Abstract
Myeloproliferative neoplasms (MPN) are a group of clonal stem cell–derived hematopoietic malignancies driven by aberrant Janus kinase-signal transducer and activator of transcription proteins (JAK/STAT) signaling. Although these are genetically simple diseases, MPNs are phenotypically heterogeneous, reflecting underlying intratumoral heterogeneity driven by the interplay of genetic and nongenetic factors. Their evolution is determined by factors that enable certain cellular subsets to outcompete others. Therefore, techniques that resolve cellular heterogeneity at the single-cell level are ideally placed to provide new insights into MPN biology. With these insights comes the potential to uncover new approaches to predict the clinical course and treat these cancers, ultimately improving outcomes for patients. MPNs present a particularly tractable model of cancer evolution, because most patients present in an early disease phase and only a small proportion progress to aggressive disease. Therefore, it is not surprising that many groundbreaking technological advances in single-cell omics have been pioneered by their application in MPNs. In this review article, we explore how single-cell approaches have provided transformative insights into MPN disease biology, which are broadly applicable across human cancers, and discuss how these studies might be swiftly translated into clinical pathways and may eventually underpin precision medicine.
Introduction
Philadelphia chromosome negative myeloproliferative neoplasms (MPNs) are a group of hematologic malignancies1,2 that result from unrestrained Janus kinase-signal transducer and activator of transcription proteins (JAK-STAT) signaling,3,4 causing increased proliferation of 1 or more myeloid blood cell lineages. Due to their long disease latency, well characterized genetic driver mutations, and ease of accessibility of serial samples, MPN have long been studied as a paradigm model of cancer evolution. Many scientific questions related to MPN biology have remained the same over decades. However, our ability to address these questions has improved dramatically with the advent of advanced cellular and molecular biology techniques. Single-cell approaches are ideally positioned to unravel the complex interplay of genetic and nongenetic elements that collectively drive MPN pathogenesis. We are witnessing an incredibly exciting time in the field, as single-cell technologies have exponentially developed in the past decade from microarrays applied to a handful of murine hematopoietic stem cells (HSCs)5 to today, where it is possible with extraordinary resolution to simultaneously to profile several “layers” of information in millions of single cells. In this review, we explore how single-cell approaches have provided unprecedented insights into MPN disease biology and how these studies are likely to be implemented into clinical pathways and, eventually, genomics-tailored precision medicine in the coming years.
MPNs: a handful of recurrent mutations lead to diverse clinical phenotypes
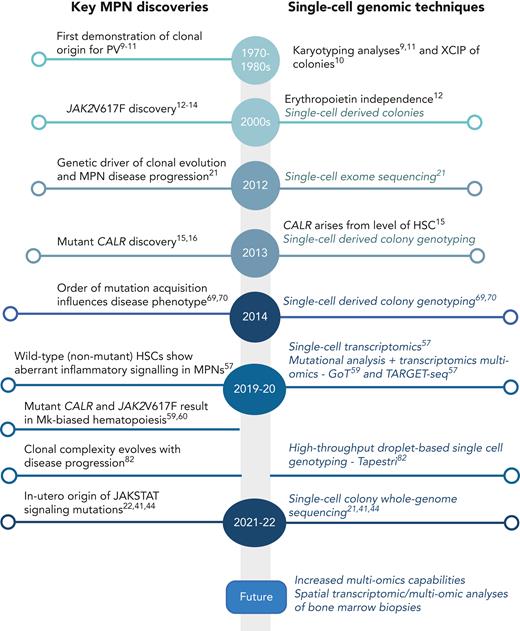
MPNs are typically initiated by the acquisition of a somatic mutation in a single HSC,6-8 leading to clonal expansion. Genomic analysis of single-cell–derived colonies has long been applied as a key methodology to study MPN, and cytogenetic analyses of single cells in patients with polycythemia vera (PV) provided one of the earliest demonstrations of clonality in these diseases (Figure 1).9-11 The most frequent mutation driving MPN development is a single nucleotide base change in exon 14 of the JAK2 gene, the JAK2V617F mutation,12-14 followed by an insertion or deletion in the calreticulin (CALR) gene15,16 and less commonly by a single nucleotide base change in the myeloproliferative leukemia virus (MPL) gene.17 Again, single-cell genomics played a role in the discovery of these mutations, with the demonstration of erythropoietin-independent single-cell–derived colony forming ability being driven by JAK2V617F (Figure 1).12 These 3 MPN driver mutations are predominantly mutually exclusive and all cause constitutive activation of JAK-STAT signaling pathways.18 Additional mutations in other genes (eg, epigenetic [TET2, DNMT3A], splicing factor [SF3B1, ZRSR2, SRSF2], metabolic [IDH1, IDH2], and other pathways [KRAS, NRAS, TP53]) are also found in MPNs, more commonly in advanced disease. Single-cell colony genotyping including cytogenetic analysis has historically provided insights into genetic heterogeneity and clonal evolution in MPNs,15,19-21 which typically heralds progression to accelerated or blast phase disease. While additional mutations are most commonly acquired sequentially within the single HSC derived clone, genomic analyses at the single-cell level have also revealed cases where separate clones arising from different HSCs have expanded in parallel.8,19,22
Timeline of key discoveries in MPNs and single-cell methodologies. Mk, megakaryocyte; XCIP, X-chromosome inactivation pattern.
Timeline of key discoveries in MPNs and single-cell methodologies. Mk, megakaryocyte; XCIP, X-chromosome inactivation pattern.
Currently, MPNs are categorized using clinical and histological criteria into essential thrombocythemia (ET), PV, and myelofibrosis (MF).1,23 Recently, a “prefibrotic” MF stage was defined: “pre-MF.”1,24 ET and PV are chronic MPNs that, for the majority of patients, are relatively indolent, albeit with a significant impact on quality of life, with an increased risk of thrombosis/hemorrhage and excess mortality compared to healthy age-matched controls.25 MF is characterized by bone marrow fibrosis, osteosclerosis, and often gross splenomegaly due to extramedullary hematopoiesis. The sequelae of MF include cytopenias and transfusion dependence, increased infections, severe constitutional symptoms, and early mortality. All MPNs are preleukemic malignancies, with an overall risk of leukemic progression of 10% to 20% for MF and 1% to 4% for PV and ET.26
In recent years, a move to integrate genomic features in MPN classification has been initiated, and this more precisely predicts clinical outcomes than clinical parameters alone.27 The majority of patients with MPNs carry the same genetic lesion: JAK2V617F. The acquisition of this mutation results in a remarkable range of clinical presentations. JAK2V617F is also detected in ∼3% of healthy individuals with normal blood counts, denoted clonal hematopoiesis (CH).28,29 In the context of CH, JAK2V617F is associated with a heightened risk of cardiovascular and overall mortality as well as the development of overt myeloid neoplasms.30 Recent mathematical modeling tools have successfully estimated the “fitness” of specific mutations.31 Clinical implementation of such tools may be helpful to identify clones that are more likely to progress to overt disease.
In a minority of individuals with CH, JAK2V617F results in MPN, which can manifest in a variety of ways, ranging from an incidental finding in an asymptomatic patient to an individual with a severe symptom burden and a significantly shortened life expectancy, such as in patients with MF. The likelihood of disease progression is determined by an interplay of factors: the relative clonal advantage of a mutant HSC and its progeny,8 the acquisition of additional mutations, germ line genetics,32 host factors (age and gender), and cell-extrinsic mediators including inflammation and environmental exposures.33,34 Single-cell technologies can be utilized to unravel these multiple factors, indeed, hematologic malignancies, particularly MPNs, are often used as a model for showcasing novel single-cell technologies. In addition to readily accessible study materials, MPNs present a window of opportunity to study cancer evolution, as the majority of patients present in an early disease phase, and only a small proportion progress to aggressive disease.
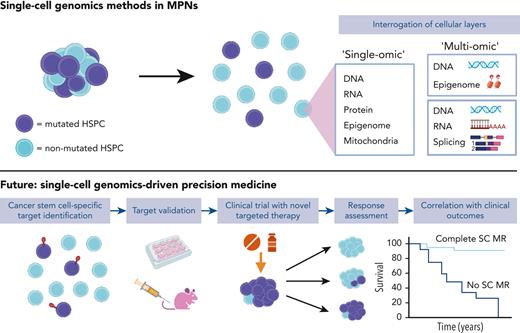
Single-cell genomics: reaching “prime-time” after a decade of rapid technological advances
The first methodologies for studying single cells date back to the 1960s, with the advent of flow cytometry, enabling the detection of surface proteins on millions of individual cells assayed simultaneously. Technologies to analyze genomic material from a single cell also have a long history, initially through karyotype analysis, a single-cell genomics technique still in routine clinical use. Single-cell genomics has advanced at an impressive pace, from single modality analysis of a particular genomic layer—such as DNA, transcriptome/gene expression or epigenome—to the capture of multiple layers of genetic, epigenetic, and also proteomic information simultaneously in “multiomic” techniques (Figure 2).35,36 This multilayered information is key to unraveling the interacting elements driving cancer development, evolution, and resistance to therapy. These experimental advancements have necessitated parallel innovations in computational methods37 to represent and integrate multimodal insights at scale.
Single-cell technologies. Layers of information in the cell can be interrogated by single-cell technologies including DNA, RNA or transcriptome, mitochondrial,38 protein (surface39 or intracellular40), epigenetic, and splicing. Technologies combining multiple modalities, “multiomic,” are highlighted and those modalities applied to study of MPNs are highlighted by an orange star.
Single-cell technologies. Layers of information in the cell can be interrogated by single-cell technologies including DNA, RNA or transcriptome, mitochondrial,38 protein (surface39 or intracellular40), epigenetic, and splicing. Technologies combining multiple modalities, “multiomic,” are highlighted and those modalities applied to study of MPNs are highlighted by an orange star.
Insights into the origins and latency of MPNs
Single-cell analyses have recently facilitated one of the most fundamental shifts in our understanding of the origins of MPNs, and a new paradigm for cancer development. Two recent studies22,41 revealed that the steady rate of mutation acquisition could be used to establish a “molecular clock,” that could trace the timing of mutation acquisition in MPN patients, similar to what was shown in a large study of somatic mutations across 36 cancer types.42 In one study, over 1000 single-cell–derived colonies from 12 MPN patients were subject to whole genome sequencing. Strikingly, the reconstruction of mutation lineage trees revealed that in many patients, the driver mutations were acquired in early postnatal life, or even in utero, with a latency between JAK2V617F mutation acquisition to clinical presentation ranging from 11 to 54 years.22 The same methodology was also applied to study 2 patients with MPN, reaching similar conclusions that driver mutations were acquired early in life with a long disease latency.41
The acquisition of somatic mutations many decades before disease onset diverges from the current dogma of MPN disease biology. Mutations in genes associated with CH, such as DNMT3A and PPM1D,43 are also thought to occur early in childhood. Rates of clonal expansion differ between patients, suggesting that additional contributors (eg, environment, gender, bone marrow niche, germ line factors, and intrinsic attributes of the cell-of-origin in which the mutation arises) are likely to influence the timing of disease emergence.
Further evidence of the key role of germ line genetics in disease latency arises from a recent study of 2 monozygotic twins who presented with MPNs in their fourth decade of life. Whole genome sequencing lineage tracing combined with single-cell–derived colony genotyping demonstrated a common clonal origin of the MPN in both twins, due to transplacental transfer of a CALR mutant clone in utero.44 First degree relatives carry a 5 to 7 fold increased risk of MPN,45 and the 46/1 JAK2 haplotype46 has long been known to predispose to JAK2-mutated and MPL-mutated MPN47 (but not CALR48). Genome-wide association studies have identified other MPN predisposing genetic variations.49,50 A recent large-scale study pairing genome-wide association studies with single-cell methodologies32 found 17 genetic risk traits that confer risk, which were collectively estimated to account for almost 20% of the familial relative risk for MPN. The identified risk variants were mapped to likely target genes, creating a 15 gene “MPN signature.” The expression of these genes was examined in 250 000 single hematopoietic cells from healthy donors, demonstrating that the MPN signature overlapped with HSC genes. This implied that MPN risk alleles may expand the HSC population and thereby increase the likelihood of a single HSC acquiring an MPN mutation. An alternative possibility is that MPN risk alleles modify the clonal advantage that an MPN driver mutation confers to an HSC.
Importantly, the finding that MPN driver mutations occur early in life and that genetic predisposition is determined at birth reveals that the window of opportunity for early intervention is wider than previously appreciated, necessitating a refocusing of our efforts to understand why, and in whom, the mutated clone will develop a competitive advantage.
Cellular heterogeneity within MPN stem cells confer therapeutic vulnerabilities
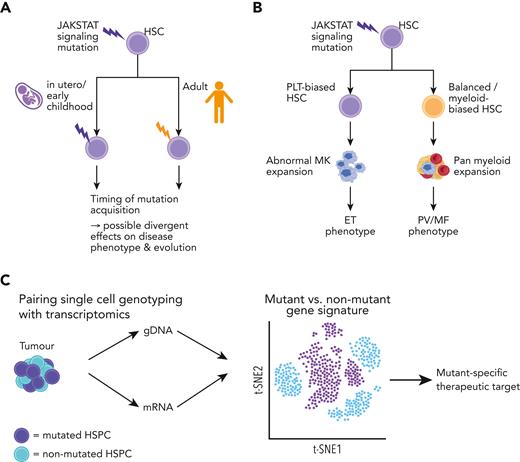
There is an increasing interest in the diversity of cellular subtypes that constitute MPN HSCs and how they differ from their normal counterparts. Substantial heterogeneity exists within healthy HSCs, with subsets biased to give rise to particular blood lineages.51 This includes the presence of platelet-biased HSCs, which have been identified using both barcoded lineage tracking52 and single-cell transplantation experiments.53,54 Platelet-biased HSCs increase in number with age,55 and may enable rapid “emergency” platelet generation.56 HSC heterogeneity may explain phenotypic heterogeneity in MPNs; for example, a mutation acquired by a fetal vs adult HSC might result in distinct clonal trajectories and disease features (Figure 3A). One intriguing hypothesis is that a JAK-STAT signaling mutation acquired in a multipotent platelet-biased HSC might lead to an ET phenotype, while acquisition in a lineage-balanced HSC might cause PV (Figure 3B). However, to date, this has not been convincingly demonstrated, partly because of the lack of availability of models to accurately study this.
Heterogeneity in the MPN stem and progenitor cell compartments. (A) Proposed model of factors affecting JAK-STAT signaling mutation acquisition in the MPN cell-of-origin (HSC) that may influence the disease phenotype/risk of evolution. The timing of mutation acquisition can be as early as fetal origin to adult onset and may affect disease evolution. (B) Mutation acquisition in a lineage-biased HSC may influence the disease phenotype; a mutation arising in a PLT-biased HSC leading to an ET phenotype, whereas in a balanced/myeloid-biased HSC may lead to a PV/MF phenotype. (C) Intrapatient single-cell multiomic analysis of mutant and nonmutant HSPC by applying methodologies (TARGET-seq,57,58 GoT59) to detect mutation information (genomic DNA) and gene expression (messenger RNA) to uncover mutant-specific gene signatures and potential therapeutic targets. ET, essential thrombocythemia; HSPCs, hematopoietic stem and progenitor cells; HSC, hematopoietic stem cell; MK, megakaryocyte; PLT, platelet; PV, polycythemia vera; MF, myelofibrosis.
Heterogeneity in the MPN stem and progenitor cell compartments. (A) Proposed model of factors affecting JAK-STAT signaling mutation acquisition in the MPN cell-of-origin (HSC) that may influence the disease phenotype/risk of evolution. The timing of mutation acquisition can be as early as fetal origin to adult onset and may affect disease evolution. (B) Mutation acquisition in a lineage-biased HSC may influence the disease phenotype; a mutation arising in a PLT-biased HSC leading to an ET phenotype, whereas in a balanced/myeloid-biased HSC may lead to a PV/MF phenotype. (C) Intrapatient single-cell multiomic analysis of mutant and nonmutant HSPC by applying methodologies (TARGET-seq,57,58 GoT59) to detect mutation information (genomic DNA) and gene expression (messenger RNA) to uncover mutant-specific gene signatures and potential therapeutic targets. ET, essential thrombocythemia; HSPCs, hematopoietic stem and progenitor cells; HSC, hematopoietic stem cell; MK, megakaryocyte; PLT, platelet; PV, polycythemia vera; MF, myelofibrosis.
Single-cell studies have also provided insights into normal hematopoiesis, which is disrupted in MPNs, through analysis of nonmutant HSCs. These cells, which are not part of the MPN clone, also show aberrant gene expression with increased expression of inflammatory pathways57 and expression of certain megakaryocyte-affiliated genes, such as vWF.60 Such insights were made possible using TARGET-seq,57,58 a highly sensitive approach enabling simultaneous mutation and RNA-sequencing analysis in individual cells, combined with cell surface proteomics using index flow cytometry. TARGET-seq requires the isolation of individual cells into 96- or 384-well plates and is therefore, lower throughput than droplet-based approaches such as Genotyping of Transcriptomes (GoT), which is based on the 10× Genomics platform.59 However, the high sensitivity enables reliable determination of “true” wild-type cells from mutant cells, where the presence of the mutation has simply not been detected due to dropouts in the sequencing reads. This precise intrapatient comparison of wild-type vs mutant hematopoietic stem and progenitor cells (HSPCs) is a powerful approach to identify mutation-specific distinct molecular signatures, and potential therapeutic vulnerabilities (Figure 3C).60
Simultaneous profiling of mutation status and transcriptomes using GoT was first applied to HSPCs from patients with CALR-mutated (CALRmut) ET.59 Mutation detection was achieved with this technique by including a gene-specific primer at the first cDNA polymerase chain reaction step using the 10× Genomics platform. CALRmut HSPCs were highly clonal, with the mutation detected throughout the HSPC compartment, with the highest frequency in megakaryocyte progenitors (MkPs). Mutant MkPs showed an increase in cell-cycling genes, suggesting that CALRmut conferred a proliferative advantage for MkPs, in keeping with its ability to specifically bind to the TPO-receptor. Mutant MkPs exhibited an upregulation of unfolded protein response genes, as identified in vitro previously61 and subsequently through single-cell RNA-sequencing analysis of murine HSCs in a CALRmut ET model.62 In comparison, CALRmut MF HSPCs showed no enrichment for the mutation in a specific population, but mutant MkPs did show higher TGFβ levels which corresponded to bone marrow fibrosis and unfolded protein response genes were upregulated specifically in mutant MkPs.59
Other studies have also highlighted the expansion of megakaryocyte-biased HSCs in MPNs. Analysis of >120 000 HSPC transcriptomes demonstrated a marked expansion of megakaryocyte-biased HSPCs in MF in patients with both JAK2V617F and CALR mutations.60 Although a proportion of MkPs from MF patients were similar in their gene signatures to healthy donor-derived MkPs, there was an expansion of MkP subtypes with gene expression signatures associated with proliferation and fibrosis, in keeping with prior data proposing MkPs as key drivers of fibrosis development in MPNs.63,64 Two subsequent studies applying single-cell genomics to HSPCs from patients with ET and PV also found expansion of megakaryocyte-biased HSCs in nonfibrotic MPNs,65,66 and a study coupling JAK2V617F genotyping of single HSPCs with gene expression in untreated patients with ET and PV noted an increase in interferon (IFN) response genes in JAK2V617F-mutated HSPCs.67 The presence of megakaryocyte-biased HSCs in MPNs has therapeutic implications. In ET, HSCs carrying the JAK2V617F mutation were hyperresponsive to IFN stimulation, enhancing megakaryocyte bias.65,66 In addition, megakaryocyte-biased HSCs were shown to have impaired long-term repopulating abilities, and IFN treatment exacerbated this shift. Chronic exposure of mice to IFN led to exhaustion of the mutant stem cell pool, which may explain why IFN therapy leads to a clonal response in some patients. In early single-cell transcriptomic analysis of HSCs in a JAK2V617F-murine model, increasing age was associated with an expansion of HSCs with a p53-related senescence signature suggestive of a decline in stem cell function, showing the utility of these methods to identify JAK2V617F-mutation associated cellular heterogeneity.68
The megakaryocyte bias of MF HSCs raises the possibility that these disease-driving stem cells may be selectively targeted by their aberrant expression of megakaryocyte-affiliated genes.60 For example, TARGET-seq analysis showed that G6B is specifically upregulated in mutant and not wild-type HSPCs in MF.
Together with other work in the field, these studies highlight the power of single-cell technologies to interrogate the molecular signatures associated with disease-driving subpopulations and uncover novel therapeutic strategies.60
Dissecting clonal hierarchies and the order of mutation acquisition and implications for MPN phenotype
In genetically complex solid tumors, understanding the order in which mutations are acquired is essential to understand cancer evolution. However, it has historically been difficult to study this using bulk genomics techniques. This is more feasible in genetically simpler malignancies such as MPNs, where single-cell colony genotyping studies have shown that the order in which mutations occur can dictate the disease phenotype. This concept was first highlighted by genotyping single-cell–derived colonies from MPN patients comutated for JAK2V617F and TET2,69 demonstrating that “JAK2-first” was associated with PV, whereas “TET2-first” was more common in ET. Similar observations in a separate study showed that patients with “DNMT3A-first” had an ET phenotype.70
In addition to colony genotyping, plate-based single-cell approaches can also resolve genetic intratumoral heterogeneity, including the order of mutations, for example, by showing that spliceosome mutations typically occur before MPN driver mutations.57 These approaches can also be used to establish zygosity, relevant for JAK2V617F and TP53, where allelic status strongly influences disease biology.57,71 High-throughput, droplet-based single-cell genotyping platforms have similarly been applied to understand clonal architecture. One example is Tapestri, a droplet-based platform that enables the targeted mutational analysis of millions of cells simultaneously. This platform was applied to study 2 unusual cases in which 2 driver mutations, JAK2/CALR and JAK2/MPL, were detected in the same patient, analyzing single-cell DNA of ∼20 000 peripheral blood mononuclear cells.72 These authors clearly showed that the mutations resided in separate clones. However, it is difficult to distinguish MPN mutation co-occurrence from a single driver mutation MPN occurring together with a “bystander” CH. Because the order of mutations as well as zygosity of certain mutations can have important clinical implications for disease classification and therapy response prediction, this is an area where single-cell genomics may have clinical utility over bulk-sequencing approaches which cannot precisely resolve mutation order or zygosity of subclones.57,69,71
MPN cells finding their niche
The bone marrow niche or microenvironment comprises cellular and structural components that are integral to the support and maintenance of hematopoietic cells. In many hematologic malignancies, including MPNs, this microenvironment is disrupted.33 In MPN, there is considerable cross talk between malignant HSPCs and nonhematopoietic cells, occurring by direct cell-cell contact as well as the release of proinflammatory/fibrotic signals, leading to a “self-reinforcing” malignant niche that promotes malignant progression.33,73 Understanding MPN clone-niche interactions is particularly important in MF, as successful treatment may require reversal of bone marrow fibrosis.
Myofibroblast cells are thought to be directly responsible for the deposition of excess collagen and abnormal matrix proteins74 in MF, other hematologic disorders, and disease-associated fibrosis in other organs in both malignant and nonmalignant pathologies.75 Bone marrow myofibroblasts can originate from mesenchymal stromal cells (MSC).76,77 Single-cell analyses of the normal murine bone marrow microenvironment discovered substantial cellular heterogeneity,78 and elucidated multiple fibroblast populations.79 Interestingly, human bone marrow studies have struggled to distinguish MSCs and fibroblasts,80 suggesting either important interspecies differences or that the isolation of fibroblasts is very difficult from the limited stromal material captured in bone biopsies.
Single-cell transcriptomic analysis of bone marrow niche cells in MPN in mice and humans81 revealed the presence of specific disease-associated MSC populations with enhanced expression of the alarmin complex S100A8/S100A9, proinflammatory molecules typically expressed only in myeloid cells and not in the stroma. Therapeutic relevance was demonstrated, as the authors showed that the treatment of JAK2V617F+ mice with tasquinimod, an inhibitor of S100A8/S100A9, improved BM fibrosis.
Progression of MPNs to accelerated phase or secondary leukemia
Increasing genetic complexity generally correlates with progression to more advanced disease stages (MF or leukemia).27 A recent study examined clonal architecture and evolution by targeted single-cell genotyping of 146 samples and ∼750 000 individual myeloid cells from individuals with CH as well as myeloid malignancies including acute myeloid leukemia (AML) and MPNs.82 This analysis found clonal complexity was higher for AML than for MPN. A significant benefit of the Tapestri platform is its throughput, which is currently limited to the analysis of DNA and protein information. Future iterations may enable simultaneous analysis of gene expression, which is required to disentangle the cellular mechanisms associated with clonal evolution.
A recent study focused on the mechanisms of leukemic progression associated with TP53,71 a tumor suppressor gene that is commonly mutated and is associated with adverse outcomes. In myeloid neoplasms, the presence of a TP53 mutation is associated with complex cytogenetic abnormalities and dire outcomes.83,84 In leukemic phase MPN, TP53 mutations are present in one-third of cases84,85 but small TP53 clones are found in a sizable cohort (15%) of chronic phase MPN86 who do not transform to AML. Biallelic loss of wild-type TP53 is frequent in leukemic phase MPN87 and other advanced myeloid neoplasms,83 suggesting that a multihit mechanism is required for leukemia development. In a recent study, ∼15 000 single HSPCs were analyzed in 14 advanced phase MPN cases. In all cases, a biallelic loss of wild-type TP53 was observed in the principal clone. The “multi-hit” mutant HSPCs contained 2 broad cellular subsets; one showing an erythroid gene signature, and the other enriched in leukemia stem cell genes. The finding of expanded erythroid-biased HSPCs was not expected, as leukemias secondary to MPNs are not typically erythroleukemia according to standard clinical criteria.26 Wild-type patient HSCs showed a reduction in cell cycle genes, suggestive of enhanced quiescence and increased expression of inflammatory genes, leading to the hypothesis that a cell-extrinsic inflammatory process mediates disease evolution and exerts a fitness advantage to the TP53 mutant clone while repressing wild-type HSPCs, potentially through DNA damage-related mechanisms. This study is another example of how single-cell multiomic studies applied in MPN can have broad relevance for human disease, given that TP53 is the most commonly mutated gene in human cancer.88
Insights into therapy response in MPN
Currently available therapies for MPN show modest effects on modifying MPN biology or mitigating disease evolution for most patients, including the first “targeted” drug, ruxolitinib. Despite substantial efficacy in terms of spleen and symptom responses,89,90 improvements in bone marrow fibrosis91 and mutation allele burdens are uncommon with ruxolitinib. The effect of ruxolitinib on the clonal architecture of HSPCs in MF has been studied.92 Targeted single-cell genotyping was performed on sequential samples in 8 patients using a multiplexed, quantitative PCR microfluidics system (Fluidigm) to assemble the HSPC clonal hierarchy per patient. JAK-STAT signaling mutations were the initial event in most patients, except in 2 cases where an SF3B1 mutation was the founder mutation. Cases with subsequent leukemic disease progression were enriched for RAS signaling pathway mutations (NRAS and KRAS) which have also been associated with adverse prognosis in MF by other groups93,94; increased risk of leukemic transformation, shortened survival and poor response to JAK inhibitors.94 This highlights the potential for single-cell analysis in studying serial samples from patients before and after therapy to provide insights into predictors of disease response, and resistance.95
The future I: new multiomic tools
Single-cell multiomics is advancing at breakneck speed. Interrogating the epigenome and spliceosome in parallel with gene expression and mutation information will be particularly powerful for dissecting the mechanisms that drive MPN evolution with improved granularity. Parallel gene expression and chromatin accessibility (ATAC-seq)96 promises to improve our understanding of cell states and regulatory programs. GoT-ChA97 combines mutation and epigenetic information (chromatin accessibility) in a high-throughput droplet-based platform to understand how mutant- and cell-specific epigenetic regulation influences cell state and differentiation. The application of this approach to study JAK2V617F CD34+ MPN HSPCs revealed a proinflammatory signature and increased myeloid/erythroid epigenetic priming in mutant HSPCs. Studying aberrant splicing in mutant cells is also essential to identify abnormal splicing signatures and potentially targeted therapies. GoT-Splice98 integrates GoT59 with long read sequencing99 to capture the full-length transcriptome and therefore combines genotyping, gene expression, and splicing information of each cell. This is particularly relevant for MPNs, where mutations in splicing factors are common and the resulting splice variants may be targetable.
The future II: spatial technologies and translation of single-cell technologies to clinical pathways
Single-cell technologies have undoubtedly led to significant advances in our understanding of MPN biology, and have identified potential druggable targets. However, many of the platforms outlined in this review are experimentally and computationally demanding and costly, impeding their routine adoption in clinical settings. In contrast, advances in histological analysis and the application of machine learning techniques to classify thousands of single cells based on morphology will be well placed for integration into routine care in the near future.
To date, histological methods for MPN diagnosis in clinical laboratories have relied on subjective assessment with significant interobserver variability.100,101 Furthermore, integrating histology with clinical and genomic parameters is essential.24 Megakaryocytes display characteristic morphological changes in MPNs and are ideal for assessment using machine learning algorithms. A need for this is clear, given the inclusion of qualitative megakaryocyte morphological features in the World Health Organization diagnostic criteria for distinguishing Pre-MF from ET and the poor interobserver concordance.102 Applying deep learning methods to digitized images holds the promise of improving the accuracy and efficiency of megakaryocyte assessment and could be widely accessed worldwide. Such techniques have shown encouraging results in solid tumor malignancies,103 and recently an automated imaging analysis method applying machine learning models was implemented to characterize megakaryocyte morphology and topography in MPNs.104 The frequency of megakaryocyte subtypes was found to correlate with the disease subtype and distinguish reactive and MPN marrows using features that are not typically discernible by standard histological assessment. This highlights the potential of single-cell spatial techniques in the clinical setting and is likely to have value beyond diagnosis, for example, identifying features that signal early disease progression and monitoring responses to therapy. Furthermore, analysis of cells in suspension loses vital information about cell-cell relationships and tissue architecture. This can be addressed with spatial transcriptomic methodologies, now becoming available, that can study cells in their native tissues.105,106 Applying techniques that maintain the 3D structure of the bone marrow is particularly crucial in diseases such as MPN where cell-cell interactions are so central to disease biology.
Beyond applying computational methodologies such as machine learning tools for improved diagnostics, mathematical modeling is increasingly recognized to be helpful in understanding disease evolution and implementation in clinical practice, for example, to improve clinical decision making and prediction of disease progression. Such modeling proved successful in predicting AML development in a cohort of patients where pretransformation samples were available.107 These tools could similarly be applied in MPNs to refine the prediction of the likelihood of disease evolution, with a potential for earlier therapeutic intervention.
Single-cell genomics in MPN: the end of the beginning
Single-cell genomics is rapidly transforming the MPN field, with a number of companies employing single-cell genomics as a key platform for therapy discovery, including MPN.108,109 Although costs and technological challenges remain prohibitive, an analogy can be drawn with whole genome sequencing, which was an expensive research tool a decade ago110 but is now offered in routine clinical care for <$1000/sample. A comparable trajectory could be expected for single-cell technologies in the coming years.
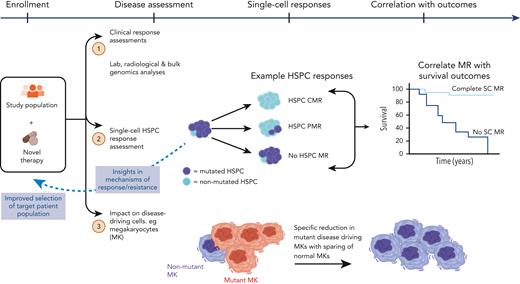
How will single-cell omics be integrated into clinical pathways? There is a clear application for improving the accuracy of diagnosis, disease stage stratification, and the prediction and monitoring of therapy response, resistance, and relapse risk. Using single-cell techniques, mutant cells can be identified and tracked during treatment, providing a more sensitive method for measuring clonal responses and clonal evolution and thereby more precisely predicting the risk of relapse or disease progression. However, in our view, the next step is to integrate exploratory end points requiring single-cell genomics in MPN clinical trials (Figure 4). Single-cell analyses have been conducted on clinical trial subjects in AML,111-113 but these analyses have not been embedded in the trial design from study inception. Disease modification is a key aim of MPN therapies in development, and ultimately, this requires the elimination of MPN stem cells, which can only be reliably ascertained with single-cell analysis of HSPCs. This will help develop an evidence-base for application in routine clinical care and realize the potential of single-cell technologies to advance personalized genomics-driven medicine.
Application of SC technologies in clinical trials. Parallel analysis of mutant HSPCs to assess the response to novel therapies and correlate with disease outcomes. HSPCs, hematopoietic stem and progenitor cells; HSPC CMR, complete molecular response (no mutation detected in HSPC compartment); HSPC PMR, partial molecular response (mutation reduction >50% in HSPC compartment); No HSPC MR, no molecular response (no mutation reduction in HSPC compartment); SC, single-cell.
Application of SC technologies in clinical trials. Parallel analysis of mutant HSPCs to assess the response to novel therapies and correlate with disease outcomes. HSPCs, hematopoietic stem and progenitor cells; HSPC CMR, complete molecular response (no mutation detected in HSPC compartment); HSPC PMR, partial molecular response (mutation reduction >50% in HSPC compartment); No HSPC MR, no molecular response (no mutation reduction in HSPC compartment); SC, single-cell.
Acknowledgment
Figures created with BioRender.com.
B.P. is funded by a Cancer Research UK Advanced Clinician Scientist Fellowship (C67633/A29034) and an Oxford BRC Senior Research Fellowship. AJM is funded by a Cancer Research UK Senior Cancer Research Fellowship (C42639/A26988)
Authorship
Contribution: J.M.O., A.J.M., and B.P. wrote the paper.
Conflict-of-interest disclosure: B.P. is a cofounder for Alethiomics, has provided paid speaking engagements or consulting, advisory services for Alethiomics, Constellation Therapeutics, Blueprint Medicines, and Novartis, and has received research funding from Alethiomics, Galecto, and Evotec. A.J.M. is a cofounder for Alethiomics, has participated in advisory boards for Alethiomics, Novartis, BMS, GSK, and CTI, and has received research funding from Alethiomics, Galecto, and Novartis. J.M.O. declares no competing financial interests.
Correspondence: Jennifer Mary O’Sullivan, Medical Research Council Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: jennifer.osullivan@rdm.ox.ac.uk; Adam J. Mead, Medical Research Council Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: adam.mead@imm.ox.ac.uk; and Bethan Psaila, Medical Research Council Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: bethan.psaila@ndcls.ox.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal