TO THE EDITOR:
Myelodysplastic syndrome (MDS) represents a heterogeneous group of disorders encompassing a wide range of pathologically distinct subtypes associated with different outcomes. Until recently, clinical and cytogenetic features have dominated the pathological classification and the prognostication of MDS.1-3 The wide use of next-generation sequencing and subsequent molecular studies have examined the impact of different genetic alterations on the pathogenetic derivation, diagnostic assignment, and disease prognosis.4-9 The latest contribution to the task of adopting molecular features into the clinical prognostic scheme is represented by the Molecular International Prognostic Scoring System (IPSS-M), a product of the International Working Group for Prognosis in MDS (IWG-PM).10 Here, we took advantage of a large cohort of patients with MDS (n = 1281, supplemental Methods, available on the Blood website) to validate the novel IPSS-M and to compare the score with the well-established revised IPSS (IPSS-R). To present our clinical experience, we used a cohort of treated and untreated patients with MDS. Informed consent was obtained according to the protocols approved by the institutional review board of the Cleveland Clinic Foundation and in accordance with the ethical principles set forth by the Declaration of Helsinki.
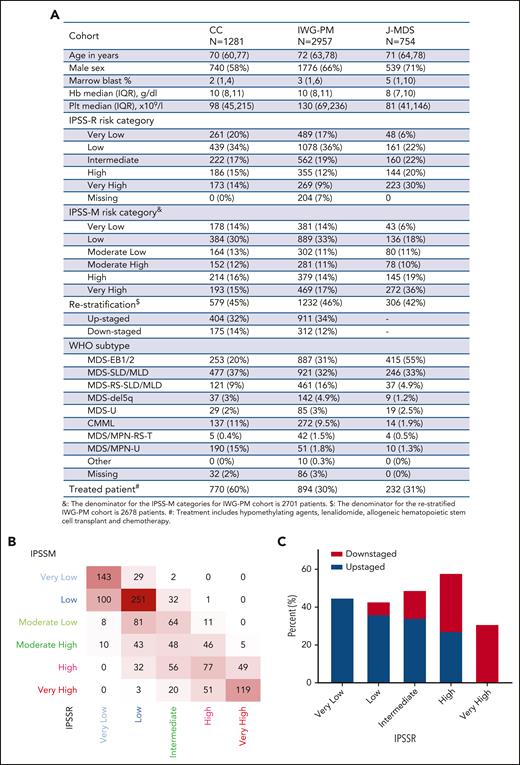
Overall, MDS–single-lineage dysplasia (SLD) and MDS–multilineage dysplasia (MLD) were the most common World Health Organization 2016 subtypes (477/1281, 37%), whereas MDS–excess blasts 1/2 (EB1/2) represented 20% (253/1281) of the cohort. The median age was 70 years (interquartile range: 62-77), and 58% (740/1281) of patients were male (Figure 1A). Somatic mutations along with karyotypic abnormalities were similar to the ones analyzed in the IWG-PM cohort. As expected, our cohort did not significantly vary from the IWG-PM cohort, but we had a higher percentage of treated patients (60% vs 30% in the IWG-PM cohort).10 According to IPSS-R, cases (n = 1281) were assigned to very-low-risk (20%, 261), low-risk (34%, 439), intermediate-risk (17%, 222), high-risk (15%, 186), and very-high-risk (14%, 173) categories. When applying IPSS-M algorithm to our cohort vs the IWG-PM, patients were restratified at similar rates: very-low-risk (14% vs 14%), low-risk (30% vs 33%), moderate-low-risk (13% vs 11%), moderate-high-risk (12% vs 11%), high-risk (16% vs 14%), and very-high-risk (15% vs 17%) categories (supplemental Figure 1A). MDS-EB1/2 cases were enriched among high-risk (28%, 60/214) and very-high-risk (42%, 81/193) IPSS-M groups (supplemental Figure 1B, upper panel). Bone marrow blasts percentage followed an incremental pattern from very-low-risk to very-high-risk subgroups (supplemental Figure 1B, lower panel). However, 60% (770/1281) of the patients were treated during the disease course with hypomethylating agents (HMA), allogeneic hematopoietic stem cell transplantation (HSCT), or lenalidomide (supplemental Figure 1C). Among the patients in the low-risk subgroup (n = 336), 38% were treated with HMA at one point of their disease (usually at progression), 16% were treated with lenalidomide, and 14% underwent allogeneic HSCT. Overall, 59% of the patients treated with HMA (n = 624) were assigned to moderate-high-risk, high-risk, or very-high-risk groups. To that end, disease-modifying treatment included HMA, lenalidomide, and allogeneic HSCT similar to what was reported in the original IWG cohort.10
Baseline characteristics of the validation cohort. (A) Characteristics of the validation cohort as compared with the original and the validation cohorts of the IPSS-M study. (B) Cross heat map for distribution of patients with MDS according to the IPSS-R (x-axis) and the IPSS-M (y-axis) scores. (C) Bar histogram shows the total percentage of patients restratified (upstaged or downstaged) according to the IPSS-M. CC, Cleveland Clinic cohort; CMML, chronic myelomonocytic leukemia; EB, MDS with excess blast; Hb, hemoglobin; IQR, interquartile range; IWG-M, International Working Group for the Prognosis of MDS Cohort; J-MDS, Japanese validation cohort; MLD, multilineage dysplasia; MPN, myeloproliferative neoplasm; Plt, platelet; RS-T, ring sideroblast-thrombocytosis; SLD, single lineage dysplasia; U, unspecified; WHO, World Health Organization.
Baseline characteristics of the validation cohort. (A) Characteristics of the validation cohort as compared with the original and the validation cohorts of the IPSS-M study. (B) Cross heat map for distribution of patients with MDS according to the IPSS-R (x-axis) and the IPSS-M (y-axis) scores. (C) Bar histogram shows the total percentage of patients restratified (upstaged or downstaged) according to the IPSS-M. CC, Cleveland Clinic cohort; CMML, chronic myelomonocytic leukemia; EB, MDS with excess blast; Hb, hemoglobin; IQR, interquartile range; IWG-M, International Working Group for the Prognosis of MDS Cohort; J-MDS, Japanese validation cohort; MLD, multilineage dysplasia; MPN, myeloproliferative neoplasm; Plt, platelet; RS-T, ring sideroblast-thrombocytosis; SLD, single lineage dysplasia; U, unspecified; WHO, World Health Organization.
Our higher-risk groups (high and very high, n = 407) were enriched in cases harboring complex karyotype (75%), TP53MT (75%), RUNX1MT (64%), and NRASMT (65%), and lower-risk groups showed higher frequencies of normal karyotype (59%, 367/620), SF3B1MT (61%, 92/151), and JAK2MT (67%, 66/98) (supplemental Figure 1D). Nearly all carriers of biallelic TP53MT (96%, 55/57) were assigned to the very-high-risk group, similar to the original cohort (91%) (supplemental Figure 2).
Overall, IPSS-M resulted in restratification of 579 (46%) patients in our cohort (Figure 1B). Of these, 70% were upstaged and 30% were downstaged, paralleling the results of the IWG-PM study (Figure 1A), in which 46% of the cases were restratified. The highest rate of restaging was among patients originally assigned to high-risk IPSS-R (59% [109/186] vs 62% in the original cohort). Remarkably, 85% (159/188) of restratified low-risk IPSS-R patients were upstaged to higher-risk groups per IPSS-M (Figure 1C).
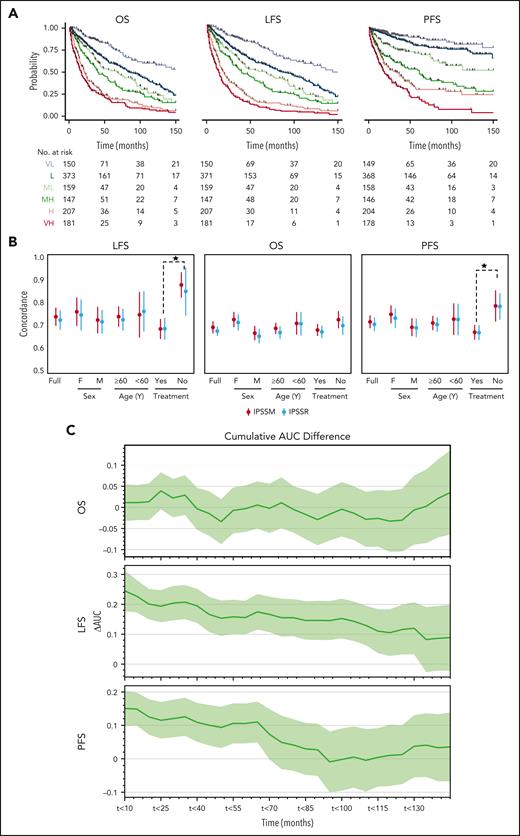
With a median follow-up time of 2.4 years, the 3-year overall survival (OS) rate was 51% (95% confidence interval [CI]: 47.6-53.8). The 3-year leukemia-free survival (LFS) and progression-free survival (PFS) rates were 47% (95% CI: 44-50) and 65% (95% CI: 61-68), respectively (supplemental Table 2). Both IPSS-M and IPSS-R risk scores showed significant differences in all outcomes (supplemental Table 1, Figure 2A, and supplemental Figure 3). Since IPSS-M aims at improving the IPSS-R with the incorporation of molecular features, we also analyzed differences in all outcomes in patients originally assigned to the same IPSS-R group. We noticed meaningful differences in patients assigned to very-low-risk, low-risk, and intermediate-risk groups (supplemental Figure 4). Performing reverse analysis, IPSS-R did not significantly restratify patients within the same IPSS-M subgroups in all clinical end points except for LFS in the very-high-risk group and PFS in the IPSS-M high-risk group (supplemental Figure 5).
Comparison of the IPSS-M and IPSS-R. (A) Stratification of our cohort based on IPSS-M for OS, LFS, and PFS. (B) Model discrimination measured by the concordance index via bootstrapping for the IPSS-R (light blue) and the IPSS-M system (red) across all the 3 outcomes end points (LFS, OS, and PFS) for the full cohort, treated vs untreated patients, males vs females, and ≥60 vs <60 years old. (C) The cumulative AUC differences (ΔAUC) based on IPSS-M between treated and untreated patients (y-axis) is plotted against the follow-up time intervals of 10 months (x-axis).
Comparison of the IPSS-M and IPSS-R. (A) Stratification of our cohort based on IPSS-M for OS, LFS, and PFS. (B) Model discrimination measured by the concordance index via bootstrapping for the IPSS-R (light blue) and the IPSS-M system (red) across all the 3 outcomes end points (LFS, OS, and PFS) for the full cohort, treated vs untreated patients, males vs females, and ≥60 vs <60 years old. (C) The cumulative AUC differences (ΔAUC) based on IPSS-M between treated and untreated patients (y-axis) is plotted against the follow-up time intervals of 10 months (x-axis).
To quantify the prognostic power of IPSS-M and IPSS-R, we used Harrell C-index bootstrapping on OS, LFS, and PFS (Figure 2B). IPSS-R and IPSS-M showed similar C-indices, regardless of treatment in our cohort. The C-indices of IPSS-M for LFS (0.73 vs 0.75) and OS (0.69 vs 0.74) for all patients in our cohort vs the IWG-PM cohort were comparable. In terms of LFS and OS, we noticed 2.0- and 2.8-point increases, respectively, in the C-index from IPSS-R to IPSS-M, as compared with 5-point increase for all clinical end points in the IWG-PM cohort. Similar observations were made when the cohort was stratified for age (≥60 vs <60) and sex (male vs female) (supplemental Figure 6). However, a general increase in C-index was observed in the untreated group, indicating that both IPSS-M and IPSS-R can provide more precise stratification in a treatment-naïve scenario. To better quantify the differences of IPSS-M performance based on treatment, we used a time-dependent receiver operating characteristic (ROC) analysis and observed a global increase of area under the curve (AUC) for LFS in the untreated (never treated) cases as compared with the treated cases. A time-dependent increase of AUC in PFS was also noticed. Nevertheless, no difference was observed for OS estimation between both groups (Figure 2C). The differences in model fit based on treatment status quantified by the C-index persisted across relevant clinical stratifications, using sex and age (Figure 2C). Remarkably, no major changes in C-index for all clinical end points between IPSS-R and IPSS-M were noticed as compared with the IWG-PM study. Finally, we repeated the time-dependent ROC analysis to better quantify the differences in IPSS-R and IPSS-M and noticed nonsignificant differences between the cumulative AUCs for all outcomes (supplemental Figure 7).
The IPSS/IPSS-R have been the standard tools for MDS prognostication in the last 2 decades.1,2 The new IPSS-M scheme recently released by the IWG-PM may establish a new standard to be used in a more rational way by assigning patients to appropriate treatment modalities and clinical trials based on genomic features. Aside from the potential usefulness of IPSS-M, a meticulous validation in diverse clinical scenarios is necessary to assure validity, facilitating its wide adoption in clinical practice. When IPSS-R is used as a benchmark, IPSS-M algorithm in the original study improves the accuracy of the prediction (C-index) of 5 points for all considered outcomes. However, in our cohort both IPSS-R and IPSS-M showed a similar C-index at all end points. Despite a persisting impact of clinical features on prognosis,2,11 our findings show that the ability of IPSS-M to further discriminate outcomes of patients within the same risk group was significant, especially for certain infrequent but impactful molecular alterations. Remarkably, in our study upstaging was registered mainly in the low-risk IPSS-R group, which is a cohort of clinical interest that needs long-term follow-up to monitor for AML transformation.
Our comprehensive validation could not find significant differences between IPSS-R and IPSS-M when calculating C-indices or time-dependent ROC for clinical outcomes. The significant added prognostic value of IPSS-M in the original study10 was likely a result of the substantial fraction (two-thirds) of untreated patients included, which might not accurately represent an actual clinical practice of more treated patients (62% in our cohort). It is unlikely that a significant proportion of low-risk patients does not eventually progress, and under current practice standards they will receive disease-modifying therapy. This is in contrast to historical cohorts in which treatment options were limited.1,2 The information on patients who never received treatment is useful to assess the need for treatment but relatively less helpful in terms of real prognosis under current clinical standard. This emphasizes the importance of validating the IPSS-M into independent cohorts to ensure reproducibility in different clinical settings. In conclusion, MDS patients should be stratified based on clinical and molecular features to establish appropriate prognostication and AML progression risk. IPSS-M score provided similar concordance indices to IPSS-R in our well-annotated cohort of patients with sufficient follow-up time. Particularly, IPSS-M better stratified low-risk patients assigned within previously similar risk groups. Multiple validation studies can further assess the novel score in clinical practice and ensure reproducibility for future application in clinical trials.
Acknowledgment
Carmelo Gurnari is a PhD candidate in Immunology, Molecular Medicine and Applied Biotechnology, University of Rome Tor Vergata, Rome, Italy, and this work is submitted in partial fulfillment of the requirement for a PhD.
Authorship
Contribution: T.K. and W.B. collected and analyzed clinical and molecular data, interpreted results, and wrote the manuscript; A.D. applied statistical methods, analyzed the data, interpreted the results, and wrote the manuscript; M.A. and O.D.O. collected clinical data and edited the manuscript; V.V., H.E.C., and M.A.S. edited the manuscript and provided helpful insights to the manuscript; C.G. and J.P.M. generated and conceived the study, provided invaluable help to the manuscript preparation, designed figures, and wrote the manuscript; and all authors participated in the critical review of the final paper and submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, 9620 Carnegie Ave, N Building, Building NE6-250, Cleveland, OH 44106; e-mail: maciejj@ccf.org.
References
Author notes
∗T.K. and W.B. contributed equally to the work.
†C.G. and J.P.M. contributed equally to the work.
Requests for additional information not provided in the main text or supplementary material should be sent to maciejj@ccf.org.
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal