top of each boxplot comparison.
Loss of B-cell–like epigenetic signature with the gain of canonical PC TFs is associated with resistance to venetoclax in t(11;14) MM
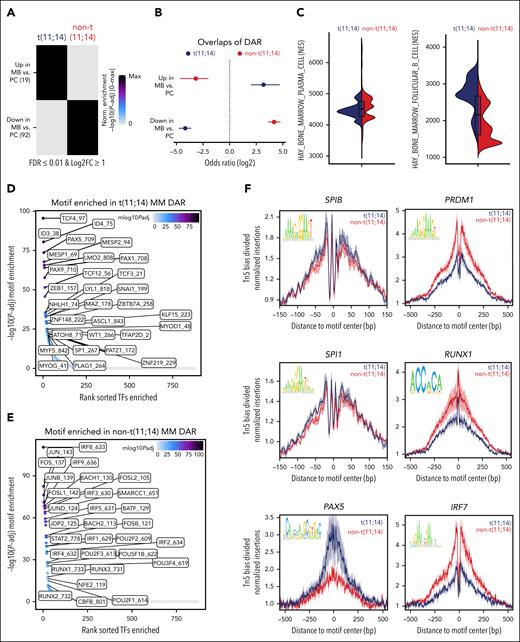
To define the contribution of epigenetic regulomes to the acquired resistance to venetoclax in t(11;14) MM, we next analyzed the chromatin-accessibility profiles of patients with t(11;14) MM, comparing prevenetoclax vs postvenetoclax samples. A distinct clustering of cells was noted (Figure 3A-B). Of interest, enrichment analysis performed on the DAR between prevenetoclax vs postvenetoclax samples showed that the regions more accessible in the postvenetoclax samples significantly overlap with regions more accessible in PCs rather than MB cells. In contrast, in the prevenetoclax samples, the chromatin-accessible regions overlapped the accessible regions present in MB cells (Figure 3C). GSEA performed on the integrated scRNA-seq data confirmed the loss of B-cell–like transcriptomic signature in postvenetoclax compared with prevenetoclax cells (Figure 3D). As expected, interpatient variability was observed when we compared individual patients’ samples. Although in patients PM01 and PM07, we observed a loss of the B-cell signature in the postvenetoclax samples at the transcriptomic (Figure 3E-F) or epigenetic levels (supplemental Figure 5), different changes were observed in patients PM09 and PM10, suggesting that mechanisms alternative to lineage plasticity might have caused resistance in these patients.
Loss of B-cell–like epigenetic signature with gain of canonical PC TFs is associated with acquired resistance to venetoclax in t(11;14) MM. (A-B) UMAP projection showing the distribution of CD138+ cells (n = 26 847 cells) collected from 4 patients with t(11;14) MM using the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked by a color code based on the different patient (A) and treatment (before vs after venetoclax) (B). (C) Odds ratios indicating the overlap of the DAR identified in the pre- (light blue) and post- (purple) venetoclax MM cells when compared with those present in the MB cells and PCs. The error bars denote the 95% confidence interval calculated using a two-sided Fisher exact test. (D) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (E-F) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells in each paired patient treated with venetoclax (PM01, PM07, PM09, and PM10). These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (E) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (F) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (G) Heat map of the motif hypergeometric enrichment-adjusted P values within the Catalog of Inferred Sequence Binding Preferences databases of each prevenetoclax and postvenetoclax MM cell from patients PM01, PM07, PM09, and PM10. The color indicates the motif enrichment (−log10[P value]) based on the hypergeometric test. The maximum mlog10Padj is indicated in parenthesis for each significant enrichment found. (H) Representation of the Tn5 bias–adjusted TF footprints for IRF4, RUNX1, ID3, and TCF4 motifs identified by the enrichment motif analysis. The lines are colored in blue and red to indicate the prevenetoclax and postvenetoclax samples for patient PM01 and in purple and green for patient PM07.
Loss of B-cell–like epigenetic signature with gain of canonical PC TFs is associated with acquired resistance to venetoclax in t(11;14) MM. (A-B) UMAP projection showing the distribution of CD138+ cells (n = 26 847 cells) collected from 4 patients with t(11;14) MM using the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked by a color code based on the different patient (A) and treatment (before vs after venetoclax) (B). (C) Odds ratios indicating the overlap of the DAR identified in the pre- (light blue) and post- (purple) venetoclax MM cells when compared with those present in the MB cells and PCs. The error bars denote the 95% confidence interval calculated using a two-sided Fisher exact test. (D) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (E-F) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells in each paired patient treated with venetoclax (PM01, PM07, PM09, and PM10). These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (E) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (F) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (G) Heat map of the motif hypergeometric enrichment-adjusted P values within the Catalog of Inferred Sequence Binding Preferences databases of each prevenetoclax and postvenetoclax MM cell from patients PM01, PM07, PM09, and PM10. The color indicates the motif enrichment (−log10[P value]) based on the hypergeometric test. The maximum mlog10Padj is indicated in parenthesis for each significant enrichment found. (H) Representation of the Tn5 bias–adjusted TF footprints for IRF4, RUNX1, ID3, and TCF4 motifs identified by the enrichment motif analysis. The lines are colored in blue and red to indicate the prevenetoclax and postvenetoclax samples for patient PM01 and in purple and green for patient PM07.
Motif enrichment analysis performed on the patients with t(11;14) revealed at the time of venetoclax resistance an enrichment of PC TFs motifs, such as IRF1, IRF4, PRDM1, and RUNX1, together with a loss of B-cell TFs motifs, such as ID3, PAX5, and TCF3 (Figure 3G). DAR footprinting of these TFs revealed an increased accessibility for IRF4 and RUNX1, whereas decreased accessibility was observed in B-cell TFs such as ID3 and TCF4 (Figure 3H). Of note, the pseudotime trajectory analysis performed in these patients confirmed the upregulation of the identified PC TFs IRF1, IRF4, RUNX1, PRDM1, and XBP1 together with the downregulation of the B-cell TFs, such as ID3, TCF4, CD79A, and MS4A1, in the postvenetoclax samples (supplemental Figure 6A-B). Of interest, we also noted the upregulation of ERN1, a kinase and endoribonuclease that induces XBP1 splicing required for PC differentiation and immunoglobulin production.25-28
Taken together, these data indicate that loss of a B-cell–like transcriptional program and a concomitant gain of canonical PC TFs occur in a significant fraction of patients who acquired resistance to venetoclax.
Integrated epigenomic and transcriptomic analysis reveals multiple coexisting changes associated with venetoclax resistance in t(11;14) MM
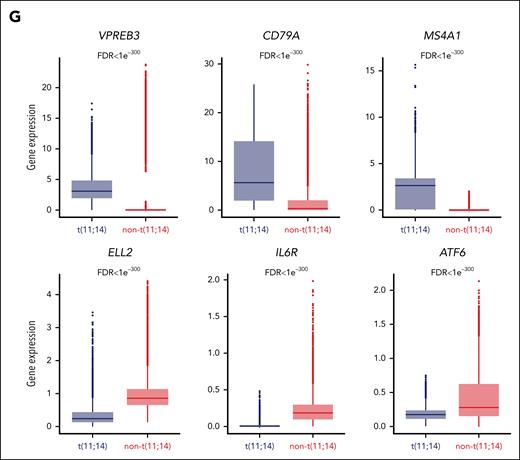
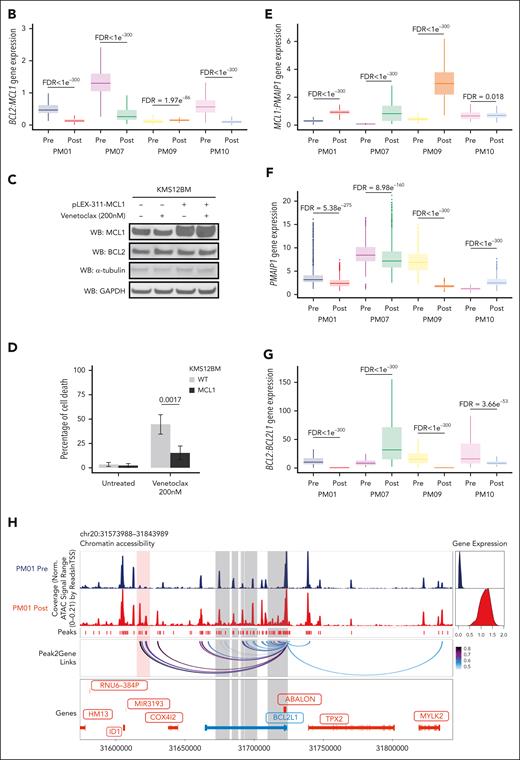
We then interrogated the changes associated with acquired venetoclax resistance by integrating epigenetic and transcriptional single-cell analysis in each patient with t(11;14). Starting with the 2 patients with a loss of B-cell phenotype at the time of relapse, we observed that in PM01, 2 large clusters separated the prevenetoclax vs postvenetoclax cells (Figure 4A). These clusters were subdivided into 12 subclusters. Of note, subclusters PM01-1 to -5 were composed of prevenetoclax cells that were sensitive to the drug and fully contracted after treatment. Seven subclusters, PM01-6 to -12, were absent in the prevenetoclax sample and emerged as de novo subclusters at the time of resistance (Figure 4A). Integration of the scRNA-seq data confirmed a B-cell–like signature in the subclusters that contracted after treatment (PM01-1 to -5), whereas the new emerging clusters (PM01-6 to -12) had a PC-like signature (Figure 4B). By interrogating the DAR motif enrichment of PM01 subclusters, we observed that almost all postvenetoclax subclusters were enriched for RUNX1, RUNX2, and their functional partner CBFB,29 whereas the others were enriched for IRF4, AP-1, and NF-κB TFs (supplemental Figure 7A). The DEG identified a B-cell signature with upregulation of MS4A1, PMAIP1, VPREB3, PAX5, and BCL6 in the venetoclax-sensitive subclusters, whereas an increase of PC markers, such as XBP1 and ERN1, was observed in the resistant subclusters (supplemental Figure 7B-D), confirming the loss of B-cell phenotype and the selection of PC phenotype subclusters at progression. Of interest, all the postvenetoclax subclusters expressed a high RUNX1 transcript (supplemental Figure 7D), suggesting a possible role of RUNX1 in venetoclax resistance. As such, in functional studies performed by overexpressing RUNX1 in the venetoclax-sensitive KMS12BM cells, we have demonstrated that the upregulation of RUNX1 reduces the sensitivity of MM cells to venetoclax through the downregulation of the NF-kB pathway (Figure 4C-D), also involved in programmed cell death. These data provide new insights into the functional role of RUNX1 in mediating resistance to venetoclax in the t(11;14) MM cells.
Integrated epigenomic and transcriptomic analysis reveals multiple coexisting changes associated with venetoclax resistance in t(11;14) MM. (A) UMAP projection showing the distribution of CD138+ cells (n = 8679 cells) identified in the t(11;14) patient PM01 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (B) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PsC (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (C) On the left, immunoblotting of parental (wild-type [WT]) and KMS12BM-RUNX1 overexpressing cells for RUNX1 and BCL2, in the presence or absence of venetoclax 200 nM for 3 hours. The bar plot (right) indicates the effect of RUNX1 on NF-kB activation in the KMS12BM overexpressing RUNX1 when compared with parental cells (WT). (D) Bar plot showing the percentage of cell death observed in parental (WT) vs RUNX1-KMS12BM cells after 24 hours of exposure to venetoclax 200 and 500 nM. The data presented are the mean ± standard error of 3 independent experiments. The adjusted P values associated are shown at the top of each bar plot. (E) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) identified in the t(11;14) patient PM07 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (F) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (G) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in PM07 subclusters. Subclusters from the prevenetoclax sample are highlighted in blue, whereas the ones from the postvenetoclax sample are in purple. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each subcluster. The dark blue indicates a lower gene expression count, and light red a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (H) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) collected from patient PM07 based on the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked with color codes based on the MCL1 amplification. The black color indicates the cells harboring the amplification of MCL1, whereas the gray or light gray indicates the cells without the amplification or with unknown information, respectively.
Integrated epigenomic and transcriptomic analysis reveals multiple coexisting changes associated with venetoclax resistance in t(11;14) MM. (A) UMAP projection showing the distribution of CD138+ cells (n = 8679 cells) identified in the t(11;14) patient PM01 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (B) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PsC (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (C) On the left, immunoblotting of parental (wild-type [WT]) and KMS12BM-RUNX1 overexpressing cells for RUNX1 and BCL2, in the presence or absence of venetoclax 200 nM for 3 hours. The bar plot (right) indicates the effect of RUNX1 on NF-kB activation in the KMS12BM overexpressing RUNX1 when compared with parental cells (WT). (D) Bar plot showing the percentage of cell death observed in parental (WT) vs RUNX1-KMS12BM cells after 24 hours of exposure to venetoclax 200 and 500 nM. The data presented are the mean ± standard error of 3 independent experiments. The adjusted P values associated are shown at the top of each bar plot. (E) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) identified in the t(11;14) patient PM07 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (F) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (G) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in PM07 subclusters. Subclusters from the prevenetoclax sample are highlighted in blue, whereas the ones from the postvenetoclax sample are in purple. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each subcluster. The dark blue indicates a lower gene expression count, and light red a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (H) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) collected from patient PM07 based on the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked with color codes based on the MCL1 amplification. The black color indicates the cells harboring the amplification of MCL1, whereas the gray or light gray indicates the cells without the amplification or with unknown information, respectively.
In patient PM07, the integration of scATAC-seq and scRNA-seq data identified 2 major clusters separating the prevenetoclax vs postvenetoclax cells (Figure 4E). These clusters were subdivided into 15 subclusters. In this case, in addition to the loss of B-cell phenotype and the enrichment of the PC-like signature observed in most of the postvenetoclax subclusters (from PM07-9 to -15) at time of progression (Figure 4F; supplemental Figure 8A), we observed a specific transcriptomic signature with upregulation of MCL1 in all the postvenetoclax cells (Figure 4G; supplemental Figure 8B-D). Copy number variation analysis obtained from scATAC-seq data identified MCL1 gene locus amplification as the driver of MCL1 transcript upregulation in the postvenetoclax subclusters PM07-11 to -13 and -15 (Figure 4H), whereas in the remaining subclusters PM07-9 and -10, MCL1 upregulation may be driven by RELB via NF-κB activation (supplemental Figure 8B-D), as previously reported in patients with CLL.30 Of interest, this patient at the time of relapse also had decreased chromatin accessibility at the enhancer region of the BBC3 (PUMA) locus, together with an associated downregulation at the mRNA level in all the postvenetoclax subclusters (supplemental Figure 9A-B). BBC3 is known to bind BCLXL31 to promote apoptosis; therefore, its downregulation could have also contributed to venetoclax resistance in this patient.
We then examined the other 2 patients who retained their B-cell–like phenotype at the time of resistance. In PM09, the integration of scATAC- and scRNA-seq data identified 2 major clusters separating the prevenetoclax vs postvenetoclax cells (supplemental Figure 10A). These clusters were subdivided into 7 subclusters. Of note, subclusters PM09-1 to -3 were composed of prevenetoclax cells that were sensitive to the drug and fully contracted after treatment. The 4 subclusters PM09-4 to -7 were not present in the prevenetoclax sample and emerged as de novo subclusters at the time of resistance. The GSEA performed in each subcluster showed a retained B-cell–like phenotype in the postvenetoclax sample (supplemental Figure 10B), suggesting that mechanisms alternative to lineage plasticity may be the causes of resistance in this patient.
Similarly, in PM10, the cause of resistance was due to additional factors, as discussed for both patients subsequently.
Genomic alterations of BCL2 members are linked to venetoclax resistance in t(11;14) MM
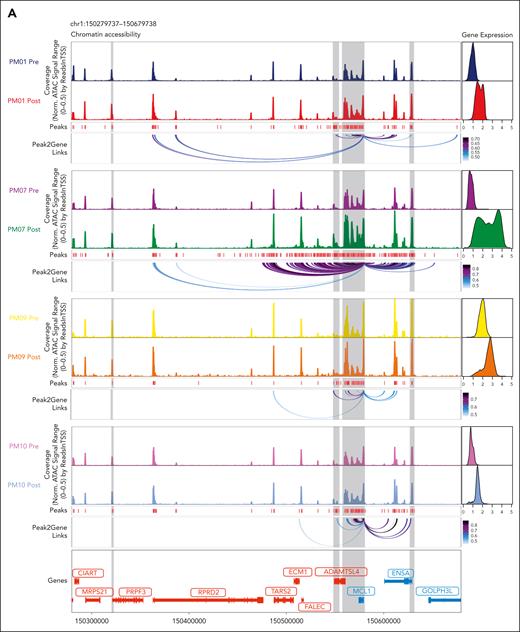
Of interest, when we examined all the patients with t(11;14), we observed increased accessibility at the promoter and enhancer regions of MCL1 in all patients (Figure 5A). These changes were associated with significant decreases in the BCL2/MCL1 ratio (Figure 5B), suggesting a role for MCL1 in the loss of BCL2 dependency. As such, the overexpression of MCL1 in the t(11;14)-sensitive MM cells KMS12BM reduces their sensitivity to venetoclax (Figure 5C-D) and plays a role in the acquired resistance to venetoclax. In addition, we found an increase in the MCL1/PMAIP1 ratio (Figure 5E) and a downregulation of PMAIP1 in most of the patients except PM10 (Figure 5F). Of interest, in this patient, our laboratory has previously discovered and functionally characterized a novel BCL2 point mutation (D111A) as the leading cause of venetoclax resistance.32 Regarding the other BCL2 family members, we found no difference in BCL2 chromatin accessibility or transcription between the prevenetoclax and postvenetoclax samples (supplemental Figure 11). However, a lower BCL2/BCL2L1 ratio was observed in all patients except PM07, who lost BBC3 expression, as previously indicated (Figure 5G; supplemental Figure 9A-B). Of interest, when we compared the chromatin accessibility at the BCL2L1 locus in the prevenetoclax and postvenetoclax samples, we found that in PM01, the significant upregulation of BCL2L1 was associated with an acquired cis-regulatory element 3′ to the BCL2L1 locus together with new peak-to-gene linkages to the BCL2L1 promoter region in the postvenetoclax sample, as highlighted in red (Figure 5H). Single-cell copy number variant analysis performed in this patient did not identify a gain at the BCL2L1 locus but rather a structural rearrangement between the IGLL5 enhancer on Chr 22 and the BCL2L1 gene locus, with the breakpoint mapping to the newly acquired cis-regulatory region on Chr 20.33 Lastly, in PM09 at the time of progression, we did not observe a decrease of BCL2 but rather an upregulation of BCL2L1 with peaks to gene linkage to BCL2L1 promoter regions (supplemental Figure 10C), together with a gain at the BCL2L1 locus on Chr 20 in all postvenetoclax cells,33 suggesting that BCL2L1 upregulation was the leading cause of venetoclax resistance in this patient.
Genomic alterations of BCL2 members are linked to venetoclax resistance in t(11;14) MM. (A) Genome accessibility track visualization of MCL1 with peak-to-gene links identified by ArchR in the prevenetoclax and postvenetoclax therapy samples for each patient (PM01, PM07, PM09, and PM10). The color of the arcs indicates the correlation strength between the peak and the gene expression, with a darker color indicating a stronger correlation. The genes are indicated in red when the gene is on the positive strand (TSS on the left) and in blue when on the negative strand (TSS on the right). The gray boxes highlighted the enhancer or promoter regions of the gene of interest. (B) Boxplots indicating the BCL2/MCL1 gene expression ratios in the prevenetoclax and postvenetoclax samples of each patient with MM treated with venetoclax (PM01, PM07, PM09, and PM10). The FDR values of the comparison prevenetoclax vs postvenetoclax sample of each patient are shown at the top of each boxplot comparison. (C) Immunoblotting of parental (WT) and MCL1-overexpressing KMS12BM cells for the indicated BCL2 family members in the presence or absence of venetoclax 200 nM for 3 hours. (D) Bar plot showing the percentage of cell death observed in parental (WT) vs MCL1-KMS12BM cells after 24 hours of exposure to venetoclax 200 nM. The data presented are the mean ± standard error of 3 independent experiments. The adjusted P values associated are shown at the top of each bar plot. (E-G) Boxplots indicating the MCL1/PMAIP1 (E), PMAIP1 (F), and BCL2/BCL2L1 (G) gene expression ratios in the prevenetoclax and postvenetoclax samples for each patient (PM01, PM07, PM09, and PM10). The FDR values associated with the comparison of prevenetoclax vs postvenetoclax cells are shown at the top of each boxplot comparison. (H) Genome accessibility track visualization of BCL2L1 with peak-to-gene links identified by ArchR in the prevenetoclax and postvenetoclax samples for patient PM01. The color of the arcs indicates the correlation strength between the peak and the gene expression with a darker color indicating a stronger correlation. The genes are indicated in red when the gene is on the positive strand (TSS on the left) and in blue when on the negative strand (TSS on the right). The gray boxes highlight the enhancer or promoter regions of the gene of interest.
Genomic alterations of BCL2 members are linked to venetoclax resistance in t(11;14) MM. (A) Genome accessibility track visualization of MCL1 with peak-to-gene links identified by ArchR in the prevenetoclax and postvenetoclax therapy samples for each patient (PM01, PM07, PM09, and PM10). The color of the arcs indicates the correlation strength between the peak and the gene expression, with a darker color indicating a stronger correlation. The genes are indicated in red when the gene is on the positive strand (TSS on the left) and in blue when on the negative strand (TSS on the right). The gray boxes highlighted the enhancer or promoter regions of the gene of interest. (B) Boxplots indicating the BCL2/MCL1 gene expression ratios in the prevenetoclax and postvenetoclax samples of each patient with MM treated with venetoclax (PM01, PM07, PM09, and PM10). The FDR values of the comparison prevenetoclax vs postvenetoclax sample of each patient are shown at the top of each boxplot comparison. (C) Immunoblotting of parental (WT) and MCL1-overexpressing KMS12BM cells for the indicated BCL2 family members in the presence or absence of venetoclax 200 nM for 3 hours. (D) Bar plot showing the percentage of cell death observed in parental (WT) vs MCL1-KMS12BM cells after 24 hours of exposure to venetoclax 200 nM. The data presented are the mean ± standard error of 3 independent experiments. The adjusted P values associated are shown at the top of each bar plot. (E-G) Boxplots indicating the MCL1/PMAIP1 (E), PMAIP1 (F), and BCL2/BCL2L1 (G) gene expression ratios in the prevenetoclax and postvenetoclax samples for each patient (PM01, PM07, PM09, and PM10). The FDR values associated with the comparison of prevenetoclax vs postvenetoclax cells are shown at the top of each boxplot comparison. (H) Genome accessibility track visualization of BCL2L1 with peak-to-gene links identified by ArchR in the prevenetoclax and postvenetoclax samples for patient PM01. The color of the arcs indicates the correlation strength between the peak and the gene expression with a darker color indicating a stronger correlation. The genes are indicated in red when the gene is on the positive strand (TSS on the left) and in blue when on the negative strand (TSS on the right). The gray boxes highlight the enhancer or promoter regions of the gene of interest.
Taken together, these data suggest that the development of venetoclax resistance in t(11;14) MM is highly complex, with multiple coexisting changes present at the genetic, epigenetic, and transcriptomic levels. These complex deregulations in 2 of 4 patients involved the loss of a B-cell–like program and the gain of canonical PC TFs. These patients also acquired a copy number gain of MCL1, and 1 had a BCL2L1 translocation. The other 2 patients, who retained their B-cell–like program despite acquiring venetoclax resistance, exhibited a copy number gain of BCL2L1 or a mutation in BCL2, suggesting that genomic alterations of BCL2 members are also linked to venetoclax resistance.
Discussion
The discovery of venetoclax has improved the treatment of hematologic malignancies, including MM.11,12,17 In several studies, venetoclax has been reported to be more effective in MM cells harboring the t(11;14).14 However, among these patients, the responses are not universal,14,34 suggesting numerous factors influence the response to venetoclax. As such, its activity may be enhanced by agents that can prime BCL2 dependency through the upregulation of NOXA and shifting of BIM loading to BCL2.20,35 Although it is evident that the unique sensitivity to venetoclax depends on the BH3 proapoptotic priming of BCL2, the biology underlying t(11;14) MM remains poorly defined.
This study provides a single-cell–level dynamic interrogation of the epigenetic and transcriptional regulators of venetoclax resistance in primary MM cells. Through longitudinal use of scATAC- and scRNA-seq, we have defined chromatin accessibility and gene expression associated with t(11;14) MM and revealed its relatedness to B-cell rather than PC biology. This B-cell–like signature was previously identified in bulk ATAC- and RNA-seq performed on venetoclax-sensitive MM cell lines predominantly harboring t(11;14).19 However, the epigenetic regulation of t(11;14) transcriptomes and its impact on apoptotic gene regulation and clinical response to venetoclax remain elusive.
In this study, we identified distinct chromatin landscapes associated with venetoclax resistance linked to specific transcriptional programs that could be targeted to restore sensitivity. By comparing prevenetoclax and postvenetoclax exposure samples of patients with t(11;14) MM, we identified a loss of the B-cell–like and gain of PC signature and canonical TFs (IRF4 and PRDM1) at the time of resistance. Of interest, we found higher expression and motif enrichment of RUNX1, a gene involved in the resistance to other anti-MM drugs36 and venetoclax efficacy in acute myeloid leukemia cells.37,38 As such, in additional functional studies, we have demonstrated that RUNX1 overexpression reduces the sensitivity of MM cells to venetoclax through the downregulation of the NF-kB pathway, also involved in programmed cell death. Further experiments are ongoing to better understand the complex interplay between RUNX1 and the BCL2 family proteins in t(11;14) MM.
Furthermore, upregulation of the proapoptotic PMAIP1 (NOXA) with a low MCL1/PMAIP1 ratio was observed in t(11;14) MM. This finding may explain t(11;14) MM sensitivity to venetoclax and its BCL2 dependency because NOXA promotes MCL1 degradation.21 Furthermore, venetoclax induces PMAIP1 transcription through the integrated stress response and ATF4/ATF3 expression.39,40 Therefore, the upregulation of PMAIP1 may be essential in the sensitivity to venetoclax, and its downregulation may have a role in resistance. Indeed, the downregulation of PMAIP1 was observed in nearly all resistant samples. Of note, the t(11;14) MM cell line U266 lacks PMAIP141 and is resistant to venetoclax.19,42 Therefore, targeting NOXA using inhibitors of the ubiquitin-proteasome system20,43,44 or CRISPR/Cas9-mediated ablation may induce BCL2 dependency and sensitivity to venetoclax. In addition, mutations affecting NOXA have been reported to affect MCL1 accumulation45-47 and have been identified in patients with CLL resistant to venetoclax.30
Moreover, at the time of relapse in all patients with t(11;14), we noted overexpression of BCL2L1 and MCL1 with increased mRNA expression and chromatin accessibility at BCL2L1 and/or MCL1 loci. These changes were due to structural copy number gains in half of the patients, consistent with a Darwinian selection of fitter clones under therapeutic pressures. Therefore, additional factors such as deregulated transcription with NF-κB–mediated translational upregulation of MCL1 or BCL2L1 may also mediate venetoclax resistance. As such, the overexpression of MCL1 in the t(11;14)-sensitive MM cells KMS12BM reduces their sensitivity to venetoclax by acting as a sink for the venetoclax-released BIM from BCL2. Therefore, dual targeting of BCL2 and MCL1 may offer a promising therapeutic approach and should be re-explored in these patients.
In summary, our single-cell, longitudinal interrogation of the epigenome, transcriptome, and genome of t(11;14) and non-t(11;14) MM identified a differential B-cell phylogeny of their PCs and revealed variable mechanisms mediating the BCL2 dependency and acquired resistance to venetoclax. MM cells are shown to adapt to the selective pressure exerted by venetoclax by transitioning from B-cell–like to mature PC-like epigenetic and transcriptomic signatures. This transition is paralleled by a loss of BCL2 dependency through PMAIP1 downregulation, RUNX1 upregulation, and an increase of MCL1 and/or BCL2L1 driven by structural changes or modified regulomes.
To our knowledge, this is the first study to date in which the integrated scATAC- and scRNA-seq analysis performed in primary t(11;14) MM cells has provided insights into the TF/regulome that could be targeted to restore B-cell–like signature and reinstate sensitivity to venetoclax.
Acknowledgments
The authors are grateful to the patients who enrolled in the study.
This work was supported by grant support (The M4 Study) from the Terry Fox Research Institute (principal investigator N.B. and joint principal investigator P.N.). B.G.B. was supported by an American Society of Hematology Scholar Award and a National Institutes of Health, National Cancer Institute Award.
Authorship
Contribution: N.L. conceptualized the study, designed and performed single-cell experiments, analyzed and interpreted the data, and wrote the paper; S.A. performed single-cell experiments, interpreted the data, and wrote the paper; R.T., M.P., R.M., H.L., and E.B. performed sample collection and interpreted the data; J.-B.A., S.S., and A.J. interpreted the data; B.G.B. and L.H.B. provided bulk ATAC-seq data from in vitro MB cells and PCs and interpreted the data; and N.B. and P.N. supervised the study and interpreted the data.
Conflict-of-interest disclosure: P.N. reports receiving speaker’s bureau honoraria from Bristol Myers Squibb (BMS), Janssen, and Sanofi and is a consultant/advisory board member for BMS and Janssen, none of which were related to this study. N.B. reports receiving speaker’s bureau honoraria from Amgen, BMS, Sanofi, Pfizer, and Janssen and is a consultant/advisory board member for BMS, Janssen, and Pfizer, none of which were related to this study. L.H.B. reports funding for research, consultancy, and honoraria from AstraZeneca and consultancy from AbbVie, none of which were related to this study. The remaining authors declare no competing financial interests.
Correspondence: Paola Neri, Division of Hematology and Bone Marrow Transplant, University of Calgary, 1403 29th St NW, Room C210, Calgary, AB T2N 2T9, Canada; email: epneri@ucalgary.ca.
References
Author notes
∗N.B. and P.N. contributed equally to this study.
Sequencing data are available via the Gene Expression Omnibus public repository (accession code GEO SuperSeries GSE199373: accession numbers GSE199268 for scATAC-seq, GSE199359 for scRNA-seq, and GSE199372 for single-cell copy number variant analysis).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Epigenetic and transcriptomic characteristics of t(11;14) MM. (A-C) Uniform manifold approximation and projection (UMAP) showing the distribution of CD138+ cells (n = 46 237 cells) collected from 15 patients with MM using the scATAC-seq and scRNA-seq data in the Latent Semantic Indexing method (LSI) space. Each point represents 1 cell. The cells are marked by a color code based on the different clusters (A), fluorescence in situ hybridization, cytogenetic (B), and patient identification (C). (D) Genome accessibility track visualization of CCND1 and MYEOV with peak-to-gene link identified by ArchR for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. The color of the arcs indicates the correlation strength between the peak and the gene expression (a darker color indicates a stronger correlation). The genes are indicated in red when the gene is on the positive strand (transcription start site [TSS] on the left) and blue when on the negative strand (TSS on the right). The gray boxes highlight the enhancer or promoter regions of the gene of interest. (E) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in each MM sample. The t(11;14) MM samples are highlighted in blue, whereas the non-t(11;14) MM samples are highlighted in red. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each patient. The dark blue indicates a lower gene expression count, and the light red indicates a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (F) Boxplots comparing the integrated expression of CCND1, BCL2, MCL1, BCL2L1, BBC3, and PMAIP1 in the t(11;14) vs non-t(11;14) MM groups. The false discovery rate (FDR) values associated with each gene are shown at the top of each boxplot comparison. (G) Genome accessibility track visualization of BCL2 with peak-to-gene links for each patient belonging to the t(11;14) (top) and non-t(11;14) (bottom) MM groups. For each group, the peaks panel highlights the different significant peaks identified by ArchR. (H-J) Boxplots indicating the BCL2/BCL2L1 (H), BCL2/MCL1 (I), and MCL1/PMAIP1 gene expression ratios (J) for each MM prevenetoclax sample (left). The boxes highlight the patients with t(11;14) MM. The comparisons between t(11;14) and non-t(11;14) MM groups are shown (right), with t(11;14) in blue and non-t(11:14) in red. The FDR values associated with each gene are shown at the top of each boxplot comparison.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/1/10.1182_blood.2023020276/4/m_blood_bld-2023-020276-gr1gj.jpeg?Expires=1769088106&Signature=stZz0hRRWTIwpPuWzVK7OTINfB~W8JN1MuQMbRBnlN20~6ZGoNRAyjJGUj4Xj0tRQWaZtUm~~jU39o1x2ee-LUO-UP1a3K95PFI15rh6~ohBYXa4yce7baqbaKTguEd4CTG8AF-2BAZbFRXNa6zjuTKw0MVP-FvXA48p3lKP-2U67vbJtTMd4sL05NemMGH78NMkJraesKwYXMd9Mf3xv8r2YoZZZYOa0JqSR16mjH4MOsW-tPdwT7P5li3WKDZ15HVkCb4STAYRnBWHaxdZJOvbj6owUrM5AKFptfsW2USB4ATmhdX2H7qjlJARzILC0eEm-RCfJF5sBo--5-OsBA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Loss of B-cell–like epigenetic signature with gain of canonical PC TFs is associated with acquired resistance to venetoclax in t(11;14) MM. (A-B) UMAP projection showing the distribution of CD138+ cells (n = 26 847 cells) collected from 4 patients with t(11;14) MM using the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked by a color code based on the different patient (A) and treatment (before vs after venetoclax) (B). (C) Odds ratios indicating the overlap of the DAR identified in the pre- (light blue) and post- (purple) venetoclax MM cells when compared with those present in the MB cells and PCs. The error bars denote the 95% confidence interval calculated using a two-sided Fisher exact test. (D) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (E-F) Violin plots of the NES obtained in the prevenetoclax vs postvenetoclax MM cells in each paired patient treated with venetoclax (PM01, PM07, PM09, and PM10). These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (E) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (F) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). (G) Heat map of the motif hypergeometric enrichment-adjusted P values within the Catalog of Inferred Sequence Binding Preferences databases of each prevenetoclax and postvenetoclax MM cell from patients PM01, PM07, PM09, and PM10. The color indicates the motif enrichment (−log10[P value]) based on the hypergeometric test. The maximum mlog10Padj is indicated in parenthesis for each significant enrichment found. (H) Representation of the Tn5 bias–adjusted TF footprints for IRF4, RUNX1, ID3, and TCF4 motifs identified by the enrichment motif analysis. The lines are colored in blue and red to indicate the prevenetoclax and postvenetoclax samples for patient PM01 and in purple and green for patient PM07.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/1/10.1182_blood.2023020276/4/m_blood_bld-2023-020276-gr3.jpeg?Expires=1769088106&Signature=zIP0sM97CEhD31oatzIN2T8mn3zx-JRzou01ZT6Rz3bbu6maKOJZ7YMG3eO-Lmf0fKhE6mu2EfMa~onhTjKevmQxnzWXiL395v4kzo92IFoJizTMx7IIu62calLlfWp0mX1GaNzgHtfN2-N3-vwaNsPTp~ujm3JK~lgPBXmCKOhjxLM-6WW4a6BxYZc0fxByxOhTVsDMj3bazp6T4bq~XdBlMRYjSy35ayNAdGRHUDFshqB9fJq5tt~CiTErucgk9g07JE-g3Cye7Beed340vhbavDJsZqWyAKnqoWwpRoDkW1sPomv5HdhEdeLTLs3hOzztdvBCLaD0UjZqZie~tw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Integrated epigenomic and transcriptomic analysis reveals multiple coexisting changes associated with venetoclax resistance in t(11;14) MM. (A) UMAP projection showing the distribution of CD138+ cells (n = 8679 cells) identified in the t(11;14) patient PM01 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (B) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PsC (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (C) On the left, immunoblotting of parental (wild-type [WT]) and KMS12BM-RUNX1 overexpressing cells for RUNX1 and BCL2, in the presence or absence of venetoclax 200 nM for 3 hours. The bar plot (right) indicates the effect of RUNX1 on NF-kB activation in the KMS12BM overexpressing RUNX1 when compared with parental cells (WT). (D) Bar plot showing the percentage of cell death observed in parental (WT) vs RUNX1-KMS12BM cells after 24 hours of exposure to venetoclax 200 and 500 nM. The data presented are the mean ± standard error of 3 independent experiments. The adjusted P values associated are shown at the top of each bar plot. (E) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) identified in the t(11;14) patient PM07 using the scATAC-seq and scRNA-seq data in the LSI space (left). Each point represents 1 cell. The cells are marked with color codes based on the different subclusters. The alluvial plot (right) shows the distribution of the cells among the prevenetoclax and postvenetoclax samples for each subcluster identified in the UMAP. (F) Violin plots of the NES obtained in each identified subcluster. These data were generated on the integrated scRNA-seq counts using the msigdb for BM PCs (left) (C8, HAY_BONE_MARROW_PLASMA_CELL) and B cells (right) (C8, HAY_BONE_MARROW_FOLLICULAR_B_CELL). Subclusters from the prevenetoclax sample are highlighted in blue, whereas those from the postvenetoclax sample are in purple. (G) Dot plot representing the scRNA-seq integrated expression of key genes involved in MM biology (CCND1, CCND2, CCND3, FGFR3, and NSD2) and BCL2 family members BCL2, BCL2L1 (BCLXL), BCL2L2 (BCLW), MCL1, BCL2A1, BCL2L11 (BIM), BID, BAX, BAK, BOK, BAD, BMF, BBC3 (PUMA), and PMAIP1 (NOXA) in PM07 subclusters. Subclusters from the prevenetoclax sample are highlighted in blue, whereas the ones from the postvenetoclax sample are in purple. The dot size represents the percentage of cells with values detected in each patient. The color represents the average gene expression in each subcluster. The dark blue indicates a lower gene expression count, and light red a higher count. Note that the average expression scales differ for CCND1 vs the other genes. (H) UMAP projection showing the distribution of CD138+ cells (n = 11 811 cells) collected from patient PM07 based on the scATAC-seq and scRNA-seq data in the LSI space. Each point represents 1 cell. The cells are marked with color codes based on the MCL1 amplification. The black color indicates the cells harboring the amplification of MCL1, whereas the gray or light gray indicates the cells without the amplification or with unknown information, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/1/10.1182_blood.2023020276/4/m_blood_bld-2023-020276-gr4.jpeg?Expires=1769088106&Signature=Owfrh0MRjmEPP2UR7OBfj7ahr6t2KXr9r0wQ12ICGpg3w1rgeTnifKY~qfU1ZIV797qwHM7yA2lrTJ4aCFhKE2PxFxTu7ujtLQL6HyLK3fDtCgQMQLmv3Us5GutsJV2dc2RzI9WF-26AtC2PXe-5VKw8TeUPWHZts3CLliLVa4Hl~HFvRdnjHdEx34hiF8L2va-6YBne-vB0BhWNNxxcUBhPdpHIW~Te5R-ebsqaRnu8HC-w956P-EscTnvAtQi9~Ck5bHKhvc0Ep4RVlW7kqSZmeUwSUzEkCkC1sPMGTEdsSepAsciobVXwy39i-mBn05xrXX~uMJwS2zKnPrygvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal