TO THE EDITOR:
Among hematologic malignancies, T-cell acute lymphoblastic leukemia (T-ALL) represents a class of aggressive tumors with dismal clinical outcome in cases of relapsed or refractory disease. Although the intensification of multiagent chemotherapy protocols has dramatically improved prognosis, refractory and relapsed cases are clinically challenging because of limited therapeutic options.1,2 p53 is a transcription factor and a master tumor suppressor gene frequently altered in cancer.3 In contrast to carcinomas and other hematologic malignancies, TP53 alterations (TP53Alt), encompassing mutations (TP53Mut), and/or pan-exon deletions (TP53Del) are remarkably rare at diagnosis in T-ALL, and their clinical implication remains elusive.4-6 Critically, TP53Alt have been reported to be acquired in up to 20% of the relapsed T-ALL cases, where they convey a deleterious prognosis.7-9 Here, we produce the first comprehensive analysis of TP53Alt and the associated oncogenetic landscape in an extensive cohort of 476 patients newly diagnosed with T-ALL.
We investigated the clinical characteristics of TP53Alt in 476 patients including 215 adults and 261 children. Adult patients were enrolled in the GRAALL-2003-2005 trial (GRAALL-2003, #NCT00222027; GRAALL-2005, #NCT00327678), and pediatric patients were enrolled in the FRALLE 2000 trial (supplemental Figure 1, available on the Blood website). Based on the DNA availability for molecular analysis, 215 adult patients out of 337 and 261 pediatric patients out of 427 were included in this study. No difference in clinical outcomes was observed between the included patients and the entire cohort (supplemental Figures 2 and 3; supplemental Table 1). Diagnostic peripheral blood or bone marrow samples were collected after informed consent was obtained, according to the Declaration of Helsinki. All samples contained ≥80% blasts, immunophenotypic of T-ALL samples, minimal residual disease (MRD) assessment, and multiplex ligation–dependent probe amplification analysis (P383 T-ALL, MRC Holland) were performed as previously described.10-12
Genomic analysis was performed using pan-exon targeted next-generation sequencing of DNA extracted from diagnostic samples, and DNA libraries were prepared using the Nextera XT kit (Illumina) and sequenced on a MiSeq (Illumina). The next-generation sequencing panel included 63 genes known to be mutated in T-ALL (supplemental Table 2). Genetic lesion co-occurrences and mutual exclusions were computed using the DISCOVER R package. We performed a computational approach previously described for the detection of copy number variants from next-generation sequencing data,13,14 including a systematic analysis of the depth of TP53 gene coverage. This method is based on variations in the depth of coverage of the aligned sequence reads using a locally developed algorithm. The copy number variants detected were confirmed by high-resolution comparative genomic hybridization and/or multiplex ligation–dependent probe amplification analysis (kits P037-CLL-1 and P038-CLL-2, MRC Holland). Diagnostic DNA was hybridized on a Cytogenetics Whole-Genome 2.7M Array (Affymetrix, Santa Clara, CA) (comparative genomic hybridization array), according to the manufacturer’s recommendations. Data analysis was performed using Chromosome Analysis Suite software (Affymetrix). Gene copy number aberrations were compared with the Database of Genomic Variants15 to study only somatic aberrations.
Comparisons of categorical and continuous variables between subgroups were performed using Fisher exact test and the Mann-Whitney test, respectively. Overall survival (OS) was calculated from the date of diagnosis to the last follow-up date, censoring patients alive. The cumulative incidence of relapse (CIR) was calculated from the complete remission date to the date of relapse, and censoring patients alive without relapse at the last follow-up date. Relapse and death in complete remission were considered to be competitive events. Univariate and multivariate analyses to assess the impact of categorical and continuous variables were performed using the Cox model. The proportional-hazards assumption was checked before conducting the multivariate analyses. In the univariate and multivariate analyses, age and log10 (white blood count) were considered continuous variables. All the analyses were stratified during the trial. Variables with P < .1 in univariate analysis were included in the multivariable models. Statistical analyses were performed using the STATA software (STATA 12.0 Corporation, College Station, TX). All P values were 2-sided, with P < .05 denoting statistical significance. Additional details are included in the supplemental Methods.
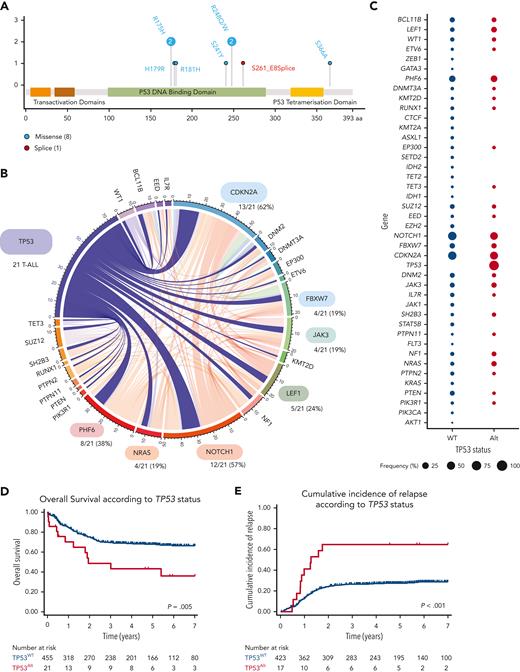
The incidence of TP53Alt at the time of diagnosis in this cohort was 4% (21/476). TP53Mut was detected in 9 patients (6 adults and 3 pediatric cases) (Figure 1A; supplemental Tables 3 and 4), TP53Del was identified in 15 patients (7 adult and 8 pediatric cases), and 3 patients harbored both TP53Mut and TP53Del. Patients with TP53Alt did not significantly differ from patients with TP53 wild-type (TP53WT) regarding sex, age, and central nervous system involvement (Table 1), but were associated with an immature phenotype (combining IM0 T-ALL; T-cell receptor δ [TCRδ] and TCRγ germ line, IMδ T-ALL; TCRδ rearranged but not TCRγ and IMg T-ALL; both TCRδ and TCRγ rearranged)12 (8/20 [40%] vs 81/399 [20%], P = .048). The oncogenetic landscape of TP53Alt was comparable to that of TP53WT T-ALLs (Figure 1B-C; supplemental Figures 4-6; supplemental Table 5). To investigate the prognostic value of TP53Alt, survival analyses were performed on the series of 476 patients. TP53Alt did not confer an increased poor prednisone response, defined by a peripheral blood blast count >1.0 × 109/L at the end of the induction phase (38% vs 45%, P = .7) (Table 1). Although TP53Alt did not significantly influence the morphological complete response rate at the end of the induction course (81% vs 93%, P = .07), patients harboring TP53Alt were associated with delayed early medullary blast clearance, as confirmed by the end of induction MRD1 assessment, with more positivity (≥10–4) in TP53Alt than in TP53WT cases (75% vs 35%, P = .01). Patients (both adult and pediatric) with TP53Alt had an inferior outcome compared with TP53WT (Table 1; Figures 1D-E; supplemental Figure 7), with an increased CIR (5-year CIR: 65% vs 27%; specific hazard ratio [HR], 3.1; 95% confidence interval [CI], 1.67-5.78; P < .001) and a shorter OS (5-year OS: 48% vs 72%; HR, 2.34; 95% CI, 1.30-4.24; P = .005). In multivariate analysis, TP53Alt predicted a statistically lower OS (HR, 2.87; 95% CI, 1.56-5.26; P = .001) and higher CIR (specific HR, 2.90; 95% CI, 1.55-5.44; P = .001) even after adjustment on the 4 genes NOTCH1/FBXW7/RAS/PTEN (NFRP) classifier, which identified patients with a poor prognosis in both GRAALL and FRALLE trials.10,16
TP53 alterations in the GRAALL03/05 and FRALLE2000 studies. (A) Lollipop plots indicating TP53 mutations in 476 patients with T-ALL. More details on the pathogenicity of the TP53 mutations are provided in supplemental Table 4. (B) Circos plots illustrating pairwise relationships across relatively common mutated genes in TP53Alt T-ALL. The width of the ribbon corresponds to the number of cases with the simultaneous presence of a first and second gene mutation. (C) Frequency of alterations per gene in TP53Alt vs TP53WT T-ALL. The width of the circles is proportional to the frequency of alterations observed in the 2 T-ALL subgroups (TP53Alt in red vs TP53WT T-ALL in blue). (D-E) Clinical impact of TP53Alt in the GRAALL0305 and FRALLE2000 studies. OS (D) and CIR (E) are shown. The red curve represents patients with TP53Alt T-ALL and the blue curve represents patients with TP53WT.
TP53 alterations in the GRAALL03/05 and FRALLE2000 studies. (A) Lollipop plots indicating TP53 mutations in 476 patients with T-ALL. More details on the pathogenicity of the TP53 mutations are provided in supplemental Table 4. (B) Circos plots illustrating pairwise relationships across relatively common mutated genes in TP53Alt T-ALL. The width of the ribbon corresponds to the number of cases with the simultaneous presence of a first and second gene mutation. (C) Frequency of alterations per gene in TP53Alt vs TP53WT T-ALL. The width of the circles is proportional to the frequency of alterations observed in the 2 T-ALL subgroups (TP53Alt in red vs TP53WT T-ALL in blue). (D-E) Clinical impact of TP53Alt in the GRAALL0305 and FRALLE2000 studies. OS (D) and CIR (E) are shown. The red curve represents patients with TP53Alt T-ALL and the blue curve represents patients with TP53WT.
Clinicobiological and outcome characteristics of adult and pediatric T-ALL (GRAALL and FRALLE protocols) according to TP53 status
| Variable . | TP53Alt . | TP53WT . | Total . | P value∗ . |
|---|---|---|---|---|
| n = 21 (4%) . | n = 455 (96%) . | N = 476 . | ||
| Male (%) | 14/21 (67) | 343/455 (75) | 357/476 (75) | .4 |
| Age, y† | 23.4 (4.0-51.8) | 15.3 (1.1-59.1) | 15.3 (1.1-59.1) | .5 |
| WBC, g/L† | 25 (5-674) | 66 (0-980) | 64 (0-980) | .01 |
| CNS involvement (%)‡ | 3/21 (14) | 48/453 (11) | 51/474 (11) | .5 |
| Immunophenotype (%) | ||||
| Early thymic precursor phenotype | 6/16 (38) | 50/291 (17) | 56/307 (18) | .09 |
| Immature (IM0/δ/γ)§ | 8/20 (40) | 81/399 (20) | 89/419 (21) | .048 |
| αβ lineage | 6/20 (30) | 205/399 (51) | 211/419 (50) | .07 |
| Mature TCRγδ | 3/20 (15) | 63/399 (16) | 66/419 (16) | >.9 |
| Oncogenetic classification (%) | ||||
| TLX1 | 1/18 (6) | 53/397 (13) | 54/415 (13) | .5 |
| TLX3 | 2/18 (11) | 70/397 (18) | 72/415 (17) | .8 |
| SIL-TAL1 | 1/18 (6) | 56/397 (14) | 57/415 (14) | .5 |
| CALM-AF10 | 0/18 (0) | 13/397 (3) | 13/415 (3) | >.9 |
| High-risk classifier|| | 13/21 (62) | 196/455 (43) | 209/476 (44) | .12 |
| Treatment response (%) | ||||
| Prednisone response | 13/21 (62) | 246/446 (55) | 259/467 (55) | .7 |
| Chemosensitivity | 12/21 (57) | 325/446 (73) | 337/467 (72) | .1 |
| MRD1 >10−4 | 9/12 (75) | 114/328 (35) | 123/340 (36) | .01 |
| Complete remission | 17/21 (81) | 423/455 (93) | 440/476 (92) | .07 |
| Allo-HSCT | 4/20 (20) | 97/436 (22) | 101/456 (22) | .7 |
| Outcome, % | ||||
| 5-y CIR (95% CI) | 65 (11-43) | 27 (23-32) | 29 (25-33) | <.001 |
| 5-y OS (95% CI) | 48 (26-67) | 72 (68-76) | 71 (67-75) | .005 |
| Variable . | TP53Alt . | TP53WT . | Total . | P value∗ . |
|---|---|---|---|---|
| n = 21 (4%) . | n = 455 (96%) . | N = 476 . | ||
| Male (%) | 14/21 (67) | 343/455 (75) | 357/476 (75) | .4 |
| Age, y† | 23.4 (4.0-51.8) | 15.3 (1.1-59.1) | 15.3 (1.1-59.1) | .5 |
| WBC, g/L† | 25 (5-674) | 66 (0-980) | 64 (0-980) | .01 |
| CNS involvement (%)‡ | 3/21 (14) | 48/453 (11) | 51/474 (11) | .5 |
| Immunophenotype (%) | ||||
| Early thymic precursor phenotype | 6/16 (38) | 50/291 (17) | 56/307 (18) | .09 |
| Immature (IM0/δ/γ)§ | 8/20 (40) | 81/399 (20) | 89/419 (21) | .048 |
| αβ lineage | 6/20 (30) | 205/399 (51) | 211/419 (50) | .07 |
| Mature TCRγδ | 3/20 (15) | 63/399 (16) | 66/419 (16) | >.9 |
| Oncogenetic classification (%) | ||||
| TLX1 | 1/18 (6) | 53/397 (13) | 54/415 (13) | .5 |
| TLX3 | 2/18 (11) | 70/397 (18) | 72/415 (17) | .8 |
| SIL-TAL1 | 1/18 (6) | 56/397 (14) | 57/415 (14) | .5 |
| CALM-AF10 | 0/18 (0) | 13/397 (3) | 13/415 (3) | >.9 |
| High-risk classifier|| | 13/21 (62) | 196/455 (43) | 209/476 (44) | .12 |
| Treatment response (%) | ||||
| Prednisone response | 13/21 (62) | 246/446 (55) | 259/467 (55) | .7 |
| Chemosensitivity | 12/21 (57) | 325/446 (73) | 337/467 (72) | .1 |
| MRD1 >10−4 | 9/12 (75) | 114/328 (35) | 123/340 (36) | .01 |
| Complete remission | 17/21 (81) | 423/455 (93) | 440/476 (92) | .07 |
| Allo-HSCT | 4/20 (20) | 97/436 (22) | 101/456 (22) | .7 |
| Outcome, % | ||||
| 5-y CIR (95% CI) | 65 (11-43) | 27 (23-32) | 29 (25-33) | <.001 |
| 5-y OS (95% CI) | 48 (26-67) | 72 (68-76) | 71 (67-75) | .005 |
| . | Univariate and multivariate analysis¶ . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| Specific HR . | 95% CI . | P value . | Specific HR . | 95% CI . | P value . | |
| CIR | ||||||
| Age, y | 1.01 | 0.98-1.03 | .57 | — | — | — |
| CNS | 1.57 | 0.85-2.59 | .08 | 1.28 | 0.77-2.13 | .34 |
| Log (WBC) | 1.62 | 1.2-2.18 | .002 | 1.62 | 1.19-2.19 | .002 |
| Prednisone response | 0.67 | 0.47-0.95 | .03 | 0.93 | 0.64-1.35 | .70 |
| High-risk classifier§ | 2.78 | 1.94-3.99 | <.001 | 2.58 | 1.78-3.74 | <.001 |
| TP53Alt | 3.11 | 1.67-5.78 | <.001 | 2.90 | 1.55-5.44 | .001 |
| . | Univariate and multivariate analysis¶ . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| Specific HR . | 95% CI . | P value . | Specific HR . | 95% CI . | P value . | |
| CIR | ||||||
| Age, y | 1.01 | 0.98-1.03 | .57 | — | — | — |
| CNS | 1.57 | 0.85-2.59 | .08 | 1.28 | 0.77-2.13 | .34 |
| Log (WBC) | 1.62 | 1.2-2.18 | .002 | 1.62 | 1.19-2.19 | .002 |
| Prednisone response | 0.67 | 0.47-0.95 | .03 | 0.93 | 0.64-1.35 | .70 |
| High-risk classifier§ | 2.78 | 1.94-3.99 | <.001 | 2.58 | 1.78-3.74 | <.001 |
| TP53Alt | 3.11 | 1.67-5.78 | <.001 | 2.90 | 1.55-5.44 | .001 |
| . | Univariate and multivariate analysis¶ . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |
| OS | ||||||
| Age | 1.03 | 1.01-1.05 | .001 | 1.05 | 1.03-1.07 | <.001 |
| CNS | 2.00 | 1.28-3.14 | .002 | 1.64 | 1.02-2.64 | .04 |
| Log (WBC) | 1.99 | 1.48-2.67 | <.001 | 2.01 | 1.51-2.86 | <.001 |
| Prednisone response | 0.54 | 0.38-0.76 | <.001 | 0.83 | 0.57-1.20 | .31 |
| High-risk classifier | 2.93 | 2.06-4.17 | <.001 | 2.90 | 2.01-4.18 | <.001 |
| TP53Alt | 2.34 | 1.30-4.24 | .005 | 2.87 | 1.56-5.26 | .001 |
| . | Univariate and multivariate analysis¶ . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |
| OS | ||||||
| Age | 1.03 | 1.01-1.05 | .001 | 1.05 | 1.03-1.07 | <.001 |
| CNS | 2.00 | 1.28-3.14 | .002 | 1.64 | 1.02-2.64 | .04 |
| Log (WBC) | 1.99 | 1.48-2.67 | <.001 | 2.01 | 1.51-2.86 | <.001 |
| Prednisone response | 0.54 | 0.38-0.76 | <.001 | 0.83 | 0.57-1.20 | .31 |
| High-risk classifier | 2.93 | 2.06-4.17 | <.001 | 2.90 | 2.01-4.18 | <.001 |
| TP53Alt | 2.34 | 1.30-4.24 | .005 | 2.87 | 1.56-5.26 | .001 |
MRD1 correspond to MRD evaluation after induction and was performed by allele-specific oligonucleotides polymerase chain reaction. TCR status and oncogenic were performed as described in supplemental Methods.
P < .05 are indicated in bold.
allo-HSCT, allogeneic hematopoietic stem cell transplantation; CNS, central nervous system; WBC, white blood count.
Statistical tests performed: Fisher exact test and Wilcoxon rank-sum test.
Statistics presented: median (minimum-maximum).
CNS involvement: CNS3 in FRALLE2000 trial, CNS2 and/or CNS3 in GRAALL2003 and GRAALL2005 trial.
T-ALL are divided into 3 subclasses as following: (1) immature (no detectable TCRβ variable diversity joining): IM0 (TCRδ and TCRγ germ line), IMδ (TCRδ rearranged but not TCRγ), and IMg (both TCRδ and TCRγ rearranged); (2) T-ALL with TCRαβ lineage (including both early-cortical IMb/pre-αβ and mature sTCRαβ+); and (3) mature sTCRγδ.12
Low-risk classifier: NOTCH1 and/or FBXW7 (N/F) mutation without N/K-RAS and PTEN (R/P) alteration. High-risk classifier: N/F mutation with R/P alteration, N/F WT with or without R/P alteration.10,16
Univariate and multivariate Cox analyses stratified on protocol.
The limited number of MRD1 assessments available for patients with TP53Alt T-ALL (12/21, 57%) did not allow us to include this end point in the multivariate analysis, justifying further studies to establish whether this alteration remains an independent prognostic factor when combined with MRD1 status in T-ALL.
This study provides the largest comprehensive analysis of TP53Alt in T-ALL, describing, for the first time, their clinical profile and, most importantly, the extremely poor prognostic impact associated with TP53Alt at diagnosis in T-ALL, urging the need to develop innovative targeted therapies for patients harboring TP53Alt.
Approximately 25% of pediatric patients and 50% of adult patients with T-ALL relapse, with a 5-year survival rate of less than 20% in both age groups.1,2 The sole therapeutic approaches with curative potential for T-ALL relapsed cases are limited to conventional chemotherapy or hematopoietic stem cell transplantation. Molecular genetic analyses and sequencing studies have recently led to the identification of recurrent T-ALL alterations associated with prognosis, allowing for the refinement of the stratification of relapse risk.10,14,17,18 However, a significant proportion of T-ALL relapses remain unpredicted, underlining the need for new predictive markers.
Negative outcomes observed in TP53Alt T-ALL are likely to be related to p53-induced therapeutic resistance previously described for other malignancies, combined with loss of p53 tumor suppressor activity and acquisition of novel functions that disrupt the DNA damage response pathway.19,20 APR-246 (eprenetapopt), a small molecule interacting with mutated p53 protein to restore its WT conformation,21,22 has recently shown promising results in myeloid malignancies and B-cell ALL,23-26 justifying further studies to explore the potential efficacy of this new drug in T-ALL TP53Mut.
Acknowledgments
The authors thank all the participants of the GRAALL-2003 and GRAALL-2005 study groups, the Société Française de lutte contre les Cancers et les leucémies de l'Enfant et de l'adolescent (SFCE) and the investigators of the 16 SFCE centers involved in the collection and provision of data and patient samples, and V. Lheritier for the collection of clinical data.
This study was supported by grants to Necker laboratory from the Cancer Research for Personalized Medicine (CARPEM), the Association pour la Recherche contre le Cancer (Equipe Labellisée), the Ligue contre le Cancer (Equipe Labellisée), and the Institut National du Cancer PRT-K 18-071. The GRAALL study was supported by grants from the Programme Hospitalier de Recherche Clinique (grants P0200701 and P030425/AOM03081), Ministère de l’Emploi et de la Solidarité, France, and the Swiss Federal Government in Switzerland. Samples were collected and processed by the Assistance Publique-Hôpitaux de Paris “Direction de Recherche Clinique” Tumor Bank at Necker-Enfants Malades. M.S. was supported by Action Leucémie, la Ligue contre le Cancer et Soutien pour la formation à la recherche translationnelle en cancérologie. G.P.A. was supported by the Fondation de France.
Authorship
Contribution: V.A., O.H., M.S., and G.P.A. conceived and designed the research and oversaw the project; M.B., C.G., N.G., J.-M.C., F.H., Y.C., Y.L.B., E.M., V.G., A.P., P.R., A.B., D.B., O.H., N.B., and V.A. provided study materials or patients; M.S., G.P.A., G.H., L.C., and V.A. performed molecular analyses; M.S. and G.P.A. collected and assembled data; N.B. and M.S. performed statistical analysis; M.S., G.P.A., R.B., O.H., V.A., and N.B. analyzed and interpreted data; and M.S., G.P.A., N.B., and V.A. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests
Correspondence: Vahid Asnafi, Laboratory of Onco-Hematology, Assistance Publique-Hôpitaux de Paris (APHP), Necker Enfants-Malades Hospital, Université de Paris, 149 rue de Sèvres, 75015 Paris, France; e-mail: vahid.asnafi@aphp.fr.
References
Author notes
∗M.S. and G.P.A. contributed equally to this work and are joint first authors.
Data are available on request from the corresponding author, Vahid Asnafi (vahid.asnafi@aphp.fr).
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal