Key Points
Adult and pediatric BL share a common pathobiology with Epstein-Barr virus, influencing the genetic and molecular profiles in both entities.
BL can be robustly divided into 3 genetic subgroups with distinct molecular underpinnings.
Abstract
Burkitt lymphoma (BL) accounts for most pediatric non-Hodgkin lymphomas, being less common but significantly more lethal when diagnosed in adults. Much of the knowledge of the genetics of BL thus far has originated from the study of pediatric BL (pBL), leaving its relationship to adult BL (aBL) and other adult lymphomas not fully explored. We sought to more thoroughly identify the somatic changes that underlie lymphomagenesis in aBL and any molecular features that associate with clinical disparities within and between pBL and aBL. Through comprehensive whole-genome sequencing of 230 BL and 295 diffuse large B-cell lymphoma (DLBCL) tumors, we identified additional significantly mutated genes, including more genetic features that associate with tumor Epstein-Barr virus status, and unraveled new distinct subgroupings within BL and DLBCL with 3 predominantly comprising BLs: DGG-BL (DDX3X, GNA13, and GNAI2), IC-BL (ID3 and CCND3), and Q53-BL (quiet TP53). Each BL subgroup is characterized by combinations of common driver and noncoding mutations caused by aberrant somatic hypermutation. The largest subgroups of BL cases, IC-BL and DGG-BL, are further characterized by distinct biological and gene expression differences. IC-BL and DGG-BL and their prototypical genetic features (ID3 and TP53) had significant associations with patient outcomes that were different among aBL and pBL cohorts. These findings highlight shared pathogenesis between aBL and pBL, and establish genetic subtypes within BL that serve to delineate tumors with distinct molecular features, providing a new framework for epidemiologic, diagnostic, and therapeutic strategies.
Introduction
Burkitt lymphoma (BL) is routinely subdivided by clinical variant status, into so-called endemic and sporadic variants,1,2 with further separation based on patient age (adult BL [aBL] and pediatric BL [pBL]), 2 divisions rooted in epidemiology rather than biology. The utility of clinical variant status was challenged by the robust differences in the frequency of driver mutations when pBLs are stratified on the basis of tumor Epstein-Barr virus (EBV) positivity rather than clinical variant status.1,3 Stratification on EBV status also showed a stronger association between aberrant somatic hypermutation (aSHM) with EBV-positive pBLs,3-5 and it is becoming accepted that EBV status is a more biologically relevant subdivision.6,7 Much of our current knowledge of the molecular etiology of BL results from the study of pBL,3,5,8 leaving its relationship to aBL and other adult B-cell Non-Hodgkin Lymphomas (B-NHLs), such as diffuse large B-cell lymphoma (DLBCL), unclear. Herein, we sought to identify any shared and distinguishing molecular and genetic features of DLBCL and the pediatric and adult forms of BL.
Genetically heterogeneous cancers can be subdivided on patterns of shared molecular or genetic features,9 theoretically resolving groupings of tumors with similar oncogenic drivers and vulnerabilities. This is exemplified by recent efforts to delineate robust genetic subgroups within DLBCL,10-12 opening the possibility for a new generation of clinical trials with treatment informed by genetics.13,14 Considering the growing appreciation of mutational heterogeneity within BL,15 the classification of patients with BL using genetic and molecular features may uncover more granular entities and could ensure accurate differential diagnosis of BL from DLBCL. Previous attempts to delineate genetic subgroups in BL have been limited by small cohort sizes, the narrow focus on individual mutation types, and sequencing panels that incompletely capture relevant significantly mutated genes (SMGs).5,16,17
To comprehensively delineate the genetic features of both aBL and pBL, we generated and assembled whole-genome sequencing (WGS) and/or transcriptome sequencing data from 281 BLs from 4 continents, including previously published pBLs and a newly sequenced cohort of 100 aBL tumors, 22 BL cell lines, and 8 tumors (“non-BL”) reclassified during pathology review. Comparing these with the genomes of 295 DLBCL tumors allowed us to identify novel genetic subgroups with characteristic genetic and molecular differences. Our analysis focused on 230 BLs with WGS data, in which we comprehensively identified simple somatic mutations (SSMs), copy number variations (CNVs), structural variations (SVs), and aSHM. This allowed for the identification of novel BL-associated mutations, genetic subgroups, and associations between genetic features and clinical outcomes in patients with both aBL and pBL.
Materials and methods
Case accrual and sequencing
Adult and pediatric cases accrued from Uganda, the United States, Brazil, France, Germany, and Canada; and samples underwent pathology consensus review. We subjected tumor and, when available, matched constitutional DNA from 181 pBL and 100 aBL cases and 22 BL cell lines to WGS and/or RNA sequencing (Table 1; supplemental Table 1, available on the Blood website). We further analyzed WGS data from 17 pBLs previously investigated by the International Cancer Genome Consortium Molecular Mechanisms in Malignant Lymphoma by Sequencing project,18-20 280 DLBCLs, including 25 HIV+ and 16 HIV– newly sequenced tumors (supplemental Table 2), 8 non-BLs, and 15 DLBCL cell lines. See supplemental Methods for details.
Summary of selected clinical and molecular characteristics of the samples in BL cohort
| Variable . | Level . | Adult (n = l00) . | Pediatric (n = 181) . | Total (n = 281) . |
|---|---|---|---|---|
| EBV status | EBV-negative | 67 (67) | 66 (36) | 133 (47) |
| EBV-positive | 33 (33) | 115 (64) | 148 (53) | |
| Sex | Female | 29 (29) | 55 (30) | 84 (30) |
| Male | 70 (70) | 123 (68) | 193 (68) | |
| Unknown | 1 (1) | 3 (2) | 4 (1) | |
| Clinical variant | Endemic | 1 (1) | 118 (65) | 119 (42) |
| Sporadic | 99 (99) | 63 (35) | 162 (58) | |
| BLIPI | 0 | 11 (11) | 34 (19) | 45 (16) |
| 1 | 25 (25) | 24 (13) | 49 (17) | |
| 2 | 19 (19) | 17 (9) | 36 (13) | |
| 3 | 6 (6) | 2 (1) | 8 (3) | |
| 4 | 3 (3) | 0 (0) | 3 (1) | |
| Unknown | 36 (36) | 104 (57) | 140 (50) | |
| HIV status | HIV-negative | 59 (59) | 146 (81) | 205 (73) |
| HIV-positive | 24 (24) | 6 (3) | 30 (11) | |
| Unknown | 17 (17) | 29 (16) | 46 (16) | |
| First-line regimen | CODOX/IVAC±R | 34 (34) | 3 (2) | 37 (13) |
| COM | 0 (0) | 39 (22) | 39 (14) | |
| COP | 0 (0) | 16 (9) | 16 (6) | |
| DA-EPOCH±R | 6 (6) | 9 (5) | 15 (5) | |
| No treatment | 0 (0) | 5 (3) | 5 (2) | |
| Other | 8 (8) | 6 (3) | 14 (5) | |
| Unknown | 52 (52) | 103 (57) | 155 (55) | |
| PFS | Median, y | 2.87 | 0.85 | 1.64 |
| No. (%) | 88 (88) | 165 (91) | 253 (90) | |
| OS | Median, y | 2.89 | 1.02 | 1.82 |
| No. (%) | 88 (88) | 165 (91) | 253 (90) |
| Variable . | Level . | Adult (n = l00) . | Pediatric (n = 181) . | Total (n = 281) . |
|---|---|---|---|---|
| EBV status | EBV-negative | 67 (67) | 66 (36) | 133 (47) |
| EBV-positive | 33 (33) | 115 (64) | 148 (53) | |
| Sex | Female | 29 (29) | 55 (30) | 84 (30) |
| Male | 70 (70) | 123 (68) | 193 (68) | |
| Unknown | 1 (1) | 3 (2) | 4 (1) | |
| Clinical variant | Endemic | 1 (1) | 118 (65) | 119 (42) |
| Sporadic | 99 (99) | 63 (35) | 162 (58) | |
| BLIPI | 0 | 11 (11) | 34 (19) | 45 (16) |
| 1 | 25 (25) | 24 (13) | 49 (17) | |
| 2 | 19 (19) | 17 (9) | 36 (13) | |
| 3 | 6 (6) | 2 (1) | 8 (3) | |
| 4 | 3 (3) | 0 (0) | 3 (1) | |
| Unknown | 36 (36) | 104 (57) | 140 (50) | |
| HIV status | HIV-negative | 59 (59) | 146 (81) | 205 (73) |
| HIV-positive | 24 (24) | 6 (3) | 30 (11) | |
| Unknown | 17 (17) | 29 (16) | 46 (16) | |
| First-line regimen | CODOX/IVAC±R | 34 (34) | 3 (2) | 37 (13) |
| COM | 0 (0) | 39 (22) | 39 (14) | |
| COP | 0 (0) | 16 (9) | 16 (6) | |
| DA-EPOCH±R | 6 (6) | 9 (5) | 15 (5) | |
| No treatment | 0 (0) | 5 (3) | 5 (2) | |
| Other | 8 (8) | 6 (3) | 14 (5) | |
| Unknown | 52 (52) | 103 (57) | 155 (55) | |
| PFS | Median, y | 2.87 | 0.85 | 1.64 |
| No. (%) | 88 (88) | 165 (91) | 253 (90) | |
| OS | Median, y | 2.89 | 1.02 | 1.82 |
| No. (%) | 88 (88) | 165 (91) | 253 (90) |
Data are given as number (percentage) of each group, unless otherwise indicated.
BLIPI, Burkitt Lymphoma International Prognostic Index; CODOX, cyclophosphamide, vincristine, doxorubicin; COM, cyclophosphamide, vincristine, methotrexate; COP, cyclophosphamide, vincristine, prednisone; DA-EPOCH, dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, and doxorubicin; IVAC, ifosfamide, etoposide, and cytarabine; OS, overall survival; PFS, progression-free survival; R, Rituximab.
Data analysis
WGS and RNA-sequencing reads were aligned to GRCh38 for BL and GRCh37 for DLBCL. Tumor EBV status was inferred from the fraction of EBV reads in the WGS data and/or the number of EBV reads aligned to Epstein-Barr virus encoded RNA 1/2 (EBER1/EBER2) in the RNA-sequencing data. We performed somatic variant calling using the Sage, LoFreq, Mutect2, and Strelka2 (SLMS-3) pipeline for SSMs, Manta and Genome Rearrangement IDentification Software Suite (GRIDSS) for SVs, and Battenberg and Control-free copy number and allelic content caller (Control-FREEC) for CNVs. Genes were defined as significantly mutated if they were identified by at least 2 of the following methods: dNdScv, MutSig2CV, HOTMAPS, and OncodriveFML. Negative matrix factorization clustering was performed in R, following the standard recommendations. Gene expression was quantified using Salmon, differential gene expression using DESeq2, and gene set enrichment using gene set variation analysis (GSVA). We conducted survival analyses to assess progression-free survival and overall survival of patients using the Kaplan-Meier method. Subgroup validation and classification were conducted using a random forest model. See supplemental Methods for details.
Results
Structural variations in aBL and pBL
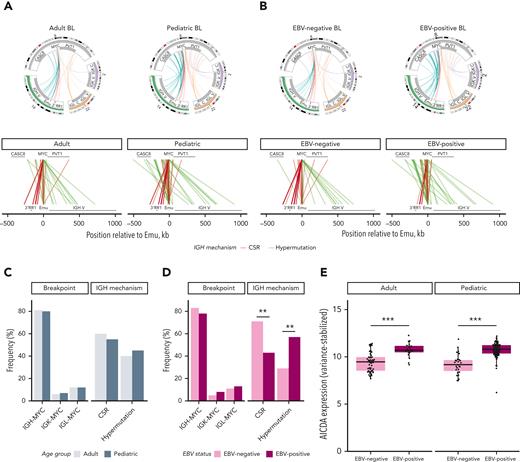
The genetic hallmark of BL is a translocation that places MYC under the regulation of a strong enhancer, typically the immunoglobulin (IG) heavy or light chain, resulting in MYC overexpression.21,22 We detected MYC translocations in 214 (93%) samples (supplemental Table 3) with an immunoglobulin heavy chain (IGH)-MYC translocation in 170 (79%) of these and 16 (7%) involving immunoglobulin light chain kappa (IGK) or 26 (12%) with IGL. Two samples harbored a BCL6-MYC translocation, and 16 (7%) cases had no MYC SV identifiable by whole-genome sequencing. Of these seemingly MYC translocation-negative genomes, 11 were positive for a MYC translocation by fluorescence in situ hybridization, 1 case had evidence of aSHM at MYC, and 2 cases had gain at or 2 megabases upstream of MYC, providing molecular evidence for the MYC aberration. Investigating the breakdown of IG-MYC translocations by EBV status and age showed no significant differences in IG-partner frequencies (Figure 1A-D).
Structural variations involving MYC in BL. Translocations between the MYC locus (chromosome 8) and the IGH (chromosome 14), IGK (chromosome 2), or IGL (chromosome 22) loci in tumors with IG-MYC breakpoints detected from WGS (N = 212). The subset of IGH-MYC breakpoints with high-confidence breakpoint positions identified are colored on the basis of their category, as determined by location within IGH (red = CSR, or blue = SHM). Bar charts on the lower left display the frequency of IG-MYC breakpoints (left) and IGH breakpoint category (right). The lower part of A and B linearly depicts IGH-MYC rearrangements colored by breakpoint category. (A) Adult (N = 80) and pediatric (N = 132) samples are shown separately. (B) EBV-negative (N = 103) and EBV-positive (N = 109) samples are shown separately. The inferred IGH breakpoint category frequencies stratified by age (C) and EBV status (D) were subjected to a Fisher exact test (∗∗P < .01). (E) AICDA expression in adult and pediatric BL tumors separated by EBV status (Wilcoxon rank sum test; ∗∗∗P < .001). AICDA, activation-induced cytidine deaminase; CSR, class-switch recombination; IGH, immunoglobulin heavy chain; IGK, immunoglobulin light chain kappa; IGL, immunoglobulin light chain lambda; SHM, somatic hypermutation.
Structural variations involving MYC in BL. Translocations between the MYC locus (chromosome 8) and the IGH (chromosome 14), IGK (chromosome 2), or IGL (chromosome 22) loci in tumors with IG-MYC breakpoints detected from WGS (N = 212). The subset of IGH-MYC breakpoints with high-confidence breakpoint positions identified are colored on the basis of their category, as determined by location within IGH (red = CSR, or blue = SHM). Bar charts on the lower left display the frequency of IG-MYC breakpoints (left) and IGH breakpoint category (right). The lower part of A and B linearly depicts IGH-MYC rearrangements colored by breakpoint category. (A) Adult (N = 80) and pediatric (N = 132) samples are shown separately. (B) EBV-negative (N = 103) and EBV-positive (N = 109) samples are shown separately. The inferred IGH breakpoint category frequencies stratified by age (C) and EBV status (D) were subjected to a Fisher exact test (∗∗P < .01). (E) AICDA expression in adult and pediatric BL tumors separated by EBV status (Wilcoxon rank sum test; ∗∗∗P < .001). AICDA, activation-induced cytidine deaminase; CSR, class-switch recombination; IGH, immunoglobulin heavy chain; IGK, immunoglobulin light chain kappa; IGL, immunoglobulin light chain lambda; SHM, somatic hypermutation.
We further annotated IGH breakpoints into 2 regions on chromosome 8: upstream of MYC and within MYC (most commonly intron 1). Comparing the breakpoint frequencies between these regions, we observed a significant difference based on EBV status (P < .001; Fisher test) with more IGH-MYC breakpoints located upstream of MYC in EBV-positive BLs and more breakpoints within MYC among EBV-negative BLs (P < .001; Fisher test). We separately annotated each breakpoint based on its location within IGH (supplemental Figure 1A-B) and categorized them as class-switch recombination (CSR; breakpoints within switch sequences), or hypermutation mediated (somatic hypermutation [SHM]–mediated breakpoints). Pairwise comparison of the frequency of breakpoints in these categories revealed that EBV-negative BLs had significantly more breakpoints attributed to CSR (P < .01; Fisher test), whereas putative SHM-mediated breakpoints were predominant among EBV-positive BLs (P < .01; Fisher test) (Figure 1D). We noted several unexpected MYC translocations within the IGH variable region and, thus, inspected breakpoints within the V and D gene segments (supplemental Figure 1A). Many of these breakpoints were internal to V genes, which is not consistent with their origin arising during recombination-activating genes (RAG)-mediated variable, diversity, and joining (VDJ) recombination. Instead, these may result from activation-induced cytidine deaminase (AID)-induced double-stranded breaks on a recombined allele. This notion is further supported by most SHM-mediated breakpoints falling within identified SHM peaks, previously reported SHM regions in IGH, AID motifs, or near large deletions bringing the breakpoints in proximity to Emu23 (supplemental Figure 1C). Consistent with the abundance of SHM-mediated breakpoints in EBV-positive cases, we found significantly higher AICDA expression among these samples in aBL and pBL (Figure 1E). No significant difference in the inferred breakpoint mechanism emerged when patients were stratified on age (Figure 1C; supplemental Figure 1B).
Distinguishing and shared genetic features between BL and DLBCL
To gain insight into the pattern of CNVs in BL (supplemental Table 4), we obtained estimates of tumor purity, ploidy, and genome-wide somatic copy number profiles from BL and DLBCL genomes (supplemental Figures 2-4). We identified a total of 94 significant “peaks” of recurrent copy number alterations (Figure 2A; supplemental Table 5), mostly representing regions previously described.
Significantly mutated genes in BL. (A) Cumulative representation of recurrent copy number aberrations across BL and DLBCL identified by Genomic Identification of Significant Targets in Cancer, version 2.0 (GISTIC2.0) (default Q value threshold). (B) EBV-positive (N = 118) and EBV-negative (N = 112) BL tumors are shown separately, and each set of genes associated with a specific pathway is separately ordered to highlight mutual exclusivity. Mutations are colored on the basis of their predicted consequence, and the frequency of each variant type is tallied in the bar plots on the right. Focal gains and deletions were defined as those <1 Mbp. Mutation prevalence in EBV-positive (N = 118) and EBV-negative (N = 112) cases was subject to a Fisher exact test with Bonferroni correction and is shown in the bar plots on the left. ∗Q < 0.1, ∗∗Q < 0.05, and ∗∗∗Q < 0.01.
Significantly mutated genes in BL. (A) Cumulative representation of recurrent copy number aberrations across BL and DLBCL identified by Genomic Identification of Significant Targets in Cancer, version 2.0 (GISTIC2.0) (default Q value threshold). (B) EBV-positive (N = 118) and EBV-negative (N = 112) BL tumors are shown separately, and each set of genes associated with a specific pathway is separately ordered to highlight mutual exclusivity. Mutations are colored on the basis of their predicted consequence, and the frequency of each variant type is tallied in the bar plots on the right. Focal gains and deletions were defined as those <1 Mbp. Mutation prevalence in EBV-positive (N = 118) and EBV-negative (N = 112) cases was subject to a Fisher exact test with Bonferroni correction and is shown in the bar plots on the left. ∗Q < 0.1, ∗∗Q < 0.05, and ∗∗∗Q < 0.01.
To identify SMGs relevant to BL while allowing for detection of genes shared with DLBCL, we analyzed SSMs (supplemental Table 6) from all BL genomes (N = 252) in conjunction with 252 DLBCLs. We detected 57 SMGs mutated in at least 2% (N = 4) of patients with BL, including 18 genes (31%) also recurrently mutated in DLBCL (supplemental Table 7). These SMGs largely represented previously identified BL-associated genes, further supporting the role of SIN3A, USP7, HIST1H1E, CHD8, and RFX7 in BL3 (Figure 2B). Not surprisingly, most of the newly identified SMGs were mutated infrequently (<5% of tumors). Mutations in some of these genes occur at similar, or higher, rates among DLBCLs (supplemental Figures 5A and 6; supplemental Table 7), including TET2, HNRNPU, BRAF, SYNCRIP, and EZH2. This could suggest a greater shared biology with DLBCL than previously appreciated. However, notably, most of these genes are mutated at drastically different frequencies between the 2 diseases. Some of the novel SMGs have patterns that imply their functional role (supplemental Figures 7-9). For example, most HNRNPU mutations are predicted to truncate the protein (supplemental Figures 7A, 8A, and 9A; supplemental Table 6), and are enriched within EBV-positive BLs (Figure 2B; supplemental Figure 9A). Although BLs exhibited the highest expression of HNRNPU across the B-cell lymphomas evaluated, we noted consistently lower abundance of HNRNPU mRNA in mutated tumors (supplemental Figure 5B-C).
Analyzing the mutational signatures in BL tumors, we identified single base substitution (SBS)1, SBS5, and SBS9 being the most predominant in BL (supplemental Figure 10; supplemental Table 8). Consistent with increased AICDA activity in EBV-positive tumors (Figure 1E), the exposure to SBS9 was also significantly increased in EBV-positive BL (supplemental Figure 10B). Comparing mutational signatures by age, we noted aBLs were enriched for SBS1 and SBS5, but with decreased SBS9 exposure relative to pBLs (supplemental Figure 10C).
We compared mutation frequencies individually to identify genes with different mutation rates when stratified on tumor EBV status (supplemental Table 9). Each of FOXO1, MIR17HG, PTEN, SMARCA4, GNAI2, CCND3, TP53, CDKN2A, SYNCRIP, FBXO11, STAT6, BCR, and PHF6 were found differentially mutated. With the exception of FOXO1 and BCR, these genes are mutated at a higher frequency in EBV-negative BLs (Figure 2B). Each of FOXO1, PCBP1, and HNRNPU appeared to exhibit distinct mutational patterns depending on EBV status (supplemental Figure 9; supplemental Table 9). When patients were stratified by age, only 2 genes had significantly different mutation rates (supplemental Table 10). ARID1A was mutated more in pBL (46% vs 26%), and TET2 had more mutations in aBL (10% vs 1.5%) (supplemental Figure 6). This further implies that EBV, rather than patient age, underlies more molecular differences within BL. To compare the oncogenic pathways mutated in BL, we assigned each SMG to a pathway and, as observed previously, stratification on EBV status identified genes related to apoptosis as more commonly mutated in EBV-negative BLs (supplemental Table 9).3
Identification and characterization of BL genetic subgroups
To identify natural subgroupings within BL and DLBCL, we applied clustering to a set of recurrent CNVs and SMGs and regions commonly affected by aSHM in either DLBCL or BL (supplemental Table 11). We first analyzed clustering of BL genomes alone, which resolved 4 clusters: 3 defined by mutations in TP53, DDX3X, and ID3 but otherwise not entirely enriched for features significantly more observed in BL (supplemental Figure 11). The fourth was enriched for features significantly more often occurring in DLBCL (supplemental Figure 11). Consensus clustering using all BLs, DLBCLs, and non-BLs (Figure 3A; supplemental Figure 12; supplemental Table 12) revealed 6 robust genetic subgroups with 3 largely representing DLBCLs (DLBCL-A, DLBCL-B, and DLBCL-C; supplemental Figure 13A), because of the heterogeneity of drivers and aSHM patterns in DLBCL. The DLBCL-predominant subgroups partially resembled those described by Wright et al12 (supplemental Figure 12), with DLBCL-A enriched for EZB (including EZH2 mutations and BCL2 translocations) DLBCLs and DLBCL-C enriched for ST2 (SGK1 and TET2 mutated).
Identification of distinct genomic subgroups in BL and DLBCL. (A) Clustering solution from non–negative matrix factorization (NMF) utilizing a combination of simple somatic mutations, CNVs, SVs, SHM patterns, and hotspot mutations as features. The proportional abundance of each feature in the clusters is shown on the left side. Each individual feature is labeled on the right side of the heat map. IGH/K/L-MYC breakpoints were considered as separate features (labeled as “MYC_IG”), and when MYC was not translocated to IGH, IGK, or IGL, it was considered separately and labeled “MYC_not_IG.” The right annotation track depicts whether the feature significantly enriched in BL, DLBCL, or neither of these entities. Alluvial plots showing distribution of different entities by EBV status (B), age (C), or sex (D) between identified clusters. For all panels except A, only patient samples with available sex information are shown. IGH, immunoglobulin heavy chain; IGK, immunoglobulin light chain kappa; IGL, immunoglobulin light chain lambda.
Identification of distinct genomic subgroups in BL and DLBCL. (A) Clustering solution from non–negative matrix factorization (NMF) utilizing a combination of simple somatic mutations, CNVs, SVs, SHM patterns, and hotspot mutations as features. The proportional abundance of each feature in the clusters is shown on the left side. Each individual feature is labeled on the right side of the heat map. IGH/K/L-MYC breakpoints were considered as separate features (labeled as “MYC_IG”), and when MYC was not translocated to IGH, IGK, or IGL, it was considered separately and labeled “MYC_not_IG.” The right annotation track depicts whether the feature significantly enriched in BL, DLBCL, or neither of these entities. Alluvial plots showing distribution of different entities by EBV status (B), age (C), or sex (D) between identified clusters. For all panels except A, only patient samples with available sex information are shown. IGH, immunoglobulin heavy chain; IGK, immunoglobulin light chain kappa; IGL, immunoglobulin light chain lambda.
The BL-enriched clusters were assigned names based on their characteristic features (supplemental Figure 12B-D): DGG-BL (DDX3X, GNA13, and GNAI2), IC-BL (ID3 and CCND3), and Q53-BL (quiet TP53). DGG-BLs included 100 tumors (85 BLs) (supplemental Figure 13A), and this cluster was enriched for hotspot mutations in FOXO1 (supplemental Figure 12B-C) and characterized by the highest frequency of HNRNPU mutations (11%). In contrast to the other clusters, both DGG-BL and DLBCL-C tumors commonly had evidence of aSHM near the transcription start site of BACH2.
IC-BL was mainly composed of BLs (N = 103/112; supplemental Figure 13A) and had the highest prevalence of mutations in ID3 and CCND3 and a paucity of mutations in genes associated with DGG-BL (Figure 3A; supplemental Figure 12B). Finally, Q53-BL was genetically lacking in driver mutations or CNVs. Other than MYC translocations, which are uniformly present in BL, Q53-BL was enriched for TP53 mutations (Figure 3A; supplemental Figure 12D). BLs represented 20 of 27 tumors in this subgroup (74%; supplemental Figure 13A). Interestingly, we noted that Q53-BLs do not resemble the TP53-deficient DLBCLs, which are characterized by aneuploidy.12 The nonsynonymous MYC mutations were the least prevalent in this subgroup (11%; supplemental Figure 12B-C). More aBLs were clustered in Q53-BL, whereas pBLs were common in IC-BL and DGG-BL (Figure 3C; supplemental Figure 13C,E), although this difference was not statistically significant. Analyzing the mutation burden in BL tumors across genetic subgroups revealed lower mutation burden in IC-BL compared with DGG-BL (P = .004; Wilcoxon test) and DLBCL-C (P < .001; Wilcoxon test). No other significant differences were found in the overall mutation burden between genetic subgroups of BL (supplemental Figure 13F).
We noted a strong association between EBV status with DGG-BL, which was predominantly composed of EBV-positive tumors (71%; Figure 3B; supplemental Figure 13B), and this proportion was significantly higher than each of Q53-BL (P < .001; Tukey honest significant difference [HSD] test) and IC-BL (P < .001; Tukey HSD test). We also observed a significant overrepresentation of male patients in DGG-BL (76% male) relative to IC-BL (P = .03; Tukey HSD test), which had more female patients (Figure 3D; supplemental Figure 13D). We attribute this to the much higher incidence of DDX3X mutations in BL among male patients (53.8%) compared with female patients (25%). Comparing the mutations in DDX3X between sexes reveals strikingly distinct patterns, with female patients having almost exclusively missense mutations and male patients having mainly truncating mutations (supplemental Figure 14), a pattern previously observed in human BL.24
Interestingly, 9% of DLBCLs were assigned to a BL cluster, specifically DGG-BL (N = 13), Q53-BL (N = 6), and IC-BL (N = 6). These had notable BL-associated features, including DDX3X mutations and BACH2 aSHM, and 14 were double-hit signature (DHITsig) positive25 (supplemental Figure 12A). Of the 25 DLBCLs assigned to one of the BL genetic subgroups, 6 were HIV-positive DLBCLs, with the remaining 19 HIV-positive cases assigned to DLBCL-predominant subgroups. Although the case number was low, this suggests that HIV-positive DLBCLs share many genetic features with HIV-negative DLBCL. Of the non-BL cases that failed central pathology review, 6 of 8 (75%) were assigned to a BL cluster, specifically IC-BL (N = 3 [38%]), DGG-BL (N = 2 [25%]), or Q53-BL (N = 1 [13%]) (supplemental Figure 13G). Despite low numbers, the rate of cases with marginal pathology clustering with BL was significantly higher than the remaining DLBCLs (P < .001; Fisher exact test). This suggests that cases that were excluded during pathology review tend to have more genetic features of BL than DLBCL. Taken together, this highlights the potential utility of genetics to resolve cases with unclear diagnosis bordering BL and DLBCL.
To assess the reproducibility of the genetic subgroups, we separately explored their representation using a machine learning approach using 3 published cohorts.26-28 Because these validation data sets only contained sequence coverage of exonic regions, we trained a random forest model to classify cases using only mutations detectable in exomes (supplemental Information). To simplify the model, we trained our classifier to separate cases into one of the BL-predominant groups (IC-BL, DGG-BL, or Q53-BL) or a unified DLBCL (ie, non-BL) subgroup. The resulting classifier had 93.3% accuracy, 94.1% sensitivity, and 92.7% specificity overall in distinguishing BL from DLBCL. When used to resolve BL and DLBCL into the 4 subgroups discussed herein, the overall error rate was 12.2% (supplemental Figure 15A). The developed classifier was applied individually to cases from 2 BL studies (Zhou et al28 and Panea et al27) and 1 DLBCL study (Schmitz et al).26 Of the samples from the study by Zhou et al, 58.9% (N = 43/73) were assigned to IC-BL, 30.1% (N = 22/73) were assigned to DGG-BL, 1.4% (N = 1/73) were assigned to Q53-BL, and the remaining 9.6% (N = 7/73) were assigned to DLBCL (Figure 3E; supplemental Figure 15B; supplemental Table 13). This distribution is consistent with results observed in our cohort, given the lack of aBL and EBV-positive samples (5.4%, N = 4/73, with 21/73 with unknown EBV status).
In contrast to the results from the cohort of Zhou et al, the representation of the 4 subgroups among cases from the study by Panea et al27 was distinct, with a surprising fraction (37.6%, N = 38/101) of their BLs classified as DLBCL (Figure 3E; supplemental Figure 15C; supplemental Table 13). This assignment was corroborated by high frequencies of BCL6, KMT2D, CREBBP, and EZH2 hotspot mutations, each of which are uncharacteristic of BL and more generally features of DLBCL (supplemental Figure 15C; supplemental Table 7). To confirm our ability to accurately differentiate BLs from DLBCLs, we applied the classifier to 470 DLBCLs from the study by Schmitz et al.26 This correctly classified 92.3% (N = 434/470) of DLBCLs (Figure 3E; supplemental Figure 15D; supplemental Table 13), which is consistent with the rate of DLBCLs clustered with BLs in our discovery data set (9%). The remaining 36 of 470 DLBCLs classified as one of the BL subgroups were, surprisingly, enriched for activated B cell-like (ABC) and did not show an enrichment for DHITsig-positive tumors. Interestingly, many of these were unclassified (or “other”), according to LymphGen. When compared with the cases classified as DLBCL, these patients had significantly shorter progression-free survival (P = .006; log-rank test). This warrants further exploration of whether some genetic features of BL contribute to more aggressive disease in DLBCL.
BL genetic subgroups are characterized by biological and clinical distinctions
To gain further insights into whether the unique BL subgroups are associated with distinct biological features, we compared the 2 largest groups (IC-BL and DGG-BL) to identify differences in gene expression. This comparison identified 71 differentially expressed genes (Figure 4A; supplemental Tables 14 and 15), with IRF4, TNFRSF13B, and SERPINA9 among the genes with the strongest differential expression (Figure 4A-B). Notably, each of these genes are components of the DLBCL cell-of-origin (COO) and DHITsig classifiers25,29 and have probes in the DLBCL90 NanoString assay.25 When samples were separated into IC-BL and DGG-BL subgroups, each of IRF4 and TNFRSF13B exhibited a striking bimodal distribution of expression with the IC-BL subgroup associated with higher expression of both genes (Figure 4 B), similar to the difference between ABC and germinal center B cell-like (GCB) DLBCL (supplemental Figure 16B).
Genetic subgroups of BL are associated with unique transcriptomic patterns. (A) The heat map displays the 71 differentially expressed genes between subgroups, with rows representing differentially expressed genes and columns representing samples. Rows and columns are clustered on the basis of Pearson correlation. The top annotations indicate subgroup membership, EBV status, age, and sex. Although the separation is incomplete, when clustered on these genes, most DGG-BL cases cluster to the left, whereas most IC-BL cases cluster to the right. (B) Variance stabilized expression of IRF4 and TNFRSF13B, the genes with the strongest differential expression between DGG-BL and IC-BL. Expression values are along the y axis, with subgroup membership indicated along the x axis. Expression values are stratified on the basis of subgroup membership, with IC-BL exhibiting significantly elevated expression of both IRF4 and TNFRSF13B (∗∗∗∗P < .001; Wilcoxon rank sum test). (C) Heat map representing the hierarchical clustering of gene sets obtained from the signatureDB database. Samples are clustered and ordered on their expression of genes within each gene set. Rows represent the gene sets, and columns represent samples. Rows and columns are clustered on the basis of euclidean distance measure.
Genetic subgroups of BL are associated with unique transcriptomic patterns. (A) The heat map displays the 71 differentially expressed genes between subgroups, with rows representing differentially expressed genes and columns representing samples. Rows and columns are clustered on the basis of Pearson correlation. The top annotations indicate subgroup membership, EBV status, age, and sex. Although the separation is incomplete, when clustered on these genes, most DGG-BL cases cluster to the left, whereas most IC-BL cases cluster to the right. (B) Variance stabilized expression of IRF4 and TNFRSF13B, the genes with the strongest differential expression between DGG-BL and IC-BL. Expression values are along the y axis, with subgroup membership indicated along the x axis. Expression values are stratified on the basis of subgroup membership, with IC-BL exhibiting significantly elevated expression of both IRF4 and TNFRSF13B (∗∗∗∗P < .001; Wilcoxon rank sum test). (C) Heat map representing the hierarchical clustering of gene sets obtained from the signatureDB database. Samples are clustered and ordered on their expression of genes within each gene set. Rows represent the gene sets, and columns represent samples. Rows and columns are clustered on the basis of euclidean distance measure.
To further characterize biological differences between IC-BL and DGG-BL, we performed gene set enrichment analyses using relevant lymphoma signatures obtained from the signatureDB database. We identified 17 differentially expressed pathways (P < .05), 2 of which involved IRF4 signaling (Figure 4C; supplemental Figure 16A,C; supplemental Table 16). The IC-BL subgroup displayed elevated expression of genes in pathways involved in IRF4 induction in ABC DLBCL, along with other pathways associated with ABC DLBCL and memory B cells (Figure 4C). More important, although NF-kB pathway activity is one of the established differences between ABC and GCB DLBCL, this pathway was not among those differentially expressed between DGG-BL and IC-BL.
Relationship between cluster-associated mutations and patient outcomes
Because SSMs were the predominant feature driving the BL subgroups, we focused on the genes affected by either coding (supplemental Figure 17) or noncoding mutations (Figure 5A) among these groups. HNRNPU and GNA13 were mutated across all DLBCL subgroups (supplemental Figure 17). Despite the existence of Q53-BL, it is also notable that many of the BLs with TP53 mutations are assigned to other subgroups.
Genetic subgroups are characterized by distinct molecular features. (A) Rates of simple somatic mutations in 1000-bp windows sliding by 500 bp within known sites affected by aSHM. Only bins with at least 20 patients harboring mutations at the particular aSHM site are included in the visualization. Only the sites mutated at differential frequency between EBV-positive and EBV-negative, or between aBL and pBL, are shown (pairwise Fisher exact test with Benjamini-Hochberg multiple test correction). The asterisk (∗) alongside aSHM site indicates its mutation rates being significantly enriched in BL compared with DLBCL. (B) Patients with BL in the genetic subgroup DLBCL-C are characterized by the highest levels of mutation at aSHM sites across common targets (using only samples from patients with BL). Each point indicates odds ratio relative to the aSHM rates for patients in Q53-BL subgroup ± SE.
Genetic subgroups are characterized by distinct molecular features. (A) Rates of simple somatic mutations in 1000-bp windows sliding by 500 bp within known sites affected by aSHM. Only bins with at least 20 patients harboring mutations at the particular aSHM site are included in the visualization. Only the sites mutated at differential frequency between EBV-positive and EBV-negative, or between aBL and pBL, are shown (pairwise Fisher exact test with Benjamini-Hochberg multiple test correction). The asterisk (∗) alongside aSHM site indicates its mutation rates being significantly enriched in BL compared with DLBCL. (B) Patients with BL in the genetic subgroup DLBCL-C are characterized by the highest levels of mutation at aSHM sites across common targets (using only samples from patients with BL). Each point indicates odds ratio relative to the aSHM rates for patients in Q53-BL subgroup ± SE.
We separately explored the density of aSHM in BL and compared these patterns with DLBCL and between BL subgroups. Surprisingly, despite a lower extent of aSHM across BL, 3 regions were significantly more frequently mutated in BL: MYC, BACH2, and TCL1A (Figure 5A). Samples belonging to Q53-BL were characterized by the lowest aSHM rates. BLs in DLBCL-C had the greatest number of mutated regions, whereas BLs in DLBCL-A harbored mutations at a limited number of sites: EBF1, FOXP1, LPP, MEF2C, and PTPN1 (Figure 5B).
To gain insights into the association of BL subgroups with survival outcomes, we performed Kaplan-Meier survival analyses on various subsets of BLs (supplemental Figures 18-21). Because of missing data and previously described batch effects,30 we excluded patients from Uganda and Brazil. As the overall survival differences were not significant between BL subgroups overall (supplemental Figure 19A), we further compared patient outcomes among the BL genetic subgroups separately within aBL and pBL. Within the aBL cases, we found the most significant differences in patient outcomes to arise when ID3 and TP53 mutations were used as alternative single-gene approximation for IC-BL and Q53-BL (supplemental Figures 19-21). However, in pBL, we found DGG-BL had the most inferior outcomes (supplemental Figure 20).
Discussion
Much of our knowledge of the genetic features of EBV-positive and EBV-negative BL was determined from pBL.1,3,5 The results from this work are consistent with many of the previous findings and highlight a limited number of genetic differences between pBL and aBL. We confirmed that tumor EBV status influences the biology of BL more strongly than patient age. Through comparing BL and DLBCL genomes, we reveal genetic subgroupings that span aBL and pBL. This includes 6 subgroups associated with unique genetic and molecular features, with 3 groups sharing a subset of genetic features with DLBCL. Using our classifier, it appears that one of the earlier genomic studies of BL was enriched for cases with genetic features of DLBCL. Similarly, a recent study31 comparing the genetics of pBL and aBL demonstrated an enrichment of DLBCL-associated mutations, including BCL2 in aBL. This is most readily explained by those cases harboring BCL2 translocations. Such variability highlights the importance of central pathology review in such studies, particularly when the differential diagnosis can lead to different treatments.
The noncoding mutations are consistent with aSHM because of aberrant activity of AID, a pattern predominate in EBV-positive BLs. Consistently, the DLBCL-predominant subgroup with the greatest enrichment for aSHM (DLBCL-C) also contained the largest proportion of EBV-positive BLs. The remaining 3 subgroups (IC-BL, DGG-BL, and Q53-BL) were dominated by BL genomes and were the focus of subsequent analyses. Although aSHM was generally lower in these 3 subgroups, AICDA expression was significantly higher in DGG-BL relative to IC-BL (supplemental Figure 1D). We tested whether the difference in aSHM rates was more strongly associated with genetic subgroup or EBV status and found a stronger association with the latter (data not shown). Taken together, we conclude that through its association with AID expression, EBV contributes to BL cases with a more pronounced aSHM pattern, influencing the coding and noncoding genetic landscape of DGG-BL. Despite this, each genetic subgroup contains EBV-positive and EBV-negative tumors, such that each cluster highlights a separate biology rather than being based on EBV status alone.
BL has been known to be associated with EBV infection and known to have different age-specific patterns,31-34 but the distinction of specific genetic profiles between aBL and pBL and their relationship to EBV status have not been extensively studied. Comparison of aBL and pBL genomes consistently showed that stratification on EBV status was associated with more distinct genetic and molecular profiles than patient age. Extending our previous findings in pBL, we support the unique genetic and molecular landscape of EBV-positive BL characterized by an overall lower number of driver mutations specifically in relation to apoptotic genes, higher aSHM rates, and AICDA activity (Figures 1E and 2). In line with previous reports,35,36 EBV-positive BLs harbor significantly more breakpoints upstream of MYC, many of which can be attributed to aberrant AID activity based on their breakpoint in IGH. In contrast, we found EBV-negative BLs to harbor significantly more oncogenic translocations attributable to CSR. These unique features imply different timing of oncogenic events between entities and further suggest that EBV has a similar influence on pBL and aBL alike.
Gene-expression–based classification of other NHLs, such as follicular lymphoma and DLBCL,29,37,38 has established prognostic significance and clinical relevance, informing on different COO and distinct underlying biology. Although the molecular signature of BL has been previously established,39,40 these studies did not consider EBV status or age and they do not inform on subgroupings within BL or different COO. Our finding that up to 9% of DLBCLs and most non-BL cases that failed central pathology review were more likely to be assigned to 1 of the BL clusters indicates ongoing ambiguity in the diagnosis of BL vs DLBCL. In view of the much higher incidence of DLBCL vs BL, significant misclassification of BL as DLBCL or other non-BL lymphomas threatens the validity of BL patterns from population-based cancer registries.41 The present study confirms the strong role of EBV infection status in BL biology and uncovers the presence of novel genetic subgroups within BL that inform on shared pathobiology in aBL and pBL. IC-BL and DGG-BL are characterized by distinct biological and transcriptomic differences that draw parallels with COO in DLBCL. Specifically, IRF4 and TNFRSF13B, which inform on ABC COO in DLBCL, are significantly overexpressed in IC-BL compared with DGG-BL subgroup (Figure 4A-B; supplemental Figure 16; supplemental Table 14), whereas SERPINA9, associated with GCB COO in DLBCL, is downregulated in IC-BL compared with DGG-BL (Figure 4A; supplemental Table 14). This is in line with previous reports of multiple myeloma oncogene 1 (MUM1) positivity in a subset of patients with BL.42 These may indicate a distinct cell of origin for DGG-BL and IC-BL cases, but this requires further exploration (Figure 4C; supplemental Figure 16). Regardless of the cause of elevated IRF4 expression, it is notable that IRF4 has been identified as an essential gene in lymphomas using genome-wide CRISPR screens, but this is inconsistent in BL cell lines,43 and our in vitro analysis identified only Thomas as the only IRF4-dependent IC-BL line (supplemental Figure 22). Intuitively, IRF4-dependent BL lines may be representative of IC-BL, and their dependency on IRF4 nominates them as a therapeutic target worthy of further exploration.
Acknowledgments
The authors thank the Foundation for Burkitt Lymphoma Research Working Group for interesting discussions. The authors also acknowledge the Information Management Systems (Silver Spring, MD), Westat, Inc (Rockville, MD), and African Field Epidemiology Network (Kampala, Uganda) for coordinating The Epidemiology of Burkitt lymphoma in East African children and minors (EMBLEM) fieldwork in Uganda. The authors also acknowledge the International Cancer Genome Consortium Molecular Mechanisms in Malignant Lymphoma by Sequencing project (https://dcc.icgc.org) for providing access to its data. Aligned reads for those genomes were obtained through a Data Access Compliance Office-approved project (to R.D.M.) using a virtual instance on the Cancer Genome Collaboratory. The data sets for validation cohorts were obtained through The European Genome-phenome Archive (data set identifiers EGAD00001005105 and EGAD00001005781) on Data Access Committee approval. The Genomic Variation in Diffuse Large B Cell Lymphomas study was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (NIH), Department of Health and Human Services. The data sets have been accessed through the NIH database for Genotypes and Phenotypes. A full list of acknowledgments can be found in the supplemental note (Schmitz et al, PMID: 29641966).26 The authors also thank the HIV Tumor Malignancy Characterization Network and the AIDS and Cancer Specimen Resource for their valuable contribution of samples to this study. The authors are grateful for contributions from various groups at Canada’s Michael Smith Genome Sciences Centre, including those from the Biospecimen, Library Construction, Sequencing, Bioinformatics, Technology Development, Quality Assurance, Laboratory Information Management System, Purchasing, and Project Management teams. The authors also thank The Biorepository of St. Jude Children’s Research Hospital (National Cancer Institute grants P30 CA021765 and R35 CA197695 to C.G.M.).
This work has been funded in part by the Foundation for Burkitt Lymphoma Research (http://www.foundationforburkittlymphoma.org) and in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health (NIH), under contract no. 75N91019D00024, task order no. 75N91020F00003, contract no. HHSN261200800001E, contract no. HHSN261201100063C, and contract no. HHSN261201100007I (Division of Cancer Epidemiology and Genetics), and in part (S.J.R.) by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. This project was also partially supported by AIDS Malignancy Consortium grant UM1CA121947 and the Intramural Research Program of the NIH, National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. This work was supported by a Terry Fox New Investigator Award (No. 1043) and by an operating grant from the Canadian Institutes for Health Research and a New Investigator Award from the Canadian Institutes for Health Research (R.D.M.). R.D.M. is a Michael Smith Foundation for Health Research Scholar, and D.W.S. is a Michael Smith Foundation for Health Research Health Professional-Investigator. M.A.M. is the recipient of the Canada Research Chair in Genome Science.
This article is dedicated to the memory of Daniela S. Gerhard.
Authorship
Contribution: N.T. and K.D. analyzed the data, produced the figures and tables, and with R.D.M., wrote the manuscript with assistance from D.S.G., J. Bethony, C.C., T.G., N.L.H., E.S.J., S.M.M., C.G.M., A.J.M., A.N., M.A.M., and D.W.S.; B.M.G., L.K.H., M.C., S.S., and J.W. helped with data analyses; D.S.G., M.A.D., N.B.G., and H.P. managed the project and coordinated data deposition; J.S.A., J. Bowen, C.C., J.M.G.-F., T.G.G., F.E.L., S.M.M., C.G.M., C.N., A.N., M.D.O., J.O., G.O., S.J.R., D.W.S., R.Y., R.H., M.N., K.B., A.O., I.M., L.R., D.H., R.M., J.C.R., P.G.R., M.S.L., S.B., E.C., S.S., G. Sissolak, S.P., R.F.A., A.C., D.P.D., and J.C.Z. contributed samples to the study; J. Bowen, J.M.G.-F., and A.S.G. collected sample metadata from tissue source sites; T.G., N.L.H., E.S.J., and S.H.S. performed consensus pathology review; C.C., T.G.G., and E.S.J. reviewed and advised on consensus anatomic site classification; D.S.G., J.D.I., J.P.M., M.-R.M., R.D.M., and L.M.S. designed the study; D.S.G., M.A.M., R.D.M., and L.M.S. directed the study; and all authors contributed to the interpretation of the data, reviewed the manuscript, and approved it for submission.
Conflict-of-interest disclosure: R.D.M. and D.W.S. are named inventors on a patent application describing the double-hit signature. C.G.M. received research funding from Pfizer and AbbVie; was an advisory board member at Illumina; and was on the speaker’s bureau at Amgen. R.Y. reports receiving research support from Celgene (now Bristol Myers Squibb) through CRADAs with the NCI. R.Y. also reports receiving drugs for clinical trials from Merck, EMD-Serano, Eli Lilly, and CTI BioPharma through CRADAs with the NCI, and he has received drug supply for laboratory research from Janssen Pharmaceuticals. R.Y. is a coinventor on US Patent 10 001 483 entitled “Methods for the treatment of Kaposi's sarcoma or KSHV-induced lymphoma using immunomodulatory compounds and uses of biomarkers.” An immediate family member of R.Y. is a coinventor on patents or patent applications related to internalization of target receptors, epigenetic analysis, and ephrin tyrosine kinase inhibitors. All rights, title, and interest to these patents have been assigned to the US Department of Health and Human Services; the government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P.L. 99-502). A.N. received research funding from Pharmacyclics/AbbVie, Kite/Gilead, and Cornerstone; was a consultant for Janssen, Morphosys, Cornerstone, Epizyme, EUSA Pharma, TG Therapeutics, ADC Therapeutics, and Astra Zeneca; and has received honoraria from Pharmacyclics/AbbVie. The remaining authors declare no competing financial interests.
Correspondence: Ryan D. Morin, Department of Molecular Biology and Biochemistry, Simon Fraser University, 8888 University Dr, Burnaby, BC V5A 1S6, Canada; e-mail: rdmorin@sfu.ca.
References
Author notes
∗N.T. and K.D. contributed equally to this study.
All molecular and clinical data used in this publication can be found on the National Cancer Institute’s Genome Data Commons Publication Page (https://gdc.cancer.gov/about-data/publications/CGCI-BLGSP-2022-1) on publication of this article. All custom bioinformatics workflows, scripts, postprocessing, and visualization functions are openly available on GitHub through repository Lymphoid Cancer Research (LCR) modules (https://github.com/LCR-BCCRC/lcr-modules), LCR scripts (https://github.com/LCR-BCCRC/lcr-scripts), and GAMBLR package (https://github.com/morinlab/GAMBLR). The random forest classifier developed in this study is openly available as part of the GAMBLR package.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal