Key Points
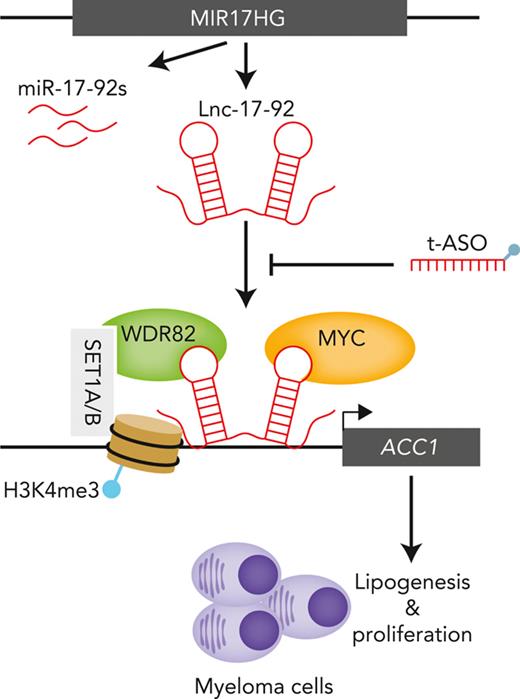
MIR17HG produces a long noncoding RNA that acts as a chromatin scaffold for protein interaction and tumor cell growth.
Targeting this long noncoding RNA with optimized antisense oligonucleotides has potent antimyeloma activity in preclinical models.
Abstract
Long noncoding RNAs (lncRNAs) can drive tumorigenesis and are susceptible to therapeutic intervention. Here, we used a large-scale CRISPR interference viability screen to interrogate cell-growth dependency to lncRNA genes in multiple myeloma (MM) and identified a prominent role for the miR-17-92 cluster host gene (MIR17HG). We show that an MIR17HG-derived lncRNA, named lnc-17-92, is the main mediator of cell-growth dependency acting in a microRNA- and DROSHA-independent manner. Lnc-17-92 provides a chromatin scaffold for the functional interaction between c-MYC and WDR82, thus promoting the expression of ACACA, which encodes the rate-limiting enzyme of de novo lipogenesis acetyl-coA carboxylase 1. Targeting MIR17HG pre-RNA with clinically applicable antisense molecules disrupts the transcriptional and functional activities of lnc-17-92, causing potent antitumor effects both in vitro and in vivo in 3 preclinical animal models, including a clinically relevant patient-derived xenograft NSG mouse model. This study establishes a novel oncogenic function of MIR17HG and provides potent inhibitors for translation to clinical trials.
Introduction
Multiple myeloma (MM) is a genetically complex malignancy of plasma cells that accounts for ∼10% of hematologic cancers and remains largely incurable.1 A growing body of evidence points to a key role played by noncoding RNA (ncRNA) networks in MM,2 suggesting that MM cells can become significantly addicted to and therapeutically susceptible to the modulation of oncogenic ncRNAs.3-8 In particular, long ncRNAs (lncRNAs) outnumber protein-coding genes in humans and are susceptible to the same oncogenic pathogenetic events.9,10 These RNA molecules, defined as transcripts greater than 200 nucleotides (nt) with no protein-coding potential, have a diverse array of functional roles, ranging from being precursor molecules for the biogenesis of mature microRNAs (miRNAs) to direct interactions with proteins and nucleic acids to regulate protein function and/or stability.11-13 With the plethora of biological functions that lncRNAs modulate to control cellular processes at multiple levels, it is not surprising that their aberrant expression and function have been implicated in the progressive gain of a malignant phenotype by tumor cells.14 Indeed, the expression of 14 lncRNAs in newly diagnosed MM patients is correlated (or anticorrelated) with progression-free survival independent of cytogenetic, international staging system, or minimal residual disease status.15 Other lncRNAs, including SMILO, also independently predict MM progression and response to therapy.16,17
To find lncRNAs that have a direct impact on MM proliferation and survival, thus providing cell-growth dependency, we conducted an lncRNA-targeted large-scale CRISPR interference (CRISPRi) viability screen. CRISPRi makes use of a catalytically inactive Cas9 (dCas9)-KRAB fusion protein to repress the expression of endogenous lncRNA genes.18,19 From this screen, we identified MIR17HG as essential in MM, and we (1) characterized its novel function as lncRNA mediating protein-protein and protein-DNA interactions and (2) developed potent inhibitors for translation to clinical trials.
Methods
Cells
Human cell lines and primary cells were grown at 37°C, 5% CO2. Detailed information is included in supplemental Methods, available on the Blood website.
RNA sequencing, microarray-based gene expression analysis, and miRNA profiling of MM patients and cell lines
These analyses were performed in purified CD138+ cells. Detailed information can be found in supplemental Methods.
CRISPRi viability screen and validation
Cell lines expressing the dCas9-KRAB fusion protein were generated as previously described.19 Detailed information on library design, guide RNA pool library production, titering of virus, primary and secondary screenings, and validation study as well as on data analysis can be found in supplemental Methods.
ASO, synthetic miRNA mimics and inhibitors, siRNAs
Long noncoding locked nucleic acid gapmeRs were custom designed and purchased from Exiqon (Vedbaek, Denmark). Sequences can be found in supplemental Methods. Synthetic miRNA mimics and inhibitors, as well as silencer select small interfering RNAs (siRNAs), were purchased from Ambion. SiRNA pool targeting WDR82 was purchased from Horizon Discovery. Design of clinically applicable antisense oligonucleotides (ASOs) is described in supplemental Table 8.
Gymnosis
Gymnotic experiments were performed as previously described.20
Transient and stable transfection of cells
Cell transfection and transduction were performed as previously described.5 Detailed information can be found in supplemental Methods.
Detection of cell proliferation and apoptosis
Cell viability was evaluated by Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies), according to the manufacturer’s instructions. Apoptosis was investigated by an Annexin V/7-AAD flow cytometry assay using FACS CANTO II (BD Biosciences).
RT and qRT-PCR
RNA extraction, reverse transcription (RT), and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) were performed as previously described.5 Detailed information can be found in supplemental Methods.
Western blot analysis
Protein extraction and Western blot analysis were performed as previously described. Detailed information can be found in supplemental Methods.
RNA fluorescence in situ hybridization and coimmunofluorescence with RNA fluorescence in situ hybridization
Luciferase reporter assay
Promoter reporter clones for human ACACA (NM_198834), ANO6 (NM_001025356), CCDC91 (NM_018318), EPT1 (NM_033505), EXT1 (NM_000127), FER (NM_001308028), and ZYG11A (NM_001004339) were cloned into the GLuc-ON Promoter Reporter Vector (GeneCopoeia, Rockville, MD). A luciferase reporter assay was performed according to the manufacturer’s instructions.
ChIRP
Lnc-17-92 and LacZ antisense DNA probes were designed using the online probe designer at singlemoleculefish.com. Oligonucleotides were biotinylated at the 3' end with an 18-carbon spacer arm. AMO1 cells were collected and subjected to chromatin isolation by RNA precipitation (ChIRP) using the EZ-Magna ChIRP RNA Interactome Kit (Millipore Sigma, Bedford, MA), according to the manufacturer’s instructions and established protocols.23
De novo lipogenesis assay
These experiments were conducted as previously described.24 Detailed information can be found in supplemental Methods.
Chromatin immunoprecipitation quantitative polymerase chain reaction
Chromatin immunoprecipitation quantitative polymerase chain reaction was performed as previously described.25 Detailed information can be found in supplemental Methods.
RNA-protein pull-down
Lnc-17-92 transcripts and truncated versions were cloned into a pBlueScript vector and sequence verified. In vitro transcription and biotinylation were performed using AmpliScribe T7-Flash Biotin-RNA Transcription Kit (Lucigen, catalog no. ASB71110), according to the manufacturer’s instructions. Cell nuclear lysates (from 1 × 107 AMO1 cells) were incubated with biotinylated RNA and streptavidin beads for RNA pull-down incubation, using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, catalog no. 20164), according to the manufacturer’s instructions. RNA-associated proteins were eluted and analyzed by Western blotting.
RNA yeast 3 hybrid
RIP quantitative polymerase chain reaction
RNA immunoprecipitation (RIP) experiments were performed using the Magna RIP RNA-binding Protein Immunoprecipitation Kit (Millipore Sigma, catalog no. 17-701), according to the manufacturer’s instructions. The anti-MYC antibody used for RIP was purchased from Abcam (ab32072). Normal rabbit immunoglobulin G was purchased from Cell Signaling Technology (catalog no. 2729). The primers used for detecting lnc-17-92 are listed in supplemental Methods.
Co-IP
Protein lysates were obtained from 1 × 107 cells (AMO1, H929, and U266MYC+, with corresponding treatments). Coimmunoprecipitation (Co-IP) was performed using the Pierce Co-Immunoprecipitation Kit (Thermo Fisher Scientific, catalog no. 26149), according to the manufacturer’s instructions. IP antibodies are listed in supplemental Methods.
BioID
Proximity-dependent biotin identification (BioID) was performed as described by Kalkat et al.26 Detailed information can be found in supplemental Methods.
Mass spectrometry
Mass spectrometry analysis of Co-IP and BioID samples was performed at the Taplin Mass Spectrometry Facility (Harvard Medical School, Boston, MA).
Animal study
Six-week-old female immunodeficient NOD.CB17-Prkdcscid/NCrCrl (NOD/SCID) mice (Charles River) or NSG mice (Jackson Laboratory) were housed in our animal facility at the Dana-Farber Cancer Institute (DFCI). All experiments were performed after approval by the Animal Ethics Committee of the DFCI and performed using institutional guidelines. Detailed information can be found in supplemental Methods.
Statistical analysis
All in vitro experiments were repeated at least 3 times and performed in triplicate. Statistical significances of differences were determined using the Student t test (unless otherwise specified), with the minimal level of significance specified as P < .05. Kaplan-Meier survival curves were compared by log-rank test. Statistical analyses were determined using GraphPad software (http://www.graphpad.com). Graphs were obtained using GraphPad software (unless otherwise specified).
Results
CRISPRi viability screens identify MIR17HG as a leading cell-growth dependency in MM
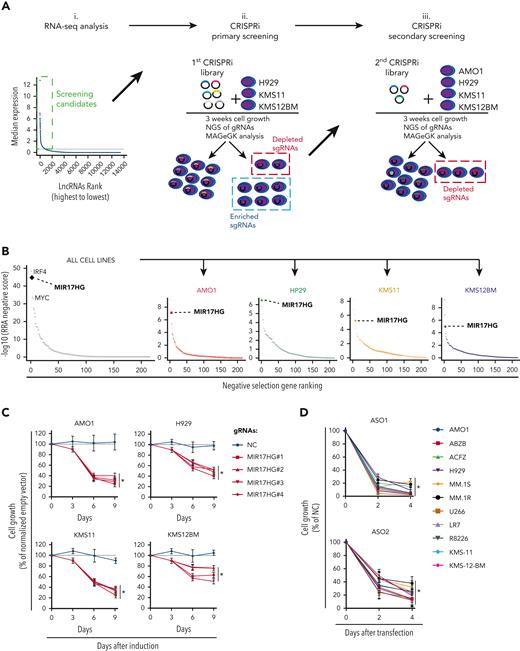
We analyzed RNA sequencing (RNA-seq) data from 360 newly diagnosed MM patients and identified 913 lncRNA transcripts expressed in primary MM cells (Figure 1Ai.) and in a panel of 70 MM cell lines (data not shown). To systematically interrogate the role of these lncRNAs in MM cell growth, we transduced 3 MM cell lines (H929, KMS-11, and KMS-12-BM) engineered to express a dCas9-KRAB fusion protein with a pooled library consisting of 7 single guide RNAs (sgRNAs) against each of the 913 transcription start sites (TSS) of the identified lncRNAs and 576 negative control sgRNAs (Figure 1Aii; supplemental Table 1). After 3 weeks, we tested for sgRNAs that were relatively depleted or enriched in the MM cell population using deep sequencing and the Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout RRA algorithm.27
CRISPRi viability screens identify MIR17HG as a leading cell growth dependency in MM. (A) Schematic of CRISPRi viability screens. (B) Robust rank algorithm (RRA)-based ranked analysis of lncRNA dependencies in the secondary screen, considering 4 MM cell lines either together or individually. The top lncRNA dependency, MIR17HG, is highlighted, along with the protein-coding genes IRF4 and MYC used as positive controls. (C) CCK-8 proliferation assay of MM cell lines (AMO1, H929, KMS11, and KMS12BM) stably expressing KRAB-dCAS9 fusion protein and transduced with lentivectors to conditionally express anti-MIR17HG sgRNAs. CCK-8 assay was performed at indicated time points after exposure to doxycycline (0.5 μg/mL). Cell proliferation is calculated compared with parental cells infected with the empty sgRNA vector and exposed to doxycycline under the same conditions. (D) CCK-8 proliferation assay of MM cell lines (n = 11) transfected with 2 different ASOs targeting the MIR17HG pre-RNA or a nontargeting ASO (NC). ASOs were used at a concentration of 25 nM. Cell viability was measured 2 and 4 days after electroporation, and it is represented as % viability compared with cells transfected with NC-ASO. Data from 1 out of 3 independent experiments are shown in panel D. Data present mean ± standard deviation in panel D. ∗P < .05 by Student t test.
CRISPRi viability screens identify MIR17HG as a leading cell growth dependency in MM. (A) Schematic of CRISPRi viability screens. (B) Robust rank algorithm (RRA)-based ranked analysis of lncRNA dependencies in the secondary screen, considering 4 MM cell lines either together or individually. The top lncRNA dependency, MIR17HG, is highlighted, along with the protein-coding genes IRF4 and MYC used as positive controls. (C) CCK-8 proliferation assay of MM cell lines (AMO1, H929, KMS11, and KMS12BM) stably expressing KRAB-dCAS9 fusion protein and transduced with lentivectors to conditionally express anti-MIR17HG sgRNAs. CCK-8 assay was performed at indicated time points after exposure to doxycycline (0.5 μg/mL). Cell proliferation is calculated compared with parental cells infected with the empty sgRNA vector and exposed to doxycycline under the same conditions. (D) CCK-8 proliferation assay of MM cell lines (n = 11) transfected with 2 different ASOs targeting the MIR17HG pre-RNA or a nontargeting ASO (NC). ASOs were used at a concentration of 25 nM. Cell viability was measured 2 and 4 days after electroporation, and it is represented as % viability compared with cells transfected with NC-ASO. Data from 1 out of 3 independent experiments are shown in panel D. Data present mean ± standard deviation in panel D. ∗P < .05 by Student t test.
The most enriched or depleted sgRNAs were further tested in secondary screens using a pooled library targeting the TSS of 224 lncRNAs, the TSS of known protein-coding oncogenes (MYC, IRF4),28,29 or tumor suppressors (TP53)30 as positive controls and 2245 nontargeting sgRNAs as negative controls (Figure 1Aiii; supplemental Table 2). In the secondary screens, 4 MM cell lines (H929, KMS11, KMS12BM, and AMO1) were used to detect and rank significantly depleted or enriched sgRNAs. As expected, sgRNAs targeting IRF4 and MYC were significantly depleted in 3 (MYC) or all (IRF4) cell lines, whereas sgRNAs targeting TP53 were significantly enriched in both TP53 wild-type (WT) cell lines (AMO1 and H929).31
Focusing on depleted sgRNAs, we identified lncRNA dependencies in MM cells that were either cell-line specific (54%) or shared (46%) (supplemental Figure 1A; supplemental Table 3). A ranked analysis of sgRNA depletion identified MIR17HG as the leading dependency, with RRA scores equal or superior to those obtained by targeting MYC or IRF4 in all cell lines tested (Figure 1B). To validate this data further, we next transduced MM cell lines expressing dCas9-KRAB fusion protein with the top 4 sgRNAs targeting MIR17HG under the regulation of a tetracycline-inducible promoter and observed reduced cell growth compared with cells infected with nontargeting sgRNAs after continued exposure to doxycycline (Figure 1C; supplemental Figure 1B). Moreover, we used 2 different locked nucleic acid gapmeR ASOs (simply referred to as ASO), which target the MIR17HG nascent RNA (pre-RNA) for ribonuclease (RNase) H–mediated degradation,32,33 to transfect 11 MM cell lines including those resistant to conventional anti-MM agents (AMO1-ABZB resistant to bortezomib; AMO1-ACFZ resistant to carfilzomib; MM.1R resistant to dexamethasone) and confirmed a significant impact on MM cell viability independent of the genetic and molecular background (Figure 1D; supplemental Figure 1C).
MIR17HG-derived lnc-17-92 mediates cell-growth dependency in an miRNA- and DROSHA-independent manner
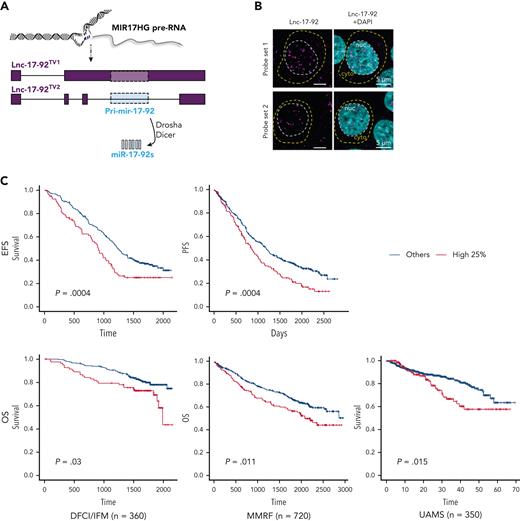
MIR17HG is the locus of the miRNA cluster miR-17-92 and an lncRNA,34,35 named here lnc-17-92. Lnc-17-92 has 2 isoforms, 1 that is ∼5000 nt long (lnc-17-92TV1) and 1 that is ∼900 nt (lnc-17-92TV2),34,35 and both have yet to be functionally explored (Figure 2A). In MM cells, both RNA-seq (supplemental Figure 2A) and qRT-PCR (supplemental Figure 2B) indicated preferential expression of lnc-17-92TV1, which is the isoform further investigated in this study and hereafter referred to as lnc-17-92. Using RNA-seq, we confirmed its expression in CD138+ cells from an additional large cohort of MM patients (MMRF/CoMMpass, n = 720) and in MM cell lines (n =60) (supplemental Figure 2C-D). In MM cell lines, we also demonstrated nuclear enrichment of lnc-17-92 using single molecule RNA fluorescence in situ hybridization (FISH) (Figure 2B) and subcellular qRT-PCR (supplemental Figure 2E). We observed that lnc-17-92 expression was higher during disease progression in 2 independent data sets from MM patients analyzed at diagnosis and/or relapse (supplemental Figures 2F-G) and that higher expression of lnc-17-92 was associated with shorter event-free survival and overall survival in 3 large cohorts of newly diagnosed MM patients (Figure 2C). Expression of lnc-17-92 did not significantly correlate with the expression of miR-17-92 miRNAs in CD138+ MM cells from 140 patients (average Spearman r = 0.16), suggesting that lnc-17-92 and miR-17-92 are under independent regulatory control and function in distinct molecular pathways mediating cell growth dependency to MIR17HG (supplemental Figure 2H).
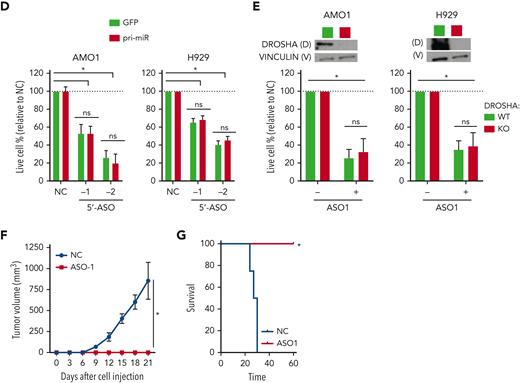
MIR17HG-derived lnc-17-92 mediates cell growth dependency in an miRNA- and DROSHA-independent manner. (A) Overview of MIR17HG locus, including both lncRNA (lnc-17-92)- and miRNA (miR-17-92)-derived transcripts. (B) Single molecule RNA FISH analysis of subcellular localization of lnc-17-92 in AMO1. Cell nuclei are stained by 4',6-diamidino-2-phenylindole (DAPI). (C) Prognostic significance (event-free survival [PFS] and overall survival [OS]) of high lnc-17-92 expression (top quartile) in 3 large cohorts of MM patients. (D) CCK-8 proliferation assay in AMO1 and H929 cells stably transduced with either a lentivector carrying pri-mir-17-92 (pri-miR) or a lentiviral vector carrying GFP as a control and transfected with 2 different ASOs targeting the 5' end (5'-ASO) of MIR17HG pre-RNA or a scrambled control (NC). Effects on cell proliferation were assessed 48 hours after transfection. (E) CCK-8 proliferation assay of DROSHA WT or KO AMO1 and H929 exposed to ASO1 (1 μM for AMO1 and 2.5 μM for H929) for 6 days. Western blot analysis of DROSHA expression in WT and KO cells. Vinculin was used as a protein-loading control. (F) Effects of lnc-17-92 depletion in a matrigel-based AMO1DR-KO xenograft in NOD SCID mice. Tumor growth of AMO1DR-KO with (ASO-1) or without (NC) lnc-17-92 depletion. (G) Survival analysis of tumor-injected mice. ∗P < .05. ns, not significant (P > .05 after Student t test).
MIR17HG-derived lnc-17-92 mediates cell growth dependency in an miRNA- and DROSHA-independent manner. (A) Overview of MIR17HG locus, including both lncRNA (lnc-17-92)- and miRNA (miR-17-92)-derived transcripts. (B) Single molecule RNA FISH analysis of subcellular localization of lnc-17-92 in AMO1. Cell nuclei are stained by 4',6-diamidino-2-phenylindole (DAPI). (C) Prognostic significance (event-free survival [PFS] and overall survival [OS]) of high lnc-17-92 expression (top quartile) in 3 large cohorts of MM patients. (D) CCK-8 proliferation assay in AMO1 and H929 cells stably transduced with either a lentivector carrying pri-mir-17-92 (pri-miR) or a lentiviral vector carrying GFP as a control and transfected with 2 different ASOs targeting the 5' end (5'-ASO) of MIR17HG pre-RNA or a scrambled control (NC). Effects on cell proliferation were assessed 48 hours after transfection. (E) CCK-8 proliferation assay of DROSHA WT or KO AMO1 and H929 exposed to ASO1 (1 μM for AMO1 and 2.5 μM for H929) for 6 days. Western blot analysis of DROSHA expression in WT and KO cells. Vinculin was used as a protein-loading control. (F) Effects of lnc-17-92 depletion in a matrigel-based AMO1DR-KO xenograft in NOD SCID mice. Tumor growth of AMO1DR-KO with (ASO-1) or without (NC) lnc-17-92 depletion. (G) Survival analysis of tumor-injected mice. ∗P < .05. ns, not significant (P > .05 after Student t test).
To test independent activity of lnc-17-92, we first established 2 MM cell lines overexpressing miR-17-92 via ectopic expression of the primary precursor pri-mir-17-92 (supplemental Figure 3A). We specifically depleted lnc-17-92 in these cell lines with ASOs targeting the 5' end of MIR17HG pre-RNA, a region not covered by pri-mir-17-92, and observed a significant inhibition of cell growth that was not rescued by ectopic pri-mir-17-92 (Figure 2D). Next, we established 2 DROSHA knockout (DR-KO) MM cell lines (AMO1DR-KO and H929DR-KO), which are unable to enzymatically digest pri-mir-17-92 and produce miR-17-92s (supplemental Figure 3B),36 and still observed strong antiproliferative activity in both DR-WT and DR-KO cell systems after lnc-17-92 depletion using gymnotic treatment with ASO1 (Figure 2E) or after transfection with 3 different ASOs (−1/−2/−3) (supplemental Figure 3C). Importantly, exposure to gymnotic ASO1 (Figure 2E-F) or transfection with ASO2 (supplemental Figure 3D) abrogated the ability of AMO1DR-KO cells to establish tumors in nonobese diabetic severe combined immunodeficiency (NOD SCID) mice, resulting in prolonged animal survival. Next, using the easy-to-transfect colorectal cancer cell line HCT-116, which is driven by MIR17HG,34 we found that ectopic expression of lnc-17-92TV1 significantly rescued the antiproliferative activity of ASOs targeting MIR17HG pre-RNA more effectively than ectopic lnc-17-92TV2 or pri-mir-17-92 (supplemental Figure 3E-F). Finally, we confirmed the independent activity of lnc-17-92 in the HCT-116 and DLD-1 colorectal cancer cell lines carrying a mutant Dicer, which confers a hypomorphic phenotype preventing the cells from enzymatically processing mature miRNAs37 (supplemental Figure 3G).
These results indicate that lnc-17-92TV1 is the main mediator of MIR17HG cancer dependency, separate from the actions and biogenesis pathway of miR-17-92.
Lnc-17-92 forms a transcriptional axis with ACACA to promote MM cell growth
The nuclear enrichment of lnc-17-92 suggests a possible role in the regulation of gene expression. We therefore depleted lnc-17-92 in DR-WT (AMO1 and H929) and DR-KO (AMO1DR-KO) MM cell lines using early exposure to gymnotic ASO1 to avoid modulation of miR-17-92 (supplemental Figure 4A) and miR-17-92’s canonical targets in DROSHA WT cells (supplemental Figure 4B-C) and identified 7 genes rapidly downregulated after depletion of lnc-17-92 in all the cell lines tested (Figure 3A). We validated these findings in CD138+ cells from 3 MM patients treated ex vivo with ASO1 (Figure 3B) and in the lymphoma cell lines Raji and Daudi (supplemental Figure 4D). Conversely, the expression of these genes was not affected by modulating individual members of miR-17-92 using synthetic mimics or inhibitors (supplemental Figure 4E-F). Moreover, we observed significant positive correlation (Spearman r > 0.3; P < .001) between lnc-17-92 and its target genes in at least 1 out of 2 large RNA-seq MM patient data sets (IFM/DFCI and MMRF/CoMMpass) (Figure 3C).
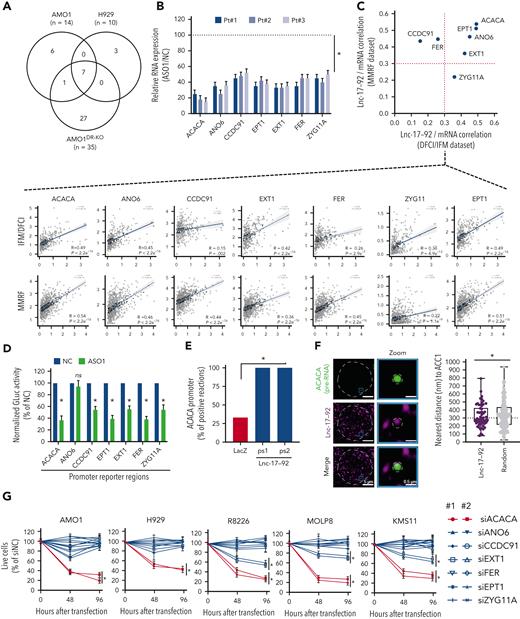
Lnc-17-92 forms a transcriptional axis with ACACA to promote proliferation and survival of MM cells. (A) Transcriptomic analysis after lnc-17-92 depletion in MM cell lines that have either DROSHA WT (AMO1, H929) or KO (AMO1DR-KO). Venn diagram of commonly downregulated genes (adjusted P < .05; log2FC < −1). Cells were exposed to ASO1 for 24 hours. (B) qRT-PCR analysis of lnc-17-92 targets in CD138+ cells from 3 MM patients exposed to ASO1 for 24 hours. The results shown are average mRNA expression levels after normalization with GAPDH and ΔΔCt calculations. RNA level in cells exposed to NC (vehicle) were set as an internal reference. (C) Correlation analysis between lnc-17-92 targets (mRNA) and lnc-17-92 in CD138+ MM patient cells from 2 large RNA-seq cohorts (DFCI/IFM, n = 360; MMRF/CoMMpass, n = 720). Spearman r obtained in DFCI/IFM (x-axis) and MMRF/CoMMpass (y-axis) data sets. Dotted red lines indicate r = 0.3. Individual correlation plots (below). (D) GLuc/SEAP dual reporter assay showing reduced activity of ACACA, ANO6, CCDC91, EPT1, EXT1, FER, and KIAA1109 promoter activity after lnc-17-92 knockdown using ASO1. The reporter vectors were cotransfected into 293T cells with either ASO1 or control ASO. Cells were harvested for the luciferase activity assay 48 hour after transfection. Results are shown as % of normalized GLuc activity in ASO1-transfected cells compared with control. (E) ChIRP-qPCR analysis showing effective amplification of ACACA promoter in chromatin purified using 2 lnc-17-92 antisense probe sets (ps1 and ps2), compared with chromatin purified using LacZ antisense probes (negative control). (F) (left) Snapshot obtained by dual RNA-FISH analysis of ACACA pre-mRNA (green) and lnc-17-92 (purple) in a representative AMO1 cell; (right) box plot showing the distance (nm) of ACACA pre-RNA spots to the nearest lnc-17-92 spots (n = 57) or to the nearest random spots (160); 300 nm was used as a cut-off determining proximity. (G) CCK-8 proliferation assay in 5 MM cells lines after transfection with siRNAs against lnc-17-92 targets. Two siRNAs were used for each target, plus a scramble siRNA (NC) as a control. Cell viability was measured at the indicated time point, represented as % of NC-transfected cells. ∗P < .05 after Student t test in panels B, D, and G or after Fisher exact test in panels E and F. Pt, patient.
Lnc-17-92 forms a transcriptional axis with ACACA to promote proliferation and survival of MM cells. (A) Transcriptomic analysis after lnc-17-92 depletion in MM cell lines that have either DROSHA WT (AMO1, H929) or KO (AMO1DR-KO). Venn diagram of commonly downregulated genes (adjusted P < .05; log2FC < −1). Cells were exposed to ASO1 for 24 hours. (B) qRT-PCR analysis of lnc-17-92 targets in CD138+ cells from 3 MM patients exposed to ASO1 for 24 hours. The results shown are average mRNA expression levels after normalization with GAPDH and ΔΔCt calculations. RNA level in cells exposed to NC (vehicle) were set as an internal reference. (C) Correlation analysis between lnc-17-92 targets (mRNA) and lnc-17-92 in CD138+ MM patient cells from 2 large RNA-seq cohorts (DFCI/IFM, n = 360; MMRF/CoMMpass, n = 720). Spearman r obtained in DFCI/IFM (x-axis) and MMRF/CoMMpass (y-axis) data sets. Dotted red lines indicate r = 0.3. Individual correlation plots (below). (D) GLuc/SEAP dual reporter assay showing reduced activity of ACACA, ANO6, CCDC91, EPT1, EXT1, FER, and KIAA1109 promoter activity after lnc-17-92 knockdown using ASO1. The reporter vectors were cotransfected into 293T cells with either ASO1 or control ASO. Cells were harvested for the luciferase activity assay 48 hour after transfection. Results are shown as % of normalized GLuc activity in ASO1-transfected cells compared with control. (E) ChIRP-qPCR analysis showing effective amplification of ACACA promoter in chromatin purified using 2 lnc-17-92 antisense probe sets (ps1 and ps2), compared with chromatin purified using LacZ antisense probes (negative control). (F) (left) Snapshot obtained by dual RNA-FISH analysis of ACACA pre-mRNA (green) and lnc-17-92 (purple) in a representative AMO1 cell; (right) box plot showing the distance (nm) of ACACA pre-RNA spots to the nearest lnc-17-92 spots (n = 57) or to the nearest random spots (160); 300 nm was used as a cut-off determining proximity. (G) CCK-8 proliferation assay in 5 MM cells lines after transfection with siRNAs against lnc-17-92 targets. Two siRNAs were used for each target, plus a scramble siRNA (NC) as a control. Cell viability was measured at the indicated time point, represented as % of NC-transfected cells. ∗P < .05 after Student t test in panels B, D, and G or after Fisher exact test in panels E and F. Pt, patient.
Using a luciferase reporter assay, performed in 293TDR-KO cells in the presence or absence of lnc-17-92 depletion, we demonstrated that the regulatory control of lnc-17-92 over these genes, except ANO6, occurs at the promoter level (Figure 3D). Consistently, we confirmed lnc-17-92 interaction at the promoter region of the top target, ACACA, by a ChIRP assay followed by qRT-PCR analysis (Figure 3E; supplemental Figure 4G-H) and showed frequent localization of lnc-17-92 to the ACACA locus by single-molecule dual RNA FISH analysis of lnc-17-92 and ACACA pre-mRNA (<300 nm to nearest lnc-17-92 spot in ∼50% of ACACA pre-RNA spots analyzed [n = 60]) (Figure 3F). Proximal localization of lnc-17-92 to the ACACA gene locus was significantly more frequent compared with random spots (Figure 3F).
Among the identified lnc-17-92 targets, ACACA had the largest impact on the proliferation and survival of MM cells (Figure 3G). ACACA encodes the rate-limiting enzyme for the de novo lipogenesis pathway ACC1, which supports tumorigenesis in different cancer contexts.38 To confirm that lnc-17-92’s control over ACACA expression has a functional effect, we depleted lnc-17-92 and found that the incorporation of C14-radiolabeled glucose into the lipid pool was significantly reduced, indicating a reduced amount of de novo lipogenesis,24 both in MM cell lines and CD138+ MM patient cells (supplemental Figure 4I). This was not observed after transfection of MM cells with synthetic inhibitors of miR-17-92s (supplemental Figure 4J). Moreover, supplementing palmitate, which is the main downstream product of ACC1 activity, significantly rescued the antiproliferative and proapoptotic effects of lnc-17-92 depletion in MM cells (supplemental Figure 4K-L).
Altogether, these data indicate lnc-17-92 is a chromatin-interacting lncRNA with transcriptional regulatory functions. We next sought to determine how it can promote transcription by searching for its protein-binding partners.
Lnc-17-92 directly interacts with c-MYC and promotes its occupancy at the ACACA promoter
The targeting of MIR17HG primarily kills c-MYC–positive (MYC+) tumor cells, including in MM.5,34,39,40 Intriguingly, MYC is known to reactivate ACACA expression and de novo lipogenesis in tumor cells,41 with MYC+ tumor cells becoming addicted to this metabolic pathway, findings that we validated in MM cells (supplemental Figure 5A-D). Therefore, we hypothesized that lnc-17-92 mediates the functional interplay between MYC and MIR17HG by directly interacting with MYC protein to promote gene expression.
We performed an RNA protein pull-down (RPPD) experiment and found that MYC forms a complex with lnc-17-92TV1 (Figure 4A). RIP assay with MYC antibody confirmed the enrichment of lnc-17-92TV1 (Figure 4B). Moreover, an RNA yeast-3-hybrid (Y3H) assay confirmed the lnc-17-92TV1–MYC interaction in an in vivo cellular model,42 as shown by yeast colony growth (Figure 4C). An analysis using truncated versions of lnc-17-92TV1 further indicated the 3'-end regions, which do not include miR-17-92, as particularly relevant for the interaction with MYC in MM cells (supplemental Figure 5E).
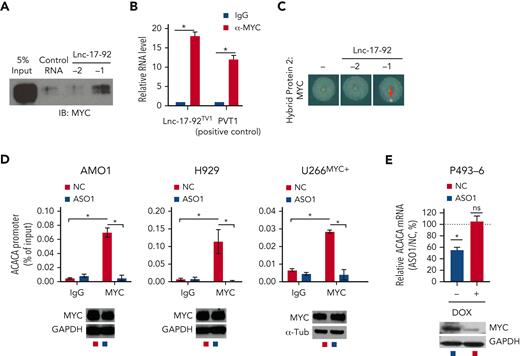
Lnc-17-92 directly interacts with c-MYC and promotes its occupancy at the ACACA promoter. (A) Western blot analysis of MYC in RPPD material precipitated with control RNA or lnc-17-92TV1 or lnc-17-92TV2. 5% input is used as a reference. (B) qRT-PCR analysis of lnc-17-92 (detecting lnc-17-92TV1) in RIP material precipitated using an anti-MYC antibody (α-MYC) or immunoglobulin G (IgG) control. LncRNA PVT1 is used as a positive control for its role as MYC interactor. (C) RNA Y3H using MYC as hybrid protein 2 and, as hybrid RNAs, a negative control RNA (−) or lnc-17-92TV1 or lnc-17-92TV2. (D) ChIP-qPCR analysis of MYC occupancy at the ACACA promoter in AMO1, H929, and U266MYC+ exposed for 24 hours to ASO1 or NC (vehicle). MYC occupancy at ACACA promoter is calculated as % of input chromatin. Western blot analysis of MYC from paired samples (below). GAPDH or α-tubulin were used as protein loading controls. (E) qRT-PCR analysis of ACACA mRNA in P493-6 cells exposed for 2 days to either doxycycline or DMSO to knock down MYC and then exposed for 2 additional days to either ASO1 or vehicle (NC) to deplete lnc-17-92. ACACA expression levels in cells exposed to NC were set as an internal reference. ∗P < .05, Student t test.
Lnc-17-92 directly interacts with c-MYC and promotes its occupancy at the ACACA promoter. (A) Western blot analysis of MYC in RPPD material precipitated with control RNA or lnc-17-92TV1 or lnc-17-92TV2. 5% input is used as a reference. (B) qRT-PCR analysis of lnc-17-92 (detecting lnc-17-92TV1) in RIP material precipitated using an anti-MYC antibody (α-MYC) or immunoglobulin G (IgG) control. LncRNA PVT1 is used as a positive control for its role as MYC interactor. (C) RNA Y3H using MYC as hybrid protein 2 and, as hybrid RNAs, a negative control RNA (−) or lnc-17-92TV1 or lnc-17-92TV2. (D) ChIP-qPCR analysis of MYC occupancy at the ACACA promoter in AMO1, H929, and U266MYC+ exposed for 24 hours to ASO1 or NC (vehicle). MYC occupancy at ACACA promoter is calculated as % of input chromatin. Western blot analysis of MYC from paired samples (below). GAPDH or α-tubulin were used as protein loading controls. (E) qRT-PCR analysis of ACACA mRNA in P493-6 cells exposed for 2 days to either doxycycline or DMSO to knock down MYC and then exposed for 2 additional days to either ASO1 or vehicle (NC) to deplete lnc-17-92. ACACA expression levels in cells exposed to NC were set as an internal reference. ∗P < .05, Student t test.
Next, we evaluated whether MYC and lnc-17-92 cooperate to promote ACACA expression in MM cells. Depletion of lnc-17-92 in MM cells indeed abrogated MYC occupancy at the ACACA promoter while not affecting MYC expression (Figure 4D) and reduced the expression of ACACA in the conditional MYC Tet-Off cell line P493-643 only in presence of high MYC levels (Figure 4E). Moreover, by coupling RNA FISH analysis of lnc-17-92TV1 and ACACA pre-RNA with immunofluorescence analysis of MYC protein, we captured the colocalization of lnc-17-92TV1 and MYC at the ACACA gene locus (supplemental Figure 5F).
These data demonstrate that lnc-17-92TV1 forms an RNA-protein complex with the transcription factor MYC to promote its chromatin occupancy and transcriptional activity at the ACACA promoter.
Lnc-17-92 mediates the assembly of an MYC-WDR82 transcriptional complex, leading to transcriptional and epigenetic activation of ACACA
MYC transcriptional activity is modulated through the interaction with transcriptional and epigenetic coregulators.41 To determine whether lnc-17-92 affects these protein-protein interactions, we integrated the results of a proximity-dependent BioID analysis (supplemental Figure 6A) with a Co-IP/MS in 3 MM cell lines (AMO1, H929, and U266MYC+), in the presence and absence of depletion of lnc-17-92. This analysis highlighted WDR82 as a very high-confidence lnc-17-92–dependent MYC interactor (Figure 5A and supplemental Tables 4-7). A direct RNA-protein interaction between lnc-17-92TV1 and WDR82 was further confirmed by both RPPD (Figure 5B) and RNA Y3H (Figure 5C) assays. Analysis using the truncated versions of lnc-17-92TV1 indicated that this interaction may involve different domains across lnc-17-92 (supplemental Figure 6B).
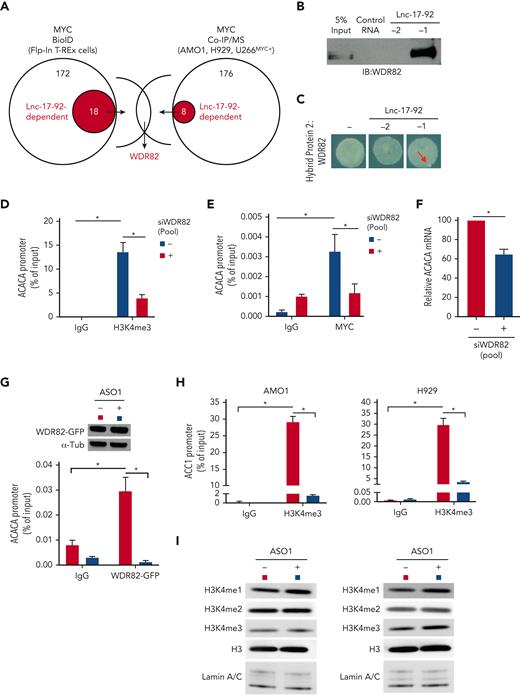
Lnc-17-92 mediates the assembly of a MYC-WDR82 transcriptional complex, leading to transcriptional and epigenetic activation of ACACA. (A) Schematic of integrated BioID and coimmunoprecipitation assay followed by mass-spectrometry analysis (Co-IP/MS) assays to explore the MYC-protein interacting network in the presence or absence of lnc-17-92 depletion. (B) Western blot analysis of WDR82 in RPPD material precipitated with lnc-17-92TV1 or lnc-17-92TV2 or with control RNA; 5% input is used as a reference. (C) RNA Y3H using WDR82 as hybrid protein 2 and, as hybrid RNAs, either a negative control RNA (−) or lnc-17-92TV1 or lnc-17-92TV2. Red arrows indicate yeast colony growth. (D) ChIP-qPCR analysis of H3K4me3 occupancy at ACACA promoter after silencing of WDR82 with a siRNA pool (n-4) in H929 (24-hour time point). Data are represented as % of input chromatin. (E) ChIP-qPCR analysis of MYC occupancy at the ACACA promoter after silencing of WDR82 with a siRNA pool (n-4) (24-hour time point). Data are represented as % of input chromatin. (F) qRT-PCR analysis of ACACA mRNA after silencing of WDR82 with a siRNA pool (n-4) (48-hour time point). Raw Ct values were normalized to GAPDH mRNA and expressed as ΔΔCt values calculated using the comparative cross threshold method. ACACA expression levels in cells transfected with NC were set as an internal reference. (G) ChIP-qPCR analysis of WDR82-GFP occupancy at the ACACA promoter in AMO1 exposed for 24 hours to gymnotic ASO1. Data are represented as % of input chromatin. Western blot analysis of WDR82-GFP from paired samples. α-Tubulin was used as the protein loading control. (H) ChIP-qPCR analysis of H3K4me3 occupancy at the ACACA promoter in AMO1 and H929 exposed for 24 hours to gymnotic ASO1. Data are represented as % of input chromatin. (I) Western blot analysis of H3, H3H3K4me1, H3H3K4me2, and H3H3K4me3 in AMO1 and H929 exposed for 24 hours to gymnotic ASO1. Lamin A/C was used as the protein loading controls (nuclear lysates). ∗P < .05, Student t test.
Lnc-17-92 mediates the assembly of a MYC-WDR82 transcriptional complex, leading to transcriptional and epigenetic activation of ACACA. (A) Schematic of integrated BioID and coimmunoprecipitation assay followed by mass-spectrometry analysis (Co-IP/MS) assays to explore the MYC-protein interacting network in the presence or absence of lnc-17-92 depletion. (B) Western blot analysis of WDR82 in RPPD material precipitated with lnc-17-92TV1 or lnc-17-92TV2 or with control RNA; 5% input is used as a reference. (C) RNA Y3H using WDR82 as hybrid protein 2 and, as hybrid RNAs, either a negative control RNA (−) or lnc-17-92TV1 or lnc-17-92TV2. Red arrows indicate yeast colony growth. (D) ChIP-qPCR analysis of H3K4me3 occupancy at ACACA promoter after silencing of WDR82 with a siRNA pool (n-4) in H929 (24-hour time point). Data are represented as % of input chromatin. (E) ChIP-qPCR analysis of MYC occupancy at the ACACA promoter after silencing of WDR82 with a siRNA pool (n-4) (24-hour time point). Data are represented as % of input chromatin. (F) qRT-PCR analysis of ACACA mRNA after silencing of WDR82 with a siRNA pool (n-4) (48-hour time point). Raw Ct values were normalized to GAPDH mRNA and expressed as ΔΔCt values calculated using the comparative cross threshold method. ACACA expression levels in cells transfected with NC were set as an internal reference. (G) ChIP-qPCR analysis of WDR82-GFP occupancy at the ACACA promoter in AMO1 exposed for 24 hours to gymnotic ASO1. Data are represented as % of input chromatin. Western blot analysis of WDR82-GFP from paired samples. α-Tubulin was used as the protein loading control. (H) ChIP-qPCR analysis of H3K4me3 occupancy at the ACACA promoter in AMO1 and H929 exposed for 24 hours to gymnotic ASO1. Data are represented as % of input chromatin. (I) Western blot analysis of H3, H3H3K4me1, H3H3K4me2, and H3H3K4me3 in AMO1 and H929 exposed for 24 hours to gymnotic ASO1. Lamin A/C was used as the protein loading controls (nuclear lysates). ∗P < .05, Student t test.
WDR82 is a regulatory component of the SET1 methyltransferase complex, which catalyzes histone H3 Lys-4 (H3K4) methylation (mono-, di-, tri-) at the transcriptional start sites of active loci,44,45 a prerequisite for MYC binding to chromatin and transactivation.46 We confirmed a global effect of silencing of WDR82 on H3K4 methylation in MM cells (supplemental Figure 6C). Consistently, depletion of WDR82 reduced the occupancy of H3K4me3 (Figure 5D and supplemental Figure 6D) and MYC (Figure 5E and supplemental Figure 6E) at the ACACA promoter and decreased ACACA mRNA expression (Figure 5F and supplemental Figure 6F) in MM cells. Furthermore, using MM cells expressing an ectopic WDR82-GFP fusion protein (supplemental Figure 6G), we demonstrated that lnc-17-92 expression is essential for WDR82 occupancy at the ACACA promoter (Figure 5G). Additionally, lnc-17-92 depletion resulted in reduced levels of H3K4me3 at the ACACA promoter (Figure 5H), without globally impacting the H3K4 methylation status (Figure 5I).
These findings suggest lnc-17-92TV1 is a chromatin scaffold mediating the assembly of the MYC-WDR82 multiprotein transcriptional complex to control the expression of ACACA and likely other genes.
Therapeutic inhibitors of MIR17HG exert potent antitumor activity in vitro and in vivo in animal models of human MM
We next explored MIR17HG as a therapeutic target, which includes both the lncRNA and miRNA factors. To develop clinically applicable inhibitors, we screened >80 fully phosphorothioated, 2'-O-methoxyethyl–modified, lipid-conjugated ASOs that could either trigger RNase H–mediated degradation of MIR17HG pre-RNA (gapmeRs) or exert function via an RNase H–independent mechanism (blockmeRs)47 (supplemental Figure 7A-B). This procedure identified an 18-mer tocopherol (T)-conjugated gapmeR G2-15b-T (“G”) and an 18-mer tocopherol (T)-conjugated steric blocker SB9-19-T (“B”) as both having strong antiproliferative effects (cell growth inhibition >50%) in a large panel of MM cell lines as well as CD138+ primary MM cells, while sparing (cell growth inhibition <50%) nonmalignant cell lines (THLE-2, HK-2, HS-5, and 293T) and peripheral blood mononuclear cells from 3 healthy donors (supplemental Figure 7C).
To assess the in vivo antitumor activity of both compounds, we first used an AMO1-based plasmacytoma xenograft model in immunocompromised NOD SCID mice. Here, we observed a significant reduction of tumor growth after a treatment cycle with either G2-15b-T (tumor growth inhibition [TGI] = 76%) or B9-19-T (TGI = 69%) (Figure 6A). Analysis of tumors retrieved from mice following this treatment confirmed reduced expression of lnc-17-92 (Figure 6B) and miR-17-92s (supplemental Figure 7D), as well as modulation of lnc-17-92’s targets (ACACA, EPT1, EXT1, CCDC91, ANO6, FER, and ZYG11A) (Figure 6C) and miR-17-92’s target BIM (also known as BCL2L11) (supplemental Figure 7E). We also observed reduced levels of tripalmitin (supplemental Figure 7F), a surrogate for the de novo lipogenesis product palmitate48 This demonstrates efficient uptake of G2-15b-T and B9-19-T by tumor cells in vivo. We observed no overt toxicity in the mice after treatment, as shown by blood cell count, clinical biochemistry (supplemental Tables 9-10), and body weight analysis (not shown).
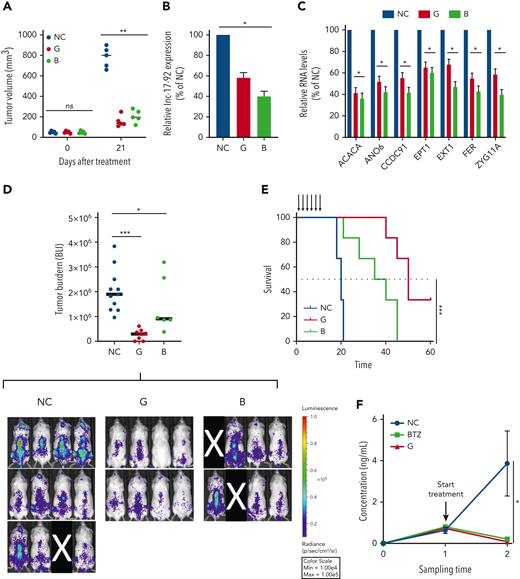
Therapeutic inhibitors of MIR17HG exert potent antitumor activity in vitro and in vivo in animal models of human MM. (A) Subcutaneous in vivo tumor growth of AMO1 cells in NOD SCID mice, 21 days after treatment with G2-15b∗-TO (G; n = 5), B9-19-TO (B; n = 5), or vehicle (NC; n = 5). (B-C) qRT-PCR analysis of lnc-17-92 (C) and lnc-17-92 targets (D) in AMO1 xenografts, retrieved from animals treated with G2-15b∗-TO (G; n = 1), B9-19-TO (B; n = 1), or vehicle (NC; n = 1) as a control. Raw Ct values were normalized to ACTB mRNA and expressed as ΔΔCt values calculated using the comparative cross threshold method. Expression levels in NC were set as an internal reference. (D) Bioluminescent imaging–based (BLI-based) measurement of in vivo tumor growth of MOLP8-luc+ in NSG mice, after treatment with G2-15b∗-TO (G; n = 8), B9-19-TO (B; n = 6), or vehicle (NC; n = 11). On the top, a scatter plot shows the analysis of bioluminescence intensity. Red bars indicate median value. Bioluminescence was measured at the end of the treatment cycle (day 15). Image acquisition (below). Mice removed from the study owing to failed IV injection of tumor cells are covered by a black rectangle. (E) Survival analysis from experiment in panel E. (F) Human κ light chain enzyme-linked immunosorbent assay–based measurement of in vivo tumor growth of MM patient cells in NSG mice (PDX-NSG), after treatment with G2-15b∗-TO (G; n = 2), bortezomib (BTZ; n = 2), or vehicle (NC; n = 3). Black arrows indicate treatments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Max, maximum; Min, minimum; ns, not significant.
Therapeutic inhibitors of MIR17HG exert potent antitumor activity in vitro and in vivo in animal models of human MM. (A) Subcutaneous in vivo tumor growth of AMO1 cells in NOD SCID mice, 21 days after treatment with G2-15b∗-TO (G; n = 5), B9-19-TO (B; n = 5), or vehicle (NC; n = 5). (B-C) qRT-PCR analysis of lnc-17-92 (C) and lnc-17-92 targets (D) in AMO1 xenografts, retrieved from animals treated with G2-15b∗-TO (G; n = 1), B9-19-TO (B; n = 1), or vehicle (NC; n = 1) as a control. Raw Ct values were normalized to ACTB mRNA and expressed as ΔΔCt values calculated using the comparative cross threshold method. Expression levels in NC were set as an internal reference. (D) Bioluminescent imaging–based (BLI-based) measurement of in vivo tumor growth of MOLP8-luc+ in NSG mice, after treatment with G2-15b∗-TO (G; n = 8), B9-19-TO (B; n = 6), or vehicle (NC; n = 11). On the top, a scatter plot shows the analysis of bioluminescence intensity. Red bars indicate median value. Bioluminescence was measured at the end of the treatment cycle (day 15). Image acquisition (below). Mice removed from the study owing to failed IV injection of tumor cells are covered by a black rectangle. (E) Survival analysis from experiment in panel E. (F) Human κ light chain enzyme-linked immunosorbent assay–based measurement of in vivo tumor growth of MM patient cells in NSG mice (PDX-NSG), after treatment with G2-15b∗-TO (G; n = 2), bortezomib (BTZ; n = 2), or vehicle (NC; n = 3). Black arrows indicate treatments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Max, maximum; Min, minimum; ns, not significant.
We next confirmed the significant anti-MM activity of G2-15b-T and B9-19-T in an aggressive model of diffused myeloma, in which tumor growth of MOLP8-luc+ MM cells is assessed by BLI. In this model, tumor growth was significantly antagonized after a treatment cycle with either G2-15b-T (TGI = 84%) or B9-19-T (TGI = 52%). Treatment with G2-15b-T resulted in tumor clearance in 2 out of 8 mice (25%) (Figure 6D). Importantly, both inhibitors significantly prolonged animal survival (Figure 6E).
Finally, we established a clinically relevant PDX-NSG mouse model by tail-vein injection of CD138+ MM cells obtained from an advanced-stage patient (PDX-NSG). In this model, tumor growth was monitored in serum samples using human κ light chain as a surrogate. Remarkably, we observed a regression of tumor growth after a treatment cycle with G2-15b-T, whose effects were comparable to bortezomib (a positive control) (Figure 6F).
Discussion
MIR17HG is often amplified and/or overexpressed in human cancer and has a driver role.34,39,49 One of its transcriptional products, pri-mir-17-92, is enzymatically digested by DROSHA into 6 precursor transcripts (pre-mir-17/−18a/−19a/−20a/−19b1/−92a) that are further processed by DICER to generate the miR-17-92 mature miRNAs (miR-17/−18a/−19a/−20a/−19b1/−92a). These miRNAs posttranscriptionally repress relevant tumor-suppressive mRNAs,34,39,40,49 such as the proapoptotic factor BIM.50 Their impact on tumorigenesis is particularly relevant when coexpressed with MYC,34,39 as there is a well-documented interplay between miR-17-92, especially miR-19b,51,52 and MYC transcriptional targets in maintaining cancer cell homeostasis.5,39,40 We have previously demonstrated that these miRNAs play a relevant role in MM by forming homeostatic feed-forward loops with MYC and BIM.5 Interestingly, our previous work also showed that depletion of mature miRNAs does not phenocopy the inhibition of MIR17HG pre-RNA, suggesting other tumor-promoting functions for this transcript.5 An alternative mechanism to explain the oncogenic role of MIR17HG has been recently identified and involves the overload of DROSHA by an overexpressed pri-mir-17-92 in B-cell lymphomas.53 Our description in this study of the miRNA-, DROSHA-, and DICER-independent function of MIR17HG, via lnc-17-92, establishes this gene as having both short (miR-17-92) and long (lnc-17-92) noncoding RNA activities, with the latter mediating tumor-promoting activity in MM and likely other cancer contexts (eg, colorectal cancer).
We described lnc-17-92 as a specific regulator of gene expression via chromatin occupancy and interaction with MYC and WDR82. Lnc-17-92 directly reduces the expression of a small subset of genes and prevents the accumulation of H3K4me3 at the ACACA promoter. These effects are in contrast to what is observed by depleting c-MYC (ie, global effect on gene expression) or WDR82 (ie, global effect on methylation of H3K4) in cancer cells, as reported in this and other studies.44,54 Our data support the emerging paradigm whereby chromatin occupancy by transcription factors like MYC may be determined through interacting with specific lncRNAs,55 in addition to protein partners.26 In a broader perspective, our observations on lnc-17-92 suggest lncRNAs are key mediators of the epigenetic and transcriptional reprogramming of MM cells. In this molecular scenario, whereas proteins act as catalytic effectors, the intrinsic structural flexibility of lncRNAs makes them good modular scaffolds able to mediate both protein-protein and protein-DNA interactions at specific chromatin regions.
We further showed that the lnc-17-92–MYC–WDR82 complex impacts tumor cell metabolism by activating the de novo lipogenesis pathway via regulation of ACACA. This anabolic pathway is primarily restricted to liver and adipose tissue in normal adults but is reactivated in cancer cells via mechanisms yet to be fully described.38,56 Notably, MYC has been implicated in the reprogramming of tumor-cell metabolism by activating that pathway via ACACA and other genes.57 In turn, lipogenesis has emerged as an essential pathway for the onset and progression of MYC-driven cancers, which are susceptible to pharmacologic inhibition of ACC1.41 This seems particularly relevant in MM, in which tumor cells need to adapt their metabolic pathways to meet the high bioenergetic and biosynthetic demand posed by the malignant cell growth coupled with unceasing production of monoclonal immunoglobulin.58,59 Nevertheless, we acknowledge that the oncogenic roles of lnc-17-92 are likely not limited to the transcriptional axis with ACACA and will require further investigation to be comprehensively elucidated.
Deletion of MIR17HG is tolerated in adult mice,60 and its haploinsufficiency is compatible with life in humans.61 The physiological role of MIR17HG seems particularly relevant only for the hematopoietic stem cell compartment,60 which continuously renews. These observations support the development of MIR17HG as a target. Thus, for translational purposes, we developed 2 therapeutic ASOs that target the MIR17HG pre-RNA via different mechanisms of action (ie, RNase H–dependent or –independent). With the recent advances in RNA medicine,62-64 the use of ASOs to therapeutically antagonize disease-driver genes is becoming increasing possible,47,65 including in MM therapy.5,66 Our optimization of design and chemistry has helped to overcome the major obstacles to the clinical use of ASOs, such as poor bioavailability,65 while limiting off-target toxicity. The inhibitors described here bear state-of-the-art chemical modifications (2'MOE, phosphorothioated-backbone, lipid conjugation) and have sufficient nucleotide length (18 mer) to ensure high specificity for MIR17HG. Inhibitors of this kind have already been tested within clinical trials, and a few are already approved by the US Food and Drug Administration for use in different human diseases.65 Our optimized ASOs targeting MIR17HG with demonstrated activity in 3 different murine models of human MM provide the rationale to now consider clinical application in MM.
Different questions remain open about the dual nature of MIR17HG and its therapeutic targeting. Important future directions will be to uncover how the splicing of MIR17HG is alternatively regulated to produce lnc-17-92 or miR-17-92 and to address the relative contribution of lnc-17-92 and miR-17-92 to the oncogenic activity of MIR17HG in other cancer models. Whether the host genes of the 2 paralogs of miR-17-92, miR-106a-363 and miR-25-106b, also retain miRNA-independent function will be an important line of investigation.
Overall, this study establishes MIR17HG with a unique lncRNA function of facilitating protein-protein and protein-DNA interactions, mediating tumor-promoting activity with therapeutic implications.
Acknowledgments
The authors gratefully acknowledge the members of their laboratories for technical advice and critical discussions. The authors thank Christina Usher (Dana-Farber Cancer Institute) for editing the manuscript and insightful comments;Dirk Eick (Helmholtz-Zentrum München, Molecular Epigenetics), Christoph Driessen (Cantonal Hospital St Gallen, St Gallen, Switzerland), and Linda Penn (Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada) for providing the relevant cellular models; Pierosandro Tagliaferri (Magna Graecia University, Catanzaro, Italy) for insightful discussions on the dual long and short nature of MIR17HG; Benjamin Izar and Johannes Melms (Columbia University, New York, NY) for insightful discussions on CRISPR viability screen methodology; and the “8th Annual Miracles for Myeloma 5K Virtual Run/Walk” organizers and attendees for the fundraising in support to this research project.
This work is supported by NIH/NCI grants SPORE-P50CA100707, R01-CA050947, R01CA207237, P01CA155258, R01-CA178264 (N.C.M., K.C.A.); by VA Healthcare System grant No. 5I01BX001584 (N.C.M.); by the Paula and Roger Riney Foundation grant (N.C.M. and K.C.A.); by NIH/NCI grants RO1CA131945, R01CA187918, P50 CA211024 and Department of Defense grants DoD PC160357 and DoD PC180582 (M.L.); by the Sheldon and Miriam Medical Research Foundation (K.C.A.); by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology–5 per mille,” 2010/15 and its Extension Program No. 9980, 2016/18 (P.T.). E.M. is supported by a Brian D. Novis Junior Grant from the International Myeloma Foundation, by a Career Enhancement Award from Dana Farber/Harvard Cancer Center SPORE in Multiple Myeloma (SPORE-P50CA100707), by a Special Fellow grant from The Leukemia & Lymphoma Society, and by a Scholar Award from the American Society of Hematology. A.G. is supported by a Fellow grant from The Leukemia & Lymphoma Society and by a Scholar Award from the American Society of Hematology. J.E.H. is supported by a NIH F32 fellowship (NCI 1F32CA254216-01). A.N. is supported by a grant from the Italian Association for Cancer Research (AIRC, IG24365). N.A. is supported by a grant from the Italian Association for Cancer Research (AIRC, IG24449). K.C.A. is an American Cancer Society Clinical Research Professor.
Authorship
Contribution: E.M. and N.C.M. conceived and designed the research studies; E.M., M.F., and N.C.M. wrote the manuscript; M.K.S., A.A.-S., and K.T. performed in silico analysis of transcriptomic data; C.F.R. performed lipidomic studies; L.W.-L. performed yeast-3-hybrid experiments; J.E.H. performed RNA FISH, dual RNA FISH, and Co-IF/dual RNA FISH; S.T. generated MM cells expressing Cas9; W.D.P. analyzed ChIP-seq data; S.G. designed t-ASOs; C.F., N.A.R., D.M., and F.S. provided support for the identification of lnc-17-92 isoforms; M.F., A.G., D.G., N.L., D.M., N.A., M.J., G.B., C.L., Y.-T.T., A.N., D.C., T.H., M.A.S., P.T., R.A.Y., K.C.A., C.D.N., and M.L. contributed to the design and interpretation of key experiments; M.L. supervised lipidomic studies; and C.D.N. supervised Y3H experiments.
Conflict-of-interest disclosure: N.C.M. serves on advisory boards of and as consultant to Takeda, BMS, Celgene, Janssen, Amgen, AbbVie, Oncopep, Karyopharm, Adaptive Biotechnology, and Novartis and holds equity ownership in Oncopep. K.C.A. serves on advisory boards to Janssen, Pfizer, Astrazeneca, Amgen, Precision Biosciences, Mana, Starton, and Raqia and is a Scientific Founder of OncoPep and C4 Therapeutics. R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. E.M., S.G., and N.C.M filed a provisional patent on MIR17HG as a target for cancer therapy. D.C. reports other support from Stemline Therapeutics, Oncopeptides, and C4 Therapeutics outside the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, Dana-Farber Cancer Institute, Jerome Lipper Myeloma Center, Harvard Medical School, 450 Brookline Ave., Boston, MA 02215; e-mail: nikhil_munshi@dfci.harvard.edu; and Eugenio Morelli, Dana-Farber Cancer Institute, Jerome Lipper Myeloma Center, Harvard Medical School, 450 Brookline Ave., Boston, MA 02215; e-mail: eugenio_morelli@dfci.harvard.edu.
References
Author notes
Data reported in this article have been deposited in the General Expression Omnibus database (accession number GSE208599).
The authors declare that all data supporting the findings of this study are available within the article and its supplemental information. Files or reagents are available from the corresponding authors, Nikhil C. Munshi and Eugenio Morelli (nikihil_munchi@dfci.harvard.edu and eugenio_morelli@dfci.harvard.edu) on request.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal