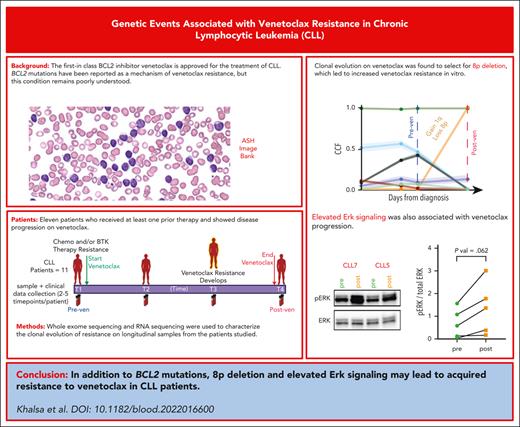
Key Points
Clonal evolution on venetoclax selects for 8p deletion, which leads to increased venetoclax resistance in vitro.
Elevated Erk signaling and constitutively high BCR signaling are associated with venetoclax progression.
Abstract
Although BCL2 mutations are reported as later occurring events leading to venetoclax resistance, many other mechanisms of progression have been reported though remain poorly understood. Here, we analyze longitudinal tumor samples from 11 patients with disease progression while receiving venetoclax to characterize the clonal evolution of resistance. All patients tested showed increased in vitro resistance to venetoclax at the posttreatment time point. We found the previously described acquired BCL2-G101V mutation in only 4 of 11 patients, with 2 patients showing a very low variant allele fraction (0.03%-4.68%). Whole-exome sequencing revealed acquired loss(8p) in 4 of 11 patients, of which 2 patients also had gain (1q21.2-21.3) in the same cells affecting the MCL1 gene. In vitro experiments showed that CLL cells from the 4 patients with loss(8p) were more resistant to venetoclax than cells from those without it, with the cells from 2 patients also carrying gain (1q21.2-21.3) showing increased sensitivity to MCL1 inhibition. Progression samples with gain (1q21.2-21.3) were more susceptible to the combination of MCL1 inhibitor and venetoclax. Differential gene expression analysis comparing bulk RNA sequencing data from pretreatment and progression time points of all patients showed upregulation of proliferation, B-cell receptor (BCR), and NF-κB gene sets including MAPK genes. Cells from progression time points demonstrated upregulation of surface immunoglobulin M and higher pERK levels compared with those from the preprogression time point, suggesting an upregulation of BCR signaling that activates the MAPK pathway. Overall, our data suggest several mechanisms of acquired resistance to venetoclax in CLL that could pave the way for rationally designed combination treatments for patients with venetoclax-resistant CLL.
Introduction
Chronic lymphocytic leukemia (CLL) remains largely incurable1 despite the development of targeted agents including BCL2 inhibitors. The first-in-class BCL2 inhibitor venetoclax2 is a highly selective US Food and Drug Administration–approved small molecule that acts as a BH3 mimetic inhibiting BCL2.3 In patients who are heavily pretreated, venetoclax given continuously showed a median progression-free survival of 28.2 months.4 However, some patients fail to respond, and others initially respond but relapse soon after.
Mechanisms of acquired resistance to venetoclax remain poorly understood. The primary identified mechanism of venetoclax resistance is mutation of the venetoclax–binding site of BCL2.5 However, these mutations were observed in patients with late disease progression, at a median of 32 months, and were only found in 7 of 15 (50%) patients, often at very low variant allele fractions (VAFs) (VAF <2% at progression in 6 of 7 patients tested positive for BCL2 p.G101V mutation). Most patients with this BCL2 mutation also have other mutations in BCL2.6,7 Other resistance mechanisms have also been proposed. A whole-exome sequencing (WES) study of 8 patients with disease progression on venetoclax with loss(17p), including several who developed Richter transformation, identified the loss of CDKN2A/B and mutations in the antiproliferative BTG1 gene as resistance-associated genetic aberrations, along with acquired nonrecurring mutations in known CLL driver genes.8,9 Other studies have identified the upregulation of MCL1 and NF-κB,10 increased BCL-XL in venetoclax-resistant lymphoma cell lines and samples from patients with CLL,11 and increased AMP-activated protein kinase signaling as mechanisms of resistance.12,13
However, detailed genomic clonal evolution analysis, which could help identify growing resistant clones, has not been performed on clinical samples collected through the course of venetoclax treatment. Hence, in this study, we characterized the clonal evolution of resistance via WES and RNA sequencing (RNA-seq) data analysis using longitudinal samples from patients with CLL who had developing progressive disease while receiving venetoclax. Furthermore, we performed in vitro validation of the key findings using CLL cells from patients with CLL collected at pre and postprogression time points on venetoclax.
Methods
Patient sample preparation
Matched peripheral blood (tumor) and saliva (healthy) samples were collected from patients who consented to our Dana-Farber Harvard Cancer Center institutional review board–approved CLL tissue banking protocol. All patients provided written informed consent before any samples were drawn.
Identification of somatic mutations
WES was performed at the Broad Institute as detailed in the supplemental Methods, available on the Blood website. High confidence somatic mutations were identified using previously published pipelines14-16 developed in the Cancer Genome Analysis group at the Broad Institute (https://software.broadinstitute.org/cancer/cga/). The somatic single nucleotide variants (sSNVs) were detected using MuTect1,15 and somatic short insertions and deletions were detected using Strelka and annotated MuTect2. The somatic mutation calls were filtered using a realignment filter, removing variants supported by ambiguously mapped reads.14 All the filtered mutation events were manually visualized and reviewed using the Integrative Genomics Viewer.17 Total copy number analysis was performed using Genome Analysis Toolkit and segmented into allelic copy number using AllelicCapSeg.18
Estimation of clonality using ABSOLUTE
The local copy number and the cancer cell fraction of each mutation (the fraction of cancer cells harboring the mutation) were estimated using ABSOLUTE,19 which calculates the sample’s purity (fraction of tumor cells) and ploidy (average DNA per cancer cell in units of haploid human genome) and local absolute copy number.
Additional methods can be found in supplemental Methods.
Results
WES and evaluation of BCL2 mutations in patient samples before and after progression with venetoclax
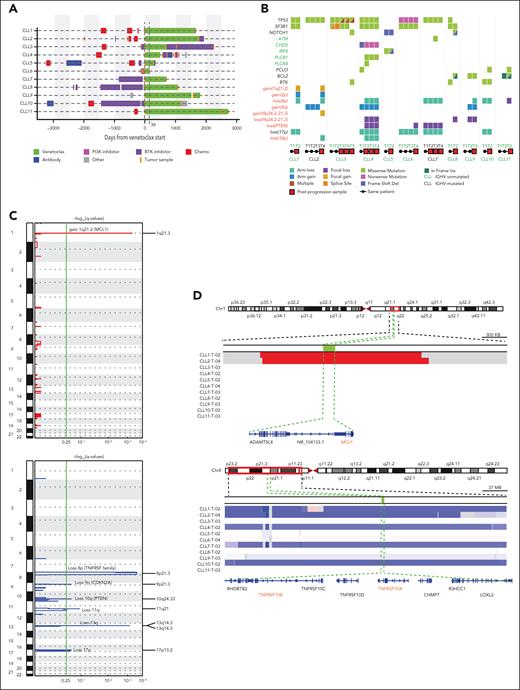
To determine whether recurrent genetic alterations result in venetoclax resistance, we evaluated matched serial CLL and germ line samples from 11 patients whose disease progressed while receiving venetoclax. Samples were taken before venetoclax and at progression from 10 patients with acquired resistance and 1 patient with primary refractory disease. A median of 3 longitudinal tumor samples (range, 2-5 samples) were sequenced per patient (Nsamples = 35). The patients had received a median of 2 prior therapies, including 8 patients with prior chemoimmunotherapy and 5 patients treated with prior Bruton tyrosine kinase (BTK) inhibitors (Figure 1A; supplemental Tables 1 and 2). Before initiation of venetoclax, del(17p) as well as complex karyotype (>3 aberrations) was observed in 4 of 11 (36%) patients (supplemental Table 3). Our cohort was also enriched in patients with CLL with TP53 (63%; 7 of 11) and SF3B1 mutations (45%; 5 of 11) (supplemental Table 3). The best response to venetoclax in all patients was partial remission per the criteria of the International Workshop on CLL, with a median time to progression of 35.76 months (range, 1.2-89.6 months) while receiving venetoclax (supplemental Table 2).
We assessed the presence of G101V mutation in the BCL2 gene.5 In our WES data, using MuTect,15 we did not identify any sSNVs in BCL2 or BCL2-family genes at baseline, as in previous reports.12,14 On further visualization of the sequencing data using the Integrative Genomics Viewer,17 2 patients (CLL8 and CLL9) had BCL2 G101V mutation at progression while receiving venetoclax. CLL3 and CLL4 also had 1 read each (1 of 63 and 1 of 71, respectively) for the mutation, in the progression samples (supplemental Figure 1A). Because of the extremely low allelic frequency of the mutation in CLL3 and CLL4, we performed a droplet digital polymerase chain reaction assay (ddPCR) using previously described probes,5 in peripheral blood mononuclear cell (PBMC) and/or bone marrow DNA from prevenetoclax and postprogression samples from our cohort. G101V mutation was detected via ddPCR in CLL8 and CLL9 at progression. In addition, G101V mutation was detected, at progression, in CLL1 and CLL4 at low VAFs of 0.03% and 4.68%, respectively (CLL1, 4 of 16064 droplets; CLL4, 640 of 13680 droplets; supplemental Table 4). We detected 1 droplet positive for G101V mutation in 2 other patients in bone marrow samples at venetoclax progression (CLL2 and CLL3). To confirm these very low frequency events, we examined postprogression PBMC RNA. We could not detect the mutations in RNA-seq data but were able to detect the G101V mutations via ddPCR using the complementary DNA from 2 patients (CLL1 and CLL4), for whom we detected mutations in DNA (supplemental Table 4). However, we could not confirm the mutations in patients CLL2 and CLL3 via ddPCR of the complementary DNA derived from the RNA. Similar to the findings from a recent report,6 we did not observe any G101V mutations in the BCL2 gene in patients who only had prior BTK inhibitor (ibrutinib) therapy (CLL6 and CLL7). Because of the absence or very low VAF of the BCL2 G101V mutations in 9 of 11 patients, other mechanisms of acquired resistance appear important.
Genetic alterations acquired while receiving venetoclax therapy
To evaluate for recurrent somatic alterations leading to venetoclax resistance, we performed WES and copy number analysis on samples from 11 patients (Nsamples = 35). MutSig analysis of WES data (q < .1) showed recurrent nonsynonymous clonal mutations in TP53 and BTK genes (Figure 1B). Three patients had PCLO (piccolo presynaptic cytomatrix protein) mutation, as previously reported,20 with CLL10 and CLL11 acquiring this mutation at progression while receiving venetoclax. (Figure 1B). Gain(8q) and loss(17p), known CLL driver events, were observed in 2 and 5 patients, respectively, before venetoclax treatment, and gain(8q) was acquired in 1 patient receiving venetoclax. Focal gain(9p24.2-21.3) affecting the CDKN2A/B gene was acquired in CLL2, whereas CLL4, CLL8, and CLL9 showed acquired loss(9p24.2-21.3).
Genetic profile of patients on venetoclax therapy. (A) Swimmer plot for 11 evaluated patients with CLL who received venetoclax, showing treatment received before and after initiation of venetoclax. Venetoclax treatment shown in green for CLL6 at 36 days is not scaled. For all other patients, days from venetoclax is per scale as defined on the x-axis. (B) Co-Mut plot representation of individual mutations and copy number aberrations present in 11 patients with CLL as derived from WES data. Half-filled squares in different colors represent distinct mutations. Mutations written in black are recurrent nonsynonymous mutations, whereas those in green are nonsynonymous mutations acquired in a single patient. (C) GISTIC plot of genomic regions with CN gain (red) or loss (blue) for 22 autosomes (y-axis) with −log(q-values) on the x-axis. (D) Visual validation of chromosomal segment loss and gains in integrative genomic viewer at the progression time point on venetoclax for all 11 patients. Blue indicates segment loss and red indicates segment gain.
Genetic profile of patients on venetoclax therapy. (A) Swimmer plot for 11 evaluated patients with CLL who received venetoclax, showing treatment received before and after initiation of venetoclax. Venetoclax treatment shown in green for CLL6 at 36 days is not scaled. For all other patients, days from venetoclax is per scale as defined on the x-axis. (B) Co-Mut plot representation of individual mutations and copy number aberrations present in 11 patients with CLL as derived from WES data. Half-filled squares in different colors represent distinct mutations. Mutations written in black are recurrent nonsynonymous mutations, whereas those in green are nonsynonymous mutations acquired in a single patient. (C) GISTIC plot of genomic regions with CN gain (red) or loss (blue) for 22 autosomes (y-axis) with −log(q-values) on the x-axis. (D) Visual validation of chromosomal segment loss and gains in integrative genomic viewer at the progression time point on venetoclax for all 11 patients. Blue indicates segment loss and red indicates segment gain.
GISTIC2.021 analysis of the copy number data from all 11 patients in this cohort detected some known CLL drivers, such as loss(17p), loss(11q), and loss(13q)14 along with loss(9p21.3)(CDKN2A/B) and loss(10q24.33)(PTEN) (Figure 1C). GISTIC also revealed 1q21.2 as significantly amplified and arm-level 8p deletion as well as focal 8p21.3 deletion as significantly deleted regions in venetoclax progression samples (Figure 1C-D; supplemental Figure 1B). Six patients showed loss(8p), of which 2 patients (CLL7 and CLL10) had this genetic event before venetoclax initiation, whereas 4 patients (CLL1, CLL2, CLL4, and CLL9), acquired it during venetoclax treatment. In addition, CLL11 appeared to acquire a focal 68-kb loss(8p) affecting the TNFRSF10A gene. Because of its subclonal nature and small size with limited probes in the region (Figure 1B-D), we could not independently confirm this event. Arm loss(8p), containing the TRAIL receptor genes, has been linked to ibrutinib resistance in CLL.20 Furthermore, acquired loss(8p) cooccurred with acquired gain(1q21.2-21.3) affecting the MCL1 gene in 2 (CLL1 and CLL2) of 4 patients. MCL1 overexpression as a mechanism of resistance to venetoclax12 has been reported recently.10,11
Patients with CLL who are resistant to venetoclax exhibit clonal evolution
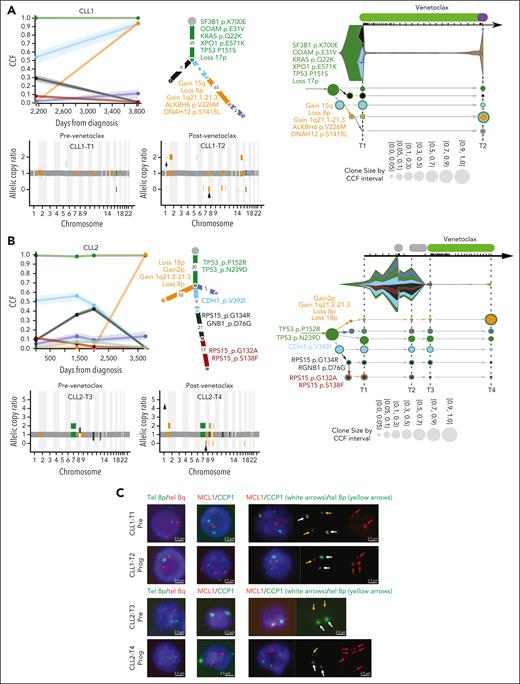
Next, we used PhylogicNDT22 and Concerti23 to study the subclonal architecture and expansion in 11 of our patients, using between 2 and 5 serial CLL samples per patient. We observed that CLL progression while receiving venetoclax was associated with marked clonal evolution in all patients (Figures 2 and 3; supplemental Figure 2). Clonal analysis of the prevenetoclax and progression time points in patients with long-term venetoclax treatment of nearly 4.5 years (CLL1 and CLL2) showed that treatment with venetoclax resulted in large clonal expansions of a clone containing arm-level loss(8p)- and gain(1q21.2)-containing MCL1 (Figure 2A-B). The prevenetoclax CLL population in CLL1 was predominantly composed of a clone harboring mutation in known CLL drivers SF3B1, TP53, KRAS, XPO1, and loss(17p), whereas in CLL2 the clonal population contained the known driver TP53 (Figure 2A-B). To validate whether loss(8p) and gain(1q21.2) were cooccurring genetic events in the same cells, we performed fluorescence in situ hybridization using telomeric 8p/8q probes and a locus-specific MCL1 hybridization probe in pre- and postprogression samples for CLL1 and CLL2 (Figure 2C; supplemental Table 5). Costaining with these probes confirms 8p loss with concomitant MCL1 gain in the same cells in accordance with the PhylogicNDT prediction.
Clonal evolution of somatic mutations in CLL patients with concomitant loss(8p) and gain (1q21.2). Subclonal structure and clonal evolution of somatic mutations derived using PhylogicNDT (left, middle) and Concerti (right) for CLL1 (A) and CLL2 (B). (Left) Each line represents the cancer cell fraction (CCF) of mutation clusters with shading representing a 95% confidence interval. Each dot represents a different time point. Phylogenetic trees (middle) represent the best estimated clonal and subclonal architecture corresponding to the same clones (represented in the same colors) as in the left panels. (Right) Time-scaled Concerti plots. Clones are sized proportionally to their prevalence and represented by the same color as in the PhylogicNDT plots. Copy number data are shown under the PhylogicNDT plots, as derived from ABSOLUTE in acquired patients with loss(8p) and gain(1q21.2) (CLL1 and CLL2). See Figure 3 and supplemental Figure 2 for the remaining patients. (C) Multiplexed fluorescence in situ hybridization performed on pre- and postprogression venetoclax samples for CLL1 and CLL2. The probe combination was tel8p (green) and tel8q (red) for the first column, MCL1 (red) and CCP1 (green) for the second column, and a combination of MCL1 (red) denoted by red arrows, CCP-1 (green) denoted by white arrows and tel8p (green) denoted by yellow arrows for the third column.
Clonal evolution of somatic mutations in CLL patients with concomitant loss(8p) and gain (1q21.2). Subclonal structure and clonal evolution of somatic mutations derived using PhylogicNDT (left, middle) and Concerti (right) for CLL1 (A) and CLL2 (B). (Left) Each line represents the cancer cell fraction (CCF) of mutation clusters with shading representing a 95% confidence interval. Each dot represents a different time point. Phylogenetic trees (middle) represent the best estimated clonal and subclonal architecture corresponding to the same clones (represented in the same colors) as in the left panels. (Right) Time-scaled Concerti plots. Clones are sized proportionally to their prevalence and represented by the same color as in the PhylogicNDT plots. Copy number data are shown under the PhylogicNDT plots, as derived from ABSOLUTE in acquired patients with loss(8p) and gain(1q21.2) (CLL1 and CLL2). See Figure 3 and supplemental Figure 2 for the remaining patients. (C) Multiplexed fluorescence in situ hybridization performed on pre- and postprogression venetoclax samples for CLL1 and CLL2. The probe combination was tel8p (green) and tel8q (red) for the first column, MCL1 (red) and CCP1 (green) for the second column, and a combination of MCL1 (red) denoted by red arrows, CCP-1 (green) denoted by white arrows and tel8p (green) denoted by yellow arrows for the third column.
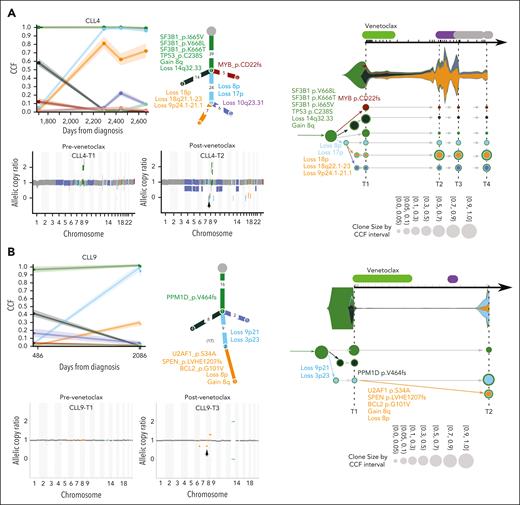
CLL4 and CLL9 are the remaining 2 of 4 patients with acquired arm-level deletion of 8p, however, in the absence of MCL1 amplification at progression (Figure 3A-B). CLL4, like CLL1, was treated with chemoimmunotherapy before venetoclax and had clonal driver mutations in SF3B1 and TP53. However, in CLL4, with relatively early relapse while receiving venetoclax (∼1.4 years), loss(8p) cooccurred with gain(8q), loss(17p), loss(18q22.1-23), loss(10q23.31), and loss(9p23-21.2) (containing the MYC, TP53, BCL2, PTEN, and CDKN2A/B genes, respectively) (Figure 3A). Venetoclax treatment in CLL9 resulted in the expansion of CLL cells with loss(9p) and loss(3p), whereas loss(8p) was acquired in a subclone also harboring BCL2 G101V mutation (Figure 3B).
Clonal evolution of somatic mutations in CLL patients with loss(8p). Subclonal structure and clonal evolution of somatic mutations derived using PhylogicNDT (left, middle) and Concerti (right) with copy number data derived from ABSOLUTE (below PhylogicNDT plots) for CLL4 (A) and CLL9 (B). For additional details, see Figure 2.
Clonal evolution of somatic mutations in CLL patients with loss(8p). Subclonal structure and clonal evolution of somatic mutations derived using PhylogicNDT (left, middle) and Concerti (right) with copy number data derived from ABSOLUTE (below PhylogicNDT plots) for CLL4 (A) and CLL9 (B). For additional details, see Figure 2.
Copy number data confirmed acquired loss(8p) in 4 patients (CLL1, CLL2, CLL4, and CLL9) in accordance with GISTIC2.0 analysis (Figure 2A-B; supplemental Figure 2A). Intriguingly, the clonal dynamics in all 4 patients with loss(8p) showed a branching evolution in which the clone harboring the loss(8p) and gain(1q21.2) in CLL1 and CLL2, and loss(8p) in CLL4 and CLL9, was selected for and became the dominant clone at progression. These clones all arose from a background clone with TP53 loss, in accordance with our previous report on loss(8p) after administration of venetoclax.12
CLL3 exhibited complex branching evolution in which several TP53 mutations were acquired in clones that expanded during venetoclax treatment as well as other subclones containing TP53 and SF3B1 mutations that initially expanded but then relatively decreased during the evolution of venetoclax resistance (supplemental Figure 2A). CLL5 exhibited linear evolution with acquired IRF8 mutations (S283C and 416fs) emerging as the dominant subclones at the time of relapse (supplemental Figure 2B). In 2 other patients (CLL6 and CLL7), who were chemo-naive with prior progression while receiving ibrutinib, the marked clonal expansion of a clone containing loss(10q23.31-24.1) that includes PTEN was observed in CLL6, whereas the PTEN clone showed a decline in CLL7 at the progression time point (supplemental Figure 2C-D).
CLL8, previously treated with ibrutinib and acalabrutinib, had TP53 and SF3B1 as truncal mutations with BTK p.C481S mutation in a major subclone that regressed upon venetoclax therapy (supplemental Figure 2E). On progression, a new subclone emerged with a new SF3B1 mutation (R775G) and NOTCH1 mutation (P2514fs). This subclone had a daughter subclone with 2 different BCL2 mutations: RRYR insertion mutation at BCL2_p109 (previously reported7,24) and BCL2_p.G101V, another NOTCH1 and BRAF mutation.
CLL7 and CLL10 showed a clonal loss(8p), possibly acquired earlier with ibrutinib resistance. CLL10 also had the ibrutinib resistance mutation at BTK p.C481S (supplemental Figure 2D,F). The emerging dominant subclone at the time of progression on venetoclax in CLL10 harbors PCLO mutation as previously reported in patients with ibrutinib-relapsed CLL.25 CLL11 had the longest time to progression on treatment (2724 days) with only 1 prior therapy (bendamustine-rituximab). CLL11, along with CLL8, had a BCL2_p109 mutation in their growing clusters at the progression time point (supplemental Figure 2E,G). Altogether, these results suggest that acquired resistance to venetoclax may result from several mechanisms including loss(8p) (36%, 4 of 11), gain(1q) (18%, 2 of 11), loss(9p21) (28%, 3 of 11), and loss(PTEN) (28%, 3 of 11). Patients in our cohort also acquired the known BCL2 mutations BCL2 p.G101V (18%, 2 of 11) and BCL2_p109 (18%, 2 of 11).
In vitro response to venetoclax is worse in a cohort of patients with loss(8p)/gain(1q)
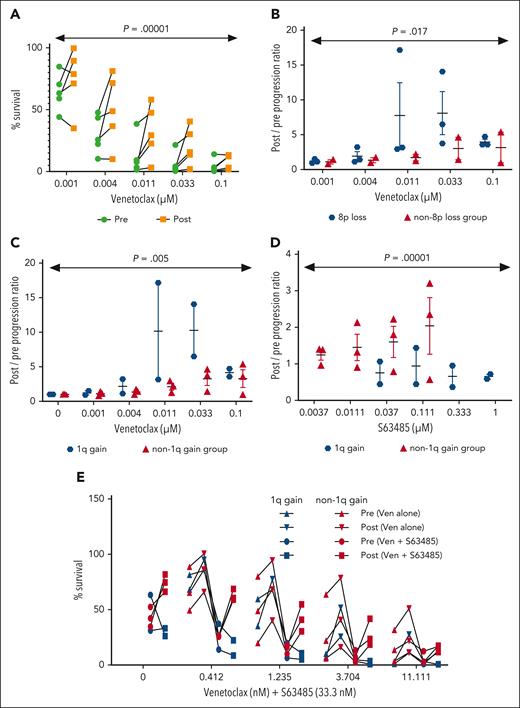
We evaluated the cell viability in prevenetoclax and progression PBMC samples treated ex vivo for 24 hours with venetoclax. We found that in the 5 patients tested (CLL1, CLL2, CLL4, CLL5, and CLL7), progression samples were significantly more resistant to venetoclax compared with corresponding prevenetoclax time points across multiple concentrations of venetoclax (n = 5; P < .001) (Figure 4A). Moreover, we found that the progression samples of patients with loss(8p) (CLL1, CLL2, and CLL4) were more viable after venetoclax treatment compared with those from patients without loss(8p) (CLL5 and CLL7) (Figure 4B), which suggests that patients with loss(8p) have more resistant disease (P < .05).
In vitro reponse to venetoclax in patient samples collected pre- and postprogression on venetoclax. Pre- and postprogression time point PBMC samples (refer to the classification column of supplemental Table 1) were treated with escalating concentrations of venetoclax (A-C) for 24 hours. (A) Representation of survival as analyzed using CellTiter-Glo and the line connects matched pre- and postprogression samples for each patient. Data from panel A was split as patients with and without loss(8p) (B) and with and without gain(1q) (C). The x-axis represents the ratio of surviving cells in postprogression/pretreatment samples. (D) Pre- and postprogression samples were treated with the MCL1 inhibitor S63485 for 24 hours and surviving cells were estimated with CellTiter-Glo. Patients with and without gain(1q) were compared. The x-axis represents the ratio of surviving pre/postprogression samples. (E) Pre- and postprogression samples were treated with 33.3 nM S63485 for 24 hours in combination with escalating concentrations of venetoclax, and surviving cells were determined by CellTiter-Glo. The line connects matched pre- and postprogression samples for each patient. Patients with and without gain(1q) were compared. The x-axis represents cell survival upon treatment with various drugs.
In vitro reponse to venetoclax in patient samples collected pre- and postprogression on venetoclax. Pre- and postprogression time point PBMC samples (refer to the classification column of supplemental Table 1) were treated with escalating concentrations of venetoclax (A-C) for 24 hours. (A) Representation of survival as analyzed using CellTiter-Glo and the line connects matched pre- and postprogression samples for each patient. Data from panel A was split as patients with and without loss(8p) (B) and with and without gain(1q) (C). The x-axis represents the ratio of surviving cells in postprogression/pretreatment samples. (D) Pre- and postprogression samples were treated with the MCL1 inhibitor S63485 for 24 hours and surviving cells were estimated with CellTiter-Glo. Patients with and without gain(1q) were compared. The x-axis represents the ratio of surviving pre/postprogression samples. (E) Pre- and postprogression samples were treated with 33.3 nM S63485 for 24 hours in combination with escalating concentrations of venetoclax, and surviving cells were determined by CellTiter-Glo. The line connects matched pre- and postprogression samples for each patient. Patients with and without gain(1q) were compared. The x-axis represents cell survival upon treatment with various drugs.
The region loss(8p) includes TRAIL receptors (TNFRSF10A/TNFRSF10B) (Figure 1D) that mediate the TRAIL apoptosis pathway and have previously been implicated in ibrutinib resistance.20 To examine whether loss(8p) results in higher resistance to TRAIL-mediated apoptosis, we compared TRAIL susceptibility in 8p-intact preprogression samples and 8p-loss postprogression samples (supplemental Figure 3). Cells were activated with CD40L for 24 hours followed by exposure to titrating concentrations of TRAIL for 24 hours. CLL8, a patient without 8p-loss, did not show any difference in cell viability between preprogression and postprogression samples. In contrast, cells from patient CLL9, who acquired 8p-loss during progression, became more resistant to TRAIL-mediated death. Interestingly, cells from patient CLL11, which may harbor a focal 8p-loss affecting TNFRSF10A, also showed a similar increased resistance to TRAIL-mediated killing at the progression time point. These data are suggestive of the direct impact of loss(8p) and increased resistance to apoptosis that might explain progression of the disease in these patients.
Gain(1q) resulting in MCL1 upregulation was previously linked with venetoclax resistance in CLL cell lines and patient cells10-12 as well as in acute myeloid leukemia (AML) cells.26 We performed RNA-seq using pre and postprogression samples for all patients and found that the MCL1 gene was upregulated at progression in 2 patients (CLL2 and CLL7), including CLL2 with gain(1q), and marginally in CLL1 (the other patient with gain[1q]), CLL8, and CLL9 (supplemental Figure 4A). We looked at the gene expression of other antiapoptotic genes BCL2, BCL-XL/BCL2L1, BCL-W/BCL2L2, and BFL1A and found that all patients had at least 1 antiapoptotic gene that was upregulated upon progression, suggesting a compensatory mechanism in response to BCL2 inhibition by venetoclax (supplemental Figure 4A). We confirmed this finding via a western blot for BCL-XL and BFL1A in which postprogression samples had higher levels of these antiapoptotic proteins (supplemental Figure 4B).
Next, we examined the association of gain(1q) with viability. We demonstrated that, after treatment for 24 hours, the gain(1q) group (CLL1 and CLL2) was more resistant to venetoclax compared with those without gain(1q) (CLL4, CLL5, and CLL7) (P < .05; Figure 4C). Because the gain(1q) region includes the MCL1 gene, we tested the viability of PBMC samples from pre- and postprogression time points after treatment with the MCL1 inhibitor, S63845, for 24 hours. Progression samples of the gain(1q) cohort showed enhanced susceptibility to the MCL1 inhibitor compared with their prevenetoclax patient-matched time points (P < .05; Figure 4D). Furthermore, we tested the combination of venetoclax and MCL1 inhibitor for 24 hours and found that progression samples of the gain(1q) cohort were more susceptible to the combination (Figure 4E). These results suggest that the combination therapy may be an effective treatment for some patients with gain(1q) venetoclax resistance, and, if used earlier, may have the potential to prevent the emergence of gain(1q), thus delaying resistance in some patients.
Transcriptomic changes associated with venetoclax progression in patients with loss(8p)
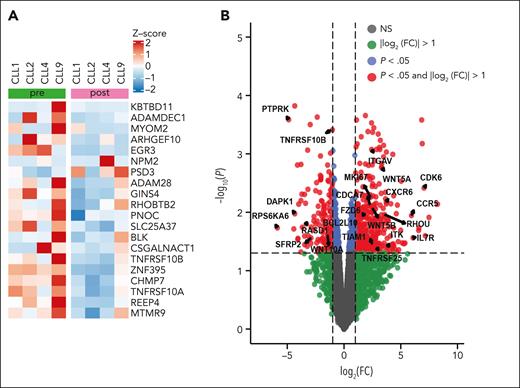
To understand the expression changes associated with venetoclax resistance in patients with loss(8p) with or without gain(1q), we looked at RNA-seq data for the genes included in the 8p deletion in 4 patients (CLL1, CLL2, CLL4, and CLL9). Postvenetoclax resistant samples from patients with loss(8p) showed an overall downregulation of genes encoded by the 8p region as compared with prevenetoclax samples (supplemental Figure 5A). The top 20 genes on 8p that showed differential gene expression (DGE) are represented in Figure 5A. RNA-seq analysis comparing prevenetoclax with postvenetoclax resistant samples from patients with loss(8p) showed a significant downregulation of the TNFRSF10A/10B genes (TRAIL-Rs; Limma-Voom, P < .1; Figure 5A-B). Gene network analysis using differentially expressed genes (P < 0.01; |logFC| > 1 [FC, fold change]) in STRING, yielded multiple significantly altered gene ontology terms related to death pathways in which TNFRSF10A/10B genes are central (supplemental Figure 5). These include the regulation of the extrinsic apoptotic signaling pathway via death domain receptors (strength = 1.07; false discovery rate [FDR] = 0.021), the apoptotic pathway (strength = 0.55; FDR = 0.024), and the apoptotic process pathway (strength = 0.41; FDR = 0.047; marked in green) (supplemental Table 6).
Transcriptomic changes associated with venetoclax progression in patients with loss(8p). (A) Normalized counts from RNA-seq data for the top 25 genes transcribed from chromosome 8p, based on mean difference in pre- and postvenetoclax progression in samples of the loss(8p) cohort (CLL1, CLL2, CLL4, and CLL9). (B) Volcano plot showing differentially expressed genes from RNA-seq data comparing samples from patients with or without loss(8p). The log2FC is plotted on the x-axis, and the negative-log10 adjusted P value (using FDR) is plotted on the y-axis. Genes marked in red show differentially expressed genes with an absolute value of log2 (FC) no less than 1 (FC = 2) and adjusted P value < .05.
Transcriptomic changes associated with venetoclax progression in patients with loss(8p). (A) Normalized counts from RNA-seq data for the top 25 genes transcribed from chromosome 8p, based on mean difference in pre- and postvenetoclax progression in samples of the loss(8p) cohort (CLL1, CLL2, CLL4, and CLL9). (B) Volcano plot showing differentially expressed genes from RNA-seq data comparing samples from patients with or without loss(8p). The log2FC is plotted on the x-axis, and the negative-log10 adjusted P value (using FDR) is plotted on the y-axis. Genes marked in red show differentially expressed genes with an absolute value of log2 (FC) no less than 1 (FC = 2) and adjusted P value < .05.
DGE analysis showed the upregulation of CDK6 that has been identified as a potential prognostic biomarker in AML27 (Figure 5B). WNT5A and FZD6, central to the noncanonical Wnt pathway, were upregulated in the loss(8p) group (Figure 5B). High levels of WNT5A have been reported in the plasma of patients with CLL, and a recent report showed that WNT5A enhances proliferation of CLL28 by binding to ROR1, which is overexpressed in CLL cells. Moreover, increased ROR1 and ROR1 signaling was observed in patients with CLL on venetoclax therapy.29 These results suggest that the poor outcome of patients with loss(8p) might be mediated via the downregulation of TRAIL-R signaling and upregulation of WNT5A signaling pathways.
Patients with disease progression on venetoclax show elevated ERK1/2 signaling
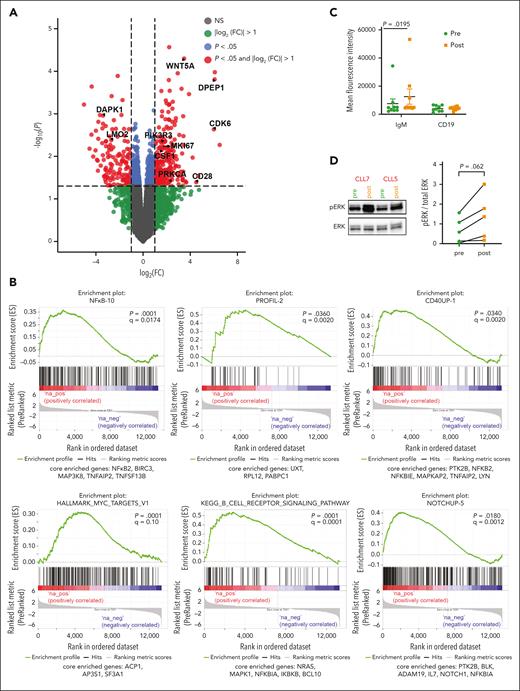
Next, we performed RNA-seq analysis of the entire cohort to evaluate transcriptomic adaptation of the cells during development of acquired resistance to venetoclax. To determine the consequence of venetoclax inhibition in vivo, we profiled 11 patients treated with venetoclax (both pre- and postvenetoclax time points) and performed DGE analysis (Figure 6A). Gene set enrichment analysis using the Staudt Lab gene set database (https://lymphochip.nih.gov/signaturedb/) (with FDR < 0.1) and MSigDB30,31 curated hallmark gene sets showed positive enrichment of several proliferation-related gene sets, suggesting a higher proliferative capacity of cells on acquiring resistance to venetoclax (supplemental Tables 7 and 8; Figure 6B). WNT5A was transcriptionally upregulated in progression samples and WNT5A is central to proliferation and upstream of extracellular signal-regulated kinase (ERK) phosphorylation (Figure 6A). Gene set enrichment analysis showed the positive enrichment of MYC targets and tumor necrosis factor α signaling via NF-κB response gene set that are both downstream of ERK signaling (supplemental Table 8). In melanoma, the ERK pathway induces c-MYC stabilization via the attenuation of its ubiquitin-mediated protein degradation mechanism.32 MYC upregulation also leads to higher proliferation. Genes related to B-cell receptor (BCR) activation and STAT-3 upregulation (supplemental Table 7), both of which lead to B-cell signaling upregulation, were also overrepresented. To test this, we compared surface immunoglobulin M (sIgM) levels in pre- and postprogression PBMCs by flow cytometry. We found consistently higher sIgM levels at progression compared with that in preprogression samples (n = 9; P = .02; Figure 6C), suggestive of constitutively higher BCR signaling. CD19 expression did not differ, suggesting that the increase in IgM surface receptor is specific (Figure 6C). The PBMC samples from pre- and postprogression time points were then profiled for downstream BCR signaling proteins. We observed higher pPLCG2 and pBTK expression in progression samples as observed through western blot, although not statistically significant because of the small sample size (supplemental Figure 6). We also observed that ERK phosphorylation (Thr 202/Tyr 204) showed a trend toward elevation in the venetoclax progression time points when compared with corresponding prevenetoclax samples for all patients tested (n = 5; P = .06; Figure 6D). ERK may be downstream of WNT5A or the BCR. In summary, these data suggest that upregulation of the BCR signaling pathway and extracellular signal-regulated kinase (ERK) phosphorylation contribute to venetoclax resistance.
Patients with disease progression on venetoclax show elevated ERK1/2 signaling. (A) Volcano plot showing differentially expressed genes between pre and postvenetoclax progression time points from RNA-seq data with adjusted P value < .05 and |log2(FC)| > 1.0. (B) Enrichment plots obtained from gene set enrichment analysis, which was performed on a list of all genes from DEG analysis and ranked based on t statistics. (C) sIgM and CD19 staining was performed on PBMCs collected from patients (n = 5) at pre- and postprogression on venetoclax time points. Mean fluorescent intensity was measured on gated CD19+ cells. (D) Representative western blot image showing pERK and ERK for PBMC lysates from pre- and postvenetoclax progression time points for 2 patients, together with quantification of pERK:ERK ratio (n = 5).
Patients with disease progression on venetoclax show elevated ERK1/2 signaling. (A) Volcano plot showing differentially expressed genes between pre and postvenetoclax progression time points from RNA-seq data with adjusted P value < .05 and |log2(FC)| > 1.0. (B) Enrichment plots obtained from gene set enrichment analysis, which was performed on a list of all genes from DEG analysis and ranked based on t statistics. (C) sIgM and CD19 staining was performed on PBMCs collected from patients (n = 5) at pre- and postprogression on venetoclax time points. Mean fluorescent intensity was measured on gated CD19+ cells. (D) Representative western blot image showing pERK and ERK for PBMC lysates from pre- and postvenetoclax progression time points for 2 patients, together with quantification of pERK:ERK ratio (n = 5).
Discussion
In this study, we identified multiple potential mechanisms of resistance in patients with CLL who progressed while receiving venetoclax. The WES of 11 such patient samples revealed no recurrent sSNV involved in the resistance to venetoclax, other than BCL2, as previously reported. However, we identified several somatic copy-number alterations showing robust clonal shifts and evolution in patients whose disease progressed while receiving venetoclax. These include the acquired loss(8p) (36%; 4 of 11) and gain(1q) (18%; 2 of 11) in particular but also acquired loss(9p21.3) affecting CDKN2A (27%, 3 of 11) and acquired loss(PTEN) (18%; 2 of 11). In patients with relapsed/refractory disease who were previously exposed to chemoimmunotherapy, loss(9p21.3) was particularly enriched, as described previously.9,33 Furthermore, we identified constitutive pERK activation across many of our samples from patients with venetoclax resistance.
Previously identified venetoclax resistance mutations in BCL2 G101V were found in 3 of 11 patients (CLL8, CLL9, and CLL11) in our cohort. We did not see these mutations in 2 patients who had received prior treatment with ibrutinib (CLL6 and CLL7), raising the possibility that the clonal diversity at start of venetoclax might be reduced in patients who have not received prior chemoimmunotherapy, although confounded by short times on venetoclax for both of these patients. The extraordinarily low VAFs of 2 of the detected mutations suggest that other mechanisms are contributing to resistance.
By evaluating PBMC viability before and after ex vivo venetoclax treatment, we found that in all the patients tested, progression samples showed significantly higher survival after venetoclax treatment compared with cells from corresponding prevenetoclax time points, consistent with acquired intrinsic resistance. Moreover, in comparing the viability of venetoclax progression samples from the loss(8p) and without loss(8p) groups, we found that progression samples from patients with loss(8p) better survived treatment with venetoclax compared with samples from patients without loss(8p) (Figure 3B), suggesting that patients with loss(8p) are more resistant and therefore likely to have a worse prognosis, as previously observed in breast cancer cells.34-36 Loss of TRAIL-Rs has also previously been linked with ibrutinib resistance in CLL.20 A recent study suggested that induction of cell death through TRAIL is a BAX-independent mechanism for overcoming venetoclax resistance; this observation is potentially consistent with our observation that deletion of TRAIL-Rs in CLL cells is an alternative mechanism of venetoclax resistance.11
Similarly, at progression, cells from patients with gain(1q) showed higher viability when compared with those from patients without gain(1q), and treatment with a MCL1 inhibitor alone or in combination with venetoclax significantly decreased viability in progression samples only in patients with gain(1q), suggesting this combination therapy as an option for treating this group of patients. These findings support and expand prior work that demonstrated 1q gain in patients who are resistant to venetoclax.12 Furthermore, our results provide rationale for initial combination therapy with venetoclax and a MCL1 inhibitor to prevent the development of resistance via this mechanism. The aforementioned combination resensitized the cells to venetoclax despite upregulation of other antiapoptotic proteins (supplemental Figure 3; Figure 3E). It is interesting that 1q gain in our cohort occurred exclusively in the same cells with 8p deletion; this mechanism of potential synergy remains to be elucidated.
Western blot analysis of pre- and postprogression samples showed elevated pERK levels at the time of progression in all the patient samples tested (CLL1, CLL2, CLL4, CLL5, and CLL7), demonstrating a potentially significant role for MAPK signaling in acquired resistance to venetoclax in CLL. Using venetoclax-resistant clones from multiple myeloma cell lines, Chakraborty et al showed that acquired resistance to venetoclax is mainly due to the activation of pERK.37 They also showed elevated expression of MCL1 and BCL-XL in the resistant clones.37 Furthermore, in venetoclax-resistant AML cell lines, Zhang et al demonstrated that resistance was primarily mediated by upregulation of MAPK signaling, resulting in increased MCL1 and BCL-XL expression.38 In follicular lymphoma cells, pERK and pBIM (S69) levels were higher in cells treated with venetoclax.39
We also observed elevated sIgM levels in patients with progressive disease, indicating that BCR signaling could be partially contributing to elevated pERK levels. Anti-IgM stimulation reportedly protects CLL cells from venetoclax-induced apoptosis through MCL1 upregulation,40 in accordance with our observation.
In summary, our findings identify potential targets to assess therapeutic opportunities for the treatment of patients with CLL who have acquired resistance to venetoclax. Our study shows that a subset of BCL2 mutations occur at very low VAFs, suggesting the existence of other mechanisms of resistance. We identified upregulation of sIgM and MAPK signaling as a mechanism of acquired resistance to venetoclax, suggesting that treatment with MEK or ERK inhibitors of patients with venetoclax-resistant disease could be beneficial. Moreover, we also observed that cells from patients with certain genetic abnormalities (ie, loss[8p] and gain[1q]) are more resistant to venetoclax than those from patients without these genetic abnormalities, possibly because of the loss of TRAIL receptors, upregulation of WNT5A, and MYC signaling. Treatment with MCL1 inhibitor either alone or with venetoclax decreased viability in progression samples in patients with gain(1q), suggesting that combining a MCL1 inhibitor with venetoclax might prevent the outgrowth of clones with this genetic abnormality. In addition, combination therapy may prevent the emergence of gain(1q), potentially delaying onset of resistance in some patients. Our results point to several future directions for combination therapy in clinical trials, with the goal of overcoming or avoiding venetoclax resistance.
Acknowledgments
The authors thank all the patients and their families who participated in the venetoclax clinical trials and were willing to donate samples for this study. The authors also thank Delphine Voisin for technical assistance and Francesca Morelli and Roberta Santos Azevedo for providing clinical data analysis support.
This research was funded by the National Institutes of Health, National Cancer Institute grant R01 CA 213442 (principal investigator, J.R.B.) and by Broad/IBM Cancer Resistance Research Project (principal investigators, G.G. and L.P.).
Authorship
Contribution: J.R.B., G.G., L.P., C.P.P., I.M., and J.K.K. designed the research; J.K.K., I.M., J.C., F.U., A.N., L.V., R.G., B.K.S., I.L., Y.K., K.V., K.R., C.L., and M.S.D. performed research and collected data; J.R.B., G.G., and L.P. contributed vital new reagents or analytical tools; I.M., J.K.K., J.C., F.U., L.L., Z.W., I.L., Y.K., K.V., K.R., C.L., and S.T. analyzed, interpreted, and performed statistical analysis; J.K.K., I.M., J.C., F.U., I.L., A.N., Y.K., K.V., S.T., S.M.F., J.-H.M., H.B., M.A., K.R., C.L., B.P.D., K.S., R.A.J., C.P.P., L.P., G.G., and J.R.B. wrote the manuscript; S.M.F., J.-H.M., H.B., M.A., B.P.D., K.S., and R.A.J. provided administrative support (ie, biobanking, managing and organizing samples, and sequencing support); and J.R.B. supervised the study.
Conflict-of-interest disclosure: J.R.B. has served as a consultant for AbbVie, Acerta/AstraZeneca, BeiGene, Genentech/Roche, Grifols Worldwide Operations, Hutchmed, iOnctura, Janssen, Kite, Loxo/Lilly, MEI Pharma, Numab Therapeutics, Pfizer, and Pharmacyclics; and received research funding from BeiGene, Gilead, iOnctura, Loxo/Lilly, MEI Pharma, and TG Therapeutics. G.G. receives research funds from IBM and Pharmacyclics; is an inventor on patent applications related to MSMuTect, MSMutSig, POLYSOLVER, TensorQTL, and MinimuMM-seq; and is a founder, consultant, and holds privately held equity in Scorpion Therapeutics. I.L. serves as a consultant for PACT Pharma Inc; has stock, is on the board, and serves as a consultant, for ennov1 LLC.; and is on the board of and holds equity in Nord Bio Inc. M.S.D. declares consultancy for AbbVie, Adaptive Biotechnologies, Ascentage Pharma, AstraZeneca, BeiGene, Bristol Myers Squibb, Eli Lilly, Genentech, Genmab, Janssen, Merck, MingSight Pharmaceuticals, Ono Pharmaceuticals, Secura Bio, Takeda, and TG Therapeutics; declares having received research funding from AbbVie, Ascentage Pharma, AstraZeneca, Genentech, Novartis, Secura Bio, and TG Therapeutics; has received honoraria from Aptitude Health, AXIS Medical Education, BioAscend, Curio Science, Medscape Education, PeerView Institute for Medical Education, Physician’s Education Resource, PlatformQ Health Education, and Plexus Communications; and declares royalties for Research to Practice Other Potential Financial Relationship. The remaining authors declare no competing financial interests.
Correspondence: Jennifer R. Brown, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: jennifer_brown@dfci.harvard.edu.
References
Author notes
∗J.K.K., J.C., F.U., A.N., and I.M. contributed equally to this study.
†L.P., G.G., and J.R.B. contributed equally to this study.
Presented in abstract form at the AACR Annual Meeting (virtual, 2021), and the International Workshop on CLL meeting (virtual, 2021).
Data are available on request from the corresponding author, Jennifer R. Brown (jennifer_brown@dfci.harvard.edu).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal