Key Points
Patterns of microbial dysbiosis can be detected in fecal samples of GI aGVHD patients peri-aGVHD onset.
Markers of microbial health pre-GVHD onset are associated with longer survival and lower risk of GVHD-related mortality after allo-HCT.
Abstract
Following allogeneic hematopoietic cell transplantation (allo-HCT), the gastrointestinal (GI) tract is frequently affected by acute graft-versus-host disease (aGVHD), the pathophysiology of which is associated with a dysbiotic microbiome. Since microbial composition varies along the length of the GI tract, the authors hypothesized that microbiome features correlate with the pattern of organ involvement after allo-HCT. We evaluated 266 allo-HCT recipients from whom 1303 stool samples were profiled by 16S ribosomal gene sequencing. Patients were classified according to which organs were affected by aGVHD. In the 20 days prior to disease onset, GVHD patients had lower abundances of members of the class Clostridia, lower counts of butyrate producers, and lower ratios of strict-to-facultative (S/F) anaerobic bacteria compared with allograft recipients who were free of GVHD. GI GVHD patients showed significant reduction in microbial diversity preonset. Patients with lower GI aGVHD had lower S/F anaerobe ratios compared with those with isolated upper GI aGVHD. In the 20 days after disease onset, dysbiosis was observed only in GVHD patients with GI involvement, particularly those with lower-tract disease. Importantly, Clostridial and butyrate-producer abundance as well as S/F anaerobe ratio were predictors of longer overall survival; higher abundance of butyrate producers and higher S/F anaerobe ratio were associated with decreased risk of GVHD-related death. These findings suggest that the intestinal microbiome can serve as a biomarker for outcomes of allo-HCT patients with GVHD.
Introduction
Graft-versus-host disease (GVHD) is a common complication after allogeneic cell transplantation (allo-HCT) that is initiated by donor immune cell activation against host tissue leading to organ damage. Acute GVHD (aGVHD) targets regions of the gastrointestinal (GI) tract, skin, and liver and may manifest in single or multiple organs. Involvement of the lower GI (LGI) tract is associated with a lower likelihood of GVHD treatment response and a higher risk of mortality.1-4 In contrast, GVHD that is isolated to the upper GI (UGI) tract or skin is commonly responsive to treatment and has negligible prognostic relevance for mortality.5,6
Although the general immunopathology of GVHD has been investigated at length, the determinants of specific organ involvement in patients are not known. In mice with GVHD and in other colitis models, dendritic cells imprint tissue-specific homing molecules on T cells that drive trafficking to the GI tract. Less is known about specific homing to the upper vs lower intestine, although expression patterns of homing molecules such as α4β7 integrin, GPR15, CCR9, and CXCR3 in various T cell subsets have been implicated.7-10 The pathways governing gut T cell trafficking in human GVHD are less clear, as are the factors that might vary between patients to explain patterns of organ involvement.
The GI tract harbors a dynamic population of microbial organisms, the composition of which increases in density and diversity along the length of the tract.11 GI microbial colonization is relevant in allo-HCT since features of the intestinal microbiome such as diversity and dominance of specific bacteria have been associated with transplant-related mortality (TRM) and GVHD outcomes.12-19 Studies evaluating microbiome disruption and diversity in the peri-engraftment period have demonstrated an association between dysbiosis and survival outcomes,13-18 which might be explained by an outgrowth of harmful organisms, a loss of beneficial or homeostatic commensals, or a combination of both. We have previously reported that higher abundance of the anaerobic intestinal commensal Blautia (member of Clostridia class) in the peri-engraftment period is associated with reduced GVHD-related mortality and prolonged overall survival (OS).15 Moreover, the loss of members of class Clostridia has also been observed in mouse models of GVHD.12 Some anaerobic commensals, among them many Clostridia (class), are producers of short-chain fatty acids (SCFA), a class of metabolites that includes butyrate, propionate, and acetate, as a byproduct of carbohydrate fermentation.20 Several groups have observed lower concentrations of SCFA, especially butyrate, in patients with GVHD at peri-engraftment and at disease onset.19,21 In addition, we observed lower circulating amounts of butyrate, among other SCFA, 3 months post-transplant in patients who went on to develop chronic GVHD.22 Microbiota-derived butyrate can modulate GI epithelial cell damage and mitigate GI aGVHD in mice by restoring intestinal epithelial cell junctional integrity and decreasing apoptosis.23-25 Nonetheless, further research on the role of butyrate is needed, as one study linked the presence of butyrogenic bacteria to GVHD severity,26 and butyrate may also be toxic to intestinal stem cells.27 These findings raise the possibility that microbial-derived metabolites have direct effects on GVHD target tissues and may affect disease outcomes. Several studies have linked intestinal injury to changes in the commensal flora in the gut28,29 characterized by an outgrowth of such facultative anaerobes as Enterococcaceae (family)14,18,25 and Gemella (genus)30,31 along with a decrease in strict anaerobes, such as Faecalibacterium prausnitzii (species).28 However, it is unknown whether this loss of anaerobiosis also happens in the context of GVHD. We hypothesized that unique patterns of microbial dysbiosis correlate with organ-specific intestinal involvement in GVHD and consequently influence GVHD outcomes after allo-HCT.
Methods
Patients and graft characteristics
Patients included in the analysis were all consecutive adult recipients of unmodified allografts at Memorial Sloan Kettering Cancer Center between January 2011 and February 2017 for the treatment of hematologic malignancies who had stool samples collected in our institutional fecal microbiome biobank. Patients provided written consent to biospecimen collection, and the analysis was approved by the institutional review board.
Study definitions
aGVHD was diagnosed with histologic confirmation as clinically appropriate. The International Bone Marrow Transplant Registry classification was used to guide aGVHD grading, except grades A through D, which were labeled as grades I through IV. Grading was reviewed by a transplant clinician panel.32 Patients were classified in 4 cohorts according to aGVHD organ involvement by day 100 post allo-HCT: (1) no GVHD in any organ, (2) non-GI: skin and/or liver involvement without any GI tract involvement, (3) UGI: UGI tract involvement without LGI tract, with or without skin/liver involvement, and (4) LGI: any LGI tract involvement with or without any other organ involvement, including UGI aGVHD (Table 1). Relapse was defined as recurrence or progression of hematologic malignancy post-HCT. Causes of death were described according to the Copelan algorithm,33 and TRM was defined as death from any cause not preceded by relapse. GVHD-related mortality included patients without disease recurrence being treated for GVHD at the time of death, including those who died of infection. Relapse, OS, TRM, and GVHD-related mortality were calculated from the time of GVHD onset to death.
GVHD clinical groups by aGVHD organ involvement
| GVHD group (no.) . | Organ involvement, no. (%) . |
|---|---|
| No GVHD (131) | None |
| Non-GI (29) | Skin: 27 (93) |
| Skin/liver: 2 (7) | |
| UGI (53) | UGI: 34 (64) |
| UGI/skin: 19 (36) | |
| LGI (53) | LGI: 7 (13) |
| LGI/UGI: 26 (49) | |
| LGI/skin: 1 (2) | |
| LGI/liver: 1 (2) | |
| LGI/UGI/skin: 17 (32) | |
| LGI/UGI/skin/liver: 1 (2) |
| GVHD group (no.) . | Organ involvement, no. (%) . |
|---|---|
| No GVHD (131) | None |
| Non-GI (29) | Skin: 27 (93) |
| Skin/liver: 2 (7) | |
| UGI (53) | UGI: 34 (64) |
| UGI/skin: 19 (36) | |
| LGI (53) | LGI: 7 (13) |
| LGI/UGI: 26 (49) | |
| LGI/skin: 1 (2) | |
| LGI/liver: 1 (2) | |
| LGI/UGI/skin: 17 (32) | |
| LGI/UGI/skin/liver: 1 (2) |
Analysis of fecal samples
Stool samples from an institutional fecal microbiome biobank were analyzed. Samples collected after a fecal microbiota transplant were removed from the analysis.34 DNA was purified using bead-beating in phenol-chloroform as previously described.15 Amplification of genomic 16S ribosomal RNA V4/V5 regions were polymerase-chain-reaction amplified and sequenced on the Illumina platform. Sequences were mapped to amplicon sequence variants using DADA235 and assigned taxonomic identity according to the National Center for Biotechnology Information 16S database. Microbial α-diversity was computed using the reciprocal Simpson index.36 Exposure to a list of gut microbiome–perturbing antibiotics34 was assessed according to the interval of sample collection (7 days prior to the first sample collected to 1 day prior to the last sample collected) (supplemental Figure 1, available on the Blood website) and classified according to published data on bacterial inhibition16,37-45 (supplemental Table 1). Organisms were classified as strict vs facultative anaerobes by collapsing assembly of gut organisms through reconstruction and analysis annotations46 into categories according to their predicted oxygen metabolism: the terms anaerobes and strict anaerobes were categorized as strict anaerobes, and the terms aerotolerants, obligate aerobes, facultative anaerobes, microaerophile/anaerobes, aerobes, nanaerobes, and facultative anaerobes were grouped as facultative anaerobes. The microbiome was classified at the genus level when all species within it belonged to the same category, whereas species-level distinctions were applied otherwise. The curated list of predicted butyrate producers was adapted from Haak et al.47 Predicted metabolic functions were computationally derived from 16S ribosomal RNA sequences using phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt).48 Metagenomic shotgun sequencing was conducted as previously described.49 In summary, samples were extracted, sheared to 650 bp, prepared using the Illumina TruSeq DNA library kit, and sequenced using the Illumina HiSeq system with coverage of 10 to 20 × 106 reads per sample and read length of 100 bp paired-end. Human sequence contaminants were removed using BMTagger and HUMAnN 3.0, and MetaCyc50,51 were used for annotation of microbial metabolic pathways and relative expression considered at counts per million.
Statistical analysis
A Wilcoxon rank-sum test was used to compare microbial associations across groups. A false discovery rate (FDR) correction was applied to these tests when comparing taxa at genus level, along with PICRUSt and shotgun associations, across GVHD-defined groups. The associations were further summarized by calculating the fold-change in microbiota features across groups. For the primary analysis of transplant outcomes, associations between microbial markers and OS, GVHD-related mortality, TRM, and relapse were estimated by first categorizing GVHD patients above and below the median value for each marker within the preonset window. As a secondary analysis, the markers were analyzed continuously and log-transformed, as appropriate. For OS, associations were estimated using Cox proportional hazards regression. The incidence of GVHD-related mortality and TRM were estimated using cumulative incidence functions, treating relapse and death unrelated to GVHD as competing events for GVHD-related mortality and relapse for TRM. A Fine and Gray model was used to further adjust for sex, age, and conditioning regimen. All statistical analyses were performed using R (version 3.6.2).
Results
Patient demographics and aGVHD
A total of 135 patients who developed grade I to IV aGVHD by day 100 after HCT had stool samples collected near the time of onset. GVHD occurred at a median onset of 32 (range, 13-86) days. The proportion of grade II to IV and III to IV aGVHD was 47% and 13%, respectively. The GI tract was the organ most commonly involved (106/135), followed by skin (67/135) and liver (2/135). Patients predominantly had grade II aGVHD. Notably, grade IV aGVHD occurred only in patients with LGI involvement. We hypothesized that the reason the LGI tract may be spared in some patients is due to protective features of the colonic microbiota. To explore this, we grouped these patients according to the GVHD target organ affected (Table 1), including non-GI (n = 29), UGI (n = 53), and LGI (n = 53). As we have previously reported that allo-HCT patients have microbiome compositions that are distinct from those of healthy volunteers before,18 during,18 and for at least 1 year following transplantation,22 we included as controls a set of 1066 fecal samples from 131 recipients of unmodified allografts who did not develop GVHD. The non-GI group had predominantly skin involvement, whereas the UGI group had isolated UGI or UGI combined with skin involvement. LGI cases had predominantly LGI combined with UGI involvement with or without skin. Table 2 summarizes the clinical variables of the groups. The no-GVHD group received more chemotherapy-based conditioning and fewer cord blood grafts, whereas the UGI group received predominantly unrelated donor and cord blood grafts. Non-GI and LGI cases were more likely to have a reduced intensity conditioning and an HLA-matched unrelated donor or cord blood graft.
Patient demographics (N = 266)
| . | No-GVHD (n = 131) . | Non-GI (n = 29) . | UGI (n = 53) . | LGI (n = 53) . | P value . |
|---|---|---|---|---|---|
| Median age (range), y | 57 (24-77) | 53 (23-72) | 55 (23-74) | 53 (21-78) | .378 |
| Male sex, no. (%) | 98 (75) | 17 (59) | 29 (52) | 27 (51) | .004 |
| Diagnosis, no. (%) | |||||
| Acute leukemia/MDS | 54 (41) | 20 (69) | 34 (64) | 25 (47.2) | .004 |
| Lymphoma/CLL/LGL/T-PLL | 74 (57) | 8 (28) | 17 (32) | 23 (43.4) | |
| CML/MM | 3 (2) | 1 (3) | 2 (4) | 5 (9.4) | |
| Conditioning regimens | |||||
| Myeloablative and reduced intensity, no. (%) | .003 | ||||
| TBI-based∗ | 40 (30.5) | 12 (41) | 29 (54.7) | 26 (49) | |
| Chemotherapy-based† | 40 (30.5) | 12 (41) | 29 (54.7) | 26 (49) | |
| Nonmyeloablative, no. (%) | 57 (43.5) | 13 (45) | 20 (37.7) | 23 (43) | |
| Cy/Flu/TBI 200 cGy | 34 (26) | 4 (14) | 4 (7.6) | 4 (8) | |
| Donor, no. (%) | |||||
| MRD | 52 (40) | 2 (7) | 11 (21%) | 5 (9.4%) | <.001 |
| MUD/MMUD | 43/3 (35) | 11/- (38) | 11/1 (22.5) | 18/5 (43.3) | |
| Haploidentical | 7 (5) | 4 (14) | 3 (5.5) | 2 (4) | |
| Cord blood | 26 (20) | 12 (41) | 27 (51) | 23 (43.3) | |
| Donor-recipient HLA-match, no. (%) | |||||
| 8/8 | 97 (74) | 13 (45) | 23 (43) | 23 (43.5) | <.001 |
| 7/8 | 3 (2) | 2 (7) | 1 (2) | 5 (9.5) | |
| <7/8 | 31 (24) | 14 (48) | 29 (55) | 25 (47) | |
| Stem cell source, no. (%) | |||||
| BM | 12 (9) | 3 (10.3) | 6 (11) | 3 (6) | <.001 |
| PBSC | 93 (71) | 14 (48.3) | 20 (38) | 27 (51) | |
| Cord blood‡ | 26 (20) | 12 (41.4) | 27 (51) | 23 (43) | |
| GVHD prophylaxis, no. (%) | |||||
| CNI/MTX/ ± siro ± other§ | 96 (73) | 13 (45) | 22 (39) | 24 (45) | <.001 |
| CSA/MMF | 26 (20) | 12 (41) | 29 (52) | 23 (43.5) | |
| PTCy based | 9 (7) | 4 (14) | 5 (9) | 6 (11.5) | |
| GVHD severity, no. (%) | |||||
| Grade I | - | 11 (38) | - | - | <.001 |
| Grade II | - | 7 (24) | 50 (94) | 32 (60) | |
| Grade III | - | 11 (38) | 3 (6) | 10 (19) | |
| Grade IV | - | - | - | 11 (21) |
| . | No-GVHD (n = 131) . | Non-GI (n = 29) . | UGI (n = 53) . | LGI (n = 53) . | P value . |
|---|---|---|---|---|---|
| Median age (range), y | 57 (24-77) | 53 (23-72) | 55 (23-74) | 53 (21-78) | .378 |
| Male sex, no. (%) | 98 (75) | 17 (59) | 29 (52) | 27 (51) | .004 |
| Diagnosis, no. (%) | |||||
| Acute leukemia/MDS | 54 (41) | 20 (69) | 34 (64) | 25 (47.2) | .004 |
| Lymphoma/CLL/LGL/T-PLL | 74 (57) | 8 (28) | 17 (32) | 23 (43.4) | |
| CML/MM | 3 (2) | 1 (3) | 2 (4) | 5 (9.4) | |
| Conditioning regimens | |||||
| Myeloablative and reduced intensity, no. (%) | .003 | ||||
| TBI-based∗ | 40 (30.5) | 12 (41) | 29 (54.7) | 26 (49) | |
| Chemotherapy-based† | 40 (30.5) | 12 (41) | 29 (54.7) | 26 (49) | |
| Nonmyeloablative, no. (%) | 57 (43.5) | 13 (45) | 20 (37.7) | 23 (43) | |
| Cy/Flu/TBI 200 cGy | 34 (26) | 4 (14) | 4 (7.6) | 4 (8) | |
| Donor, no. (%) | |||||
| MRD | 52 (40) | 2 (7) | 11 (21%) | 5 (9.4%) | <.001 |
| MUD/MMUD | 43/3 (35) | 11/- (38) | 11/1 (22.5) | 18/5 (43.3) | |
| Haploidentical | 7 (5) | 4 (14) | 3 (5.5) | 2 (4) | |
| Cord blood | 26 (20) | 12 (41) | 27 (51) | 23 (43.3) | |
| Donor-recipient HLA-match, no. (%) | |||||
| 8/8 | 97 (74) | 13 (45) | 23 (43) | 23 (43.5) | <.001 |
| 7/8 | 3 (2) | 2 (7) | 1 (2) | 5 (9.5) | |
| <7/8 | 31 (24) | 14 (48) | 29 (55) | 25 (47) | |
| Stem cell source, no. (%) | |||||
| BM | 12 (9) | 3 (10.3) | 6 (11) | 3 (6) | <.001 |
| PBSC | 93 (71) | 14 (48.3) | 20 (38) | 27 (51) | |
| Cord blood‡ | 26 (20) | 12 (41.4) | 27 (51) | 23 (43) | |
| GVHD prophylaxis, no. (%) | |||||
| CNI/MTX/ ± siro ± other§ | 96 (73) | 13 (45) | 22 (39) | 24 (45) | <.001 |
| CSA/MMF | 26 (20) | 12 (41) | 29 (52) | 23 (43.5) | |
| PTCy based | 9 (7) | 4 (14) | 5 (9) | 6 (11.5) | |
| GVHD severity, no. (%) | |||||
| Grade I | - | 11 (38) | - | - | <.001 |
| Grade II | - | 7 (24) | 50 (94) | 32 (60) | |
| Grade III | - | 11 (38) | 3 (6) | 10 (19) | |
| Grade IV | - | - | - | 11 (21) |
P values compare the distribution of covariates (rows) across all cohorts with the exception of GVHD severity, which only includes those with GVHD and were obtained using a Fisher exact test for categorical variables and Kruskal-Wallis for age as a continuous variable.
BM, bone marrow; CLL, chornic lymphocytic leukemia; CNI, calcineurin inhibitor; CML, chronic myeloid leukemia; CSA, cyclosporine-A; HLA, human leukocyte antigen; LGL, large granular lymphocytic;MA, myeloablative; MDS, myelodysplastic syndrome; MM, multiple myeloma; MMF, mycophenolate mofetil; MMUD, mismatched-unrelated donor; MRD, matched-related donor; MTX, methotrexate; MUD, matched-unrelated donor; N/A, not applicable; NMA, non-myeloablative; PBSC, peripheral blood stem cell; PTCy, post-transplant cyclophosphamide; RI, reduced intensity; siro, sirolimus; TBI, total body irradiation; T-PLL, T-cell prolymphocytic leukemia.
Includes cyclophosphamide/TBI 1375 cGy, fludarabine/TBI 1375 cGy, cyclophosphamide/thiotepa/TBI 1375 cGy, fludarabine/cyclophosphamide/TBI 1320 to 1375 cGy, cyclophosphamide/fludarabine/thiotepa/TBI 400 cGy.
Includes melphalan/fludarabine, fludarabine/busulfan, melphalan/thiotepa/fludarabine, busulfan/melphalan, busulfan/fludarabine/cyclophosphamide, clofarabine/thiotepa /melphalan, busulfan/cyclophosphamide.
Includes 33 patients who received cord blood graft combined with a haploidentical ex vivo CD34+ selected T-cell depleted graft as a myeloid bridge as part of protocol (NCT01682226).
Other includes bortezomib, maraviroc, and mycophenolate mofetil. Five patients did not receive methotrexate but tacrolimus/sirolimus/MMF (n = 1), tacrolimus/MMF (n = 2), and tacrolimus/sirolimus (n = 2).
Genomic 16S sequences were generated from 1303 stool specimens from 266 patients (average, 4.89 samples per patient). Stool specimens were grouped into preonset (day −20 to day −1 relative to onset of GVHD) and postonset (day 0 to day 20 relative to GVHD onset) bins for the analyses; this resulted in an overall sampling range of day −3 to day 102 relative to HCT. For patients without GVHD, samples were included when collected in the same overall sampling window relative to HCT (supplemental Figure 1A-C). When patients had samples collected on multiple days within a sampling window, the average of each microbiome feature across all samples was taken. In the event that more than 1 sample was collected on the same day from the same patient, 1 sample from that day was randomly selected for the analysis and others omitted.
Dysbiosis in GVHD
To map patterns of microbial injury to specific organ involvement in aGVHD, we first characterized microbial differences between cohorts both pre- and postonset. Analysis of microbial composition at genus level in samples collected preonset revealed distinct bacterial compositions between those with and without GVHD. Microbial disruption in samples from patients with GVHD consisted mainly of a lower abundance of anaerobes of the class Clostridia. The genus Blautia was 3.4-fold and 3.6-fold lower in the UGI and LGI groups, respectively, vs the no-GVHD group (P ≤ .001 for both), whereas the genus Anaerostipes was 37.2-fold, 4.6-fold, and 26.4-fold lower in the non-GI, UGI, and LGI groups, respectively, vs the no-GVHD group (P ≤ .001 for all) (Figure 1A-C; supplemental Table 3). Compared with the no-GVHD group, GVHD patients also had lower abundances of other Clostridia, including the genera Eubacterium, Coprococcus, and Ruminococcus. In contrast, among these preonset samples there were no significant differences in genus-level abundances between patients with GVHD (supplemental Figure 2A-C).
Allo-HCT patients with GVHD developed microbial disruption preonset GVHD. (A-C) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of preonset GVHD (day −20 to −1 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean relative abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). No-GVHD patients had higher abundance of commensals belonging to the class Clostridia when compared with non-GI, UGI, and LGI cohort. p, phylum; o, order; f, family; g, genus; Proteo, Proteobacteria.
Allo-HCT patients with GVHD developed microbial disruption preonset GVHD. (A-C) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of preonset GVHD (day −20 to −1 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean relative abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). No-GVHD patients had higher abundance of commensals belonging to the class Clostridia when compared with non-GI, UGI, and LGI cohort. p, phylum; o, order; f, family; g, genus; Proteo, Proteobacteria.
Microbial disparities continued for GI GVHD patients 20 days postonset compared with their no-GVHD counterparts (Figure 2A-B; supplemental Table 4). Although non-GI patients had a similar flora composition compared with the no-GVHD group at postonset (supplemental Figure 2D), UGI patients showed a respective 3-log-fold and 14.2-fold reduction of Coprococcus and Parabacteroides compared with no-GVHD controls (P ≤ .001) (Figure 2A). The LGI cohort continued to show features of microbiome disruption postonset when compared with the no-GVHD group, including decreased abundance of various Clostridia genera, such as Blautia and Erysipelatoclostridium (5.5-fold and 9.2-fold, respectively; P ≤ .001 for both) (Figure 2B). Although non-GI and UGI groups had similar microbial composition (supplemental Figure 2E), non-GI patients had higher relative abundances of Erysipelatoclostridium, Blautia, and Anaerostipes compared with the LGI group (11.3-fold, 8.2-fold, and 3.5-fold, respectively; P < .05 for all) (Figure 2C). Similarly, the UGI cohort microbiome was distinct in comparison with the LGI group with increased relative abundance of Blautia and Erysipelatoclostridium (5.2-fold and 6.6-fold, respectively; P < .01 for both) (Figure 2D). Taken together, these observations indicate distinct changes in the microbiota correlate with organ involvement both pre- and post-GVHD onset.
Allo-HCT patients with GI GVHD developed microbial disruption postonset GVHD. (A-D) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of postonset GVHD (day 0 to +20 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). (A-B) No-GVHD compared with UGI and LGI patients had increased abundance of class Clostridia commensals Coprococcus and Blautia, respectively, post-GVHD onset. (C-D) Non-GI and UGI patients also had increased relative abundance of Blautia and Erysipelatoclostridium when compared with LGI patients. f, family; g, genus; p, phylum; Proteo, Proteobacteria.
Allo-HCT patients with GI GVHD developed microbial disruption postonset GVHD. (A-D) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of postonset GVHD (day 0 to +20 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). (A-B) No-GVHD compared with UGI and LGI patients had increased abundance of class Clostridia commensals Coprococcus and Blautia, respectively, post-GVHD onset. (C-D) Non-GI and UGI patients also had increased relative abundance of Blautia and Erysipelatoclostridium when compared with LGI patients. f, family; g, genus; p, phylum; Proteo, Proteobacteria.
Summary markers of microbiome composition and aGVHD onset
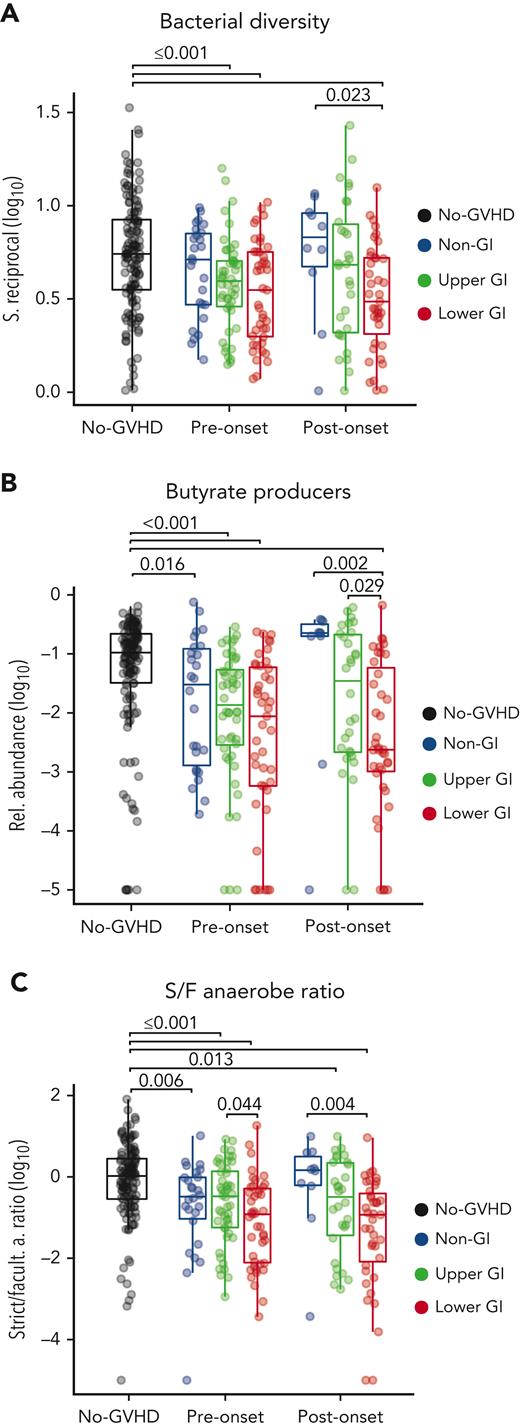
Although specific attributes of microbial composition vary widely between patients after transplantation, several studies have described similar microbial shifts in GVHD, such as a loss in members of the class Clostridia and reduction of diversity.12,14,17,19,34 We sought to determine whether summary indices of microbial composition could serve as indicators of dysbiosis peri-GVHD onset. Prior to aGVHD onset, intestinal α-diversity was significantly lower in patients with GI tract involvement (P ≤ .001) compared with the no-GVHD cohort, whereas the non-GI group had a similar microbiota diversity to the no-GVHD group (P = .241) (Figure 3A). Postonset, LGI patients exhibited significantly lower diversity when compared with no-GVHD and non-GI patients (P < .001 and P = .023, respectively).
GVHD patients have reduced diversity, abundance of butyrate producers, and loss of anaerobiosis pre- and post-GVHD onset. (A) Pre- and postonset aGVHD microbial analysis show reduced diversity in GI GVHD patients. (B) GI GVHD patients also had lower relative abundance of predicted butyrate-producing strains when compared with the other cohorts, and this was also found for LGI patients when compared with non-GI and UGI patients postonset. (C) GVHD patients also had lower strict-to-facultative (S/F) anaerobe ratio in comparison with no-GVHD patients, whereas the LGI patients, specifically, showed lower ratio when compared with UGI and non-GI patients, respectively pre- and postonset.
GVHD patients have reduced diversity, abundance of butyrate producers, and loss of anaerobiosis pre- and post-GVHD onset. (A) Pre- and postonset aGVHD microbial analysis show reduced diversity in GI GVHD patients. (B) GI GVHD patients also had lower relative abundance of predicted butyrate-producing strains when compared with the other cohorts, and this was also found for LGI patients when compared with non-GI and UGI patients postonset. (C) GVHD patients also had lower strict-to-facultative (S/F) anaerobe ratio in comparison with no-GVHD patients, whereas the LGI patients, specifically, showed lower ratio when compared with UGI and non-GI patients, respectively pre- and postonset.
Because butyrate has been implicated in the pathophysiology of GVHD, we next analyzed the aggregated abundance of predicted butyrate producers (supplemental Table 5) in fecal samples of patients with or without GVHD. No-GVHD patients had a higher abundance of predicted butyrate producers when compared with all GVHD groups at preonset, including the non-GI (P = .016), UGI, and LGI (P < .001, for both) groups (Figure 3B). Furthermore, after onset, only patients with LGI GVHD maintained lower abundance of butyrate producers when compared with the other cohorts, including no-GVHD (P < .001), non-GI (P = .002), and UGI (P = .029).
A common feature of the normal human intestinal microbiome is a predominance of obligate anaerobic bacteria,52,53 whereas expansion of facultative anaerobes has been linked to GI inflammatory conditions.54 We and others have observed expansion of facultative anaerobes, for example those from genus Enterococcus, as a significant risk factor for the development of aGVHD and GVHD-related mortality.14,25,55 Thus, we sought to determine the ratio of S/F anaerobes among GVHD groups, defined by annotating the taxa in this data set against a reference-predicted metagenomic functional classification.46 All GVHD groups at preonset had significantly lower S/F anaerobe ratio when compared with the no-GVHD group—non-GI (P = .006), UGI (P = .001), and LGI (P ≤.001) cohorts (Figure 3C)—which was driven both by a low relative abundance of strict anaerobes and high relative abundance of facultative anaerobes (supplemental Figure 3A-C). Notably, LGI patients also had a lower S/F anaerobe ratio (P = .044) (Figure 3C) and less strict anaerobes (P = .039) (supplemental Figure 3B) prior to onset when compared with UGI patients, indicating increased severity of dysbiosis in LGI patients preonset when compared with their UGI counterparts. At postonset UGI and LGI patients maintained a decreased S/F anaerobe ratio compared with no-GVHD controls (P = .013 and P < .001, respectively), and LGI patients also had a lower S/F anaerobe ratio compared with their non-GI counterparts (P = .004).
We also investigated potential cofounders to microbial composition within our data such as antibiotic exposure, infection, and nutritional health. Given the heterogeneity in the different antibiotics to which the patients were exposed, we classified the antibiotics as having medium to high vs low microbiome-disrupting potential close to sample collection (“Methods”) (supplemental Table 1). Nearly all patients received some type of microbiome-disrupting antibiotic during the sampling windows (supplemental Figure 4). The proportion of patients exposed to antibiotics with medium-to-high-microbiome perturbation potential was lower in the no-GVHD cohort compared with those with GVHD preonset, whereas postonset the proportion of non-GI GVHD patients exposed to these antibiotics was lower compared with those with LGI GVHD (supplemental Table 6). Exposure to this group of antibiotics was also associated with reductions in α-diversity, relative abundance of butyrate producers, and S/F anaerobe ratio (supplemental Figure 5A-C), which were independent of the occurrence of bloodstream infection prior to GVHD onset (supplemental Figure 5D-G; supplemental Table 7). Finally, we also explored other potential confounding clinical variables, including nutritional status, modus of nutrition, viral enteritis, and C difficile infection (supplemental Tables 8-11). Although all groups displayed similar patterns of weight loss and associated decrease in body mass index after HCT, patients with GI GVHD had a greater decline in nutritional status as assessed by the nutritional risk index56-58 after GVHD onset (supplemental Figure 6A-F). This index correlated only weakly with microbial features in samples (Spearman R = 0.3) (supplemental Figure 6G-I). Exposure to total parenteral nutrition was associated with lower diversity and lower S/F anaerobe ratio within the no-GVHD group and lower diversity and relative abundance of butyrate producers in the UGI GVHD group prior to GVHD onset (supplemental Figure 6J-L). Notably, in a subset analysis that excluded any patient who received total parenteral nutrition, those with GVHD had lower diversity, lower relative abundance of butyrate producers, and lower S/F anaerobe ratios when compared with no-GVHD controls (supplemental Figure 6M-O; supplemental Table 11). We observed no significant association between C difficile infection prior to GVHD onset with microbiota features in GVHD patients (supplemental Figure 5H-J; supplemental Table 9).
Together, our data suggest that summary indices of pre- and postonset microbial injury, such as bacterial diversity, abundance of butyrate producers, and S/F anaerobe ratio, are sensitive markers of organ-specific GVHD.
Metagenomic pathways analysis
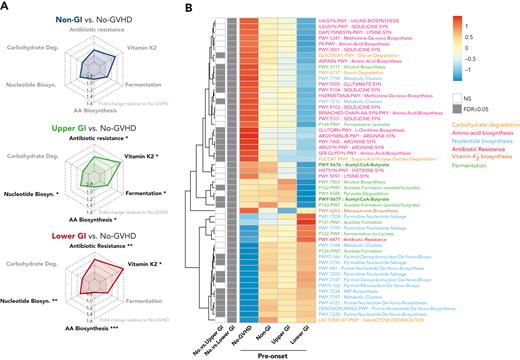
We next explored functional features of the fecal microbiome by extrapolating metabolic pathway abundances from 16S-based taxonomic profiles using the PICRUSt algorithm. We recovered 6 main metabolic pathways that were classified according to the MetaCyc database51: carbohydrate degradation, nucleotide biosynthesis, amino acid biosynthesis, antibiotic resistance, vitamin K2 biosynthesis, and fermentation including SCFA metabolism (Figures 4-5). Patients with GI GVHD had increased abundance of pathways associated with vitamin K2 biosynthesis, nucleotide biosynthesis, antibiotic resistance and reduced abundance of pathways related to amino acid biosynthesis when compared with no-GVHD patients at preonset (P < .05 for all) (Figure 4A-B; supplemental Table 12). Interestingly, although UGI GVHD patients showed increased presence of pathways broadly associated with fermentation prior to onset (P < .05), LGI GVHD patients had lower abundance of butyrate-specific pathways when compared with no-GVHD controls (Figure 4B, bold green), which is consistent with our findings of composition at the genus level and aggregated abundance of butyrate producers.
GVHD patients present distinct preonset predicted PICRUSt pathways. (A) Radar chart representation of PICRUSt predicted functional pathway relative abundance from patients without GVHD and GVHD patients at preonset. Relevant pathways belonged to 6 main metabolic categories: carbohydrate degradation, nucleotide biosynthesis, amino acid biosynthesis, antibiotic resistance, vitamin K2 biosynthesis, and fermentation. Axis represents fold change relative to no-GVHD patients. (B) Heat map with hierarchical clustering of statistically significant (FDR P ≤ .05) unique PICRUSt pathways. Figure displays pathways found in a minimum of 5% samples within all groups. UGI and LGI GVHD patients show increased pathways associated with general antibiotic resistance, vitamin K2 metabolism, and nucleotide biosynthesis, with reduced representation related to amino acid biosynthesis compared with no-GVHD patients. There was reduced presence of butyrate-producing specific pathways in LGI patients (bold). ∗FDR P ≤ .05; ∗∗FDR P ≤ .01; ∗∗∗FDR P ≤ .001. NS, not significant.
GVHD patients present distinct preonset predicted PICRUSt pathways. (A) Radar chart representation of PICRUSt predicted functional pathway relative abundance from patients without GVHD and GVHD patients at preonset. Relevant pathways belonged to 6 main metabolic categories: carbohydrate degradation, nucleotide biosynthesis, amino acid biosynthesis, antibiotic resistance, vitamin K2 biosynthesis, and fermentation. Axis represents fold change relative to no-GVHD patients. (B) Heat map with hierarchical clustering of statistically significant (FDR P ≤ .05) unique PICRUSt pathways. Figure displays pathways found in a minimum of 5% samples within all groups. UGI and LGI GVHD patients show increased pathways associated with general antibiotic resistance, vitamin K2 metabolism, and nucleotide biosynthesis, with reduced representation related to amino acid biosynthesis compared with no-GVHD patients. There was reduced presence of butyrate-producing specific pathways in LGI patients (bold). ∗FDR P ≤ .05; ∗∗FDR P ≤ .01; ∗∗∗FDR P ≤ .001. NS, not significant.
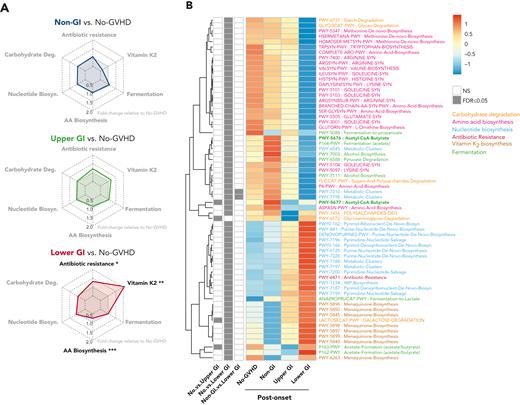
GVHD patients present distinct postonset predicted PICRUSt pathways. (A) Radar chart representation of PICRUSt predicted functional pathways belonging to 6 main metabolic pathways: carbohydrate degradation, nucleotide biosynthesis, amino acid biosynthesis, antibiotic resistance, vitamin K2 biosynthesis, and fermentation for GVHD patients post-GVHD onset relative to no-GVHD controls. Axis represents fold change relative to no-GVHD patients. (B) Heat map with hierarchical clustering of statistically significant (FDR P ≤ .05) unique PICRUSt pathways. Figure displays pathways found in a minimum of 5% of samples within all groups. LGI GVHD patients show increased abundance of pathways associated with general antibiotic resistance and vitamin K2 metabolism with lower abundance of pathways linked to amino acid biosynthesis relative to no-GVHD patients. Lower abundance of pathways specific to butyrate production was also found in LGI patients compared with no-GVHD controls (bold). ∗FDR P ≤ .05; ∗∗FDR P ≤ .01; ∗∗∗FDR P ≤ .001. NS, not significant.
GVHD patients present distinct postonset predicted PICRUSt pathways. (A) Radar chart representation of PICRUSt predicted functional pathways belonging to 6 main metabolic pathways: carbohydrate degradation, nucleotide biosynthesis, amino acid biosynthesis, antibiotic resistance, vitamin K2 biosynthesis, and fermentation for GVHD patients post-GVHD onset relative to no-GVHD controls. Axis represents fold change relative to no-GVHD patients. (B) Heat map with hierarchical clustering of statistically significant (FDR P ≤ .05) unique PICRUSt pathways. Figure displays pathways found in a minimum of 5% of samples within all groups. LGI GVHD patients show increased abundance of pathways associated with general antibiotic resistance and vitamin K2 metabolism with lower abundance of pathways linked to amino acid biosynthesis relative to no-GVHD patients. Lower abundance of pathways specific to butyrate production was also found in LGI patients compared with no-GVHD controls (bold). ∗FDR P ≤ .05; ∗∗FDR P ≤ .01; ∗∗∗FDR P ≤ .001. NS, not significant.
PICRUSt analysis of samples at postonset suggested that microbial metabolic dysfunction was most apparent in LGI patients (Figure 5A-B; supplemental Table 13). LGI GVHD patients continued to show increased abundance of metabolic pathways linked to antibiotic resistance (P = .02) and vitamin K2 (P =.005) and lower abundance of amino acid biosynthesis pathways (P < .001) when compared with no-GVHD patients (Figure 5A-B). Moreover, we also found reduced abundance of butyrate-specific pathways in LGI GVHD patients at postonset when compared with no-GVHD controls (Figure 5B, bold green).
To confirm these PICRUSt observations, we next analyzed the metabolic genome profiles of samples from a subset of 131 patients belonging to no-GVHD (n = 68), non-GI (n = 8), UGI (n = 23), and LGI (n = 32) cohorts who had undergone whole metagenomic shotgun sequencing (supplemental Figures 1, 7, and 8). Prior to onset, the microbiome of LGI GVHD patients had higher overall abundance of pathways associated with carbohydrate degradation and lower abundance of those associated with amino acid biosynthesis (supplemental Figure 7A-C; supplemental Table 14), which partially corroborated our PICRUSt data. Post-GVHD onset, the LGI group had increased abundance of broad gene pathways associated with carbohydrate degradation and decreased abundance of genes associated with amino acid biosynthesis and various fermentation pathways linked to butyrate production (supplemental Figure 8A-B; supplemental Table 15). As such, metagenomics confirmed a pattern of compromised butyrate production and abundance differences in various other metabolism-related pathways in the intestinal microbiota, particularly in LGI patients postonset.
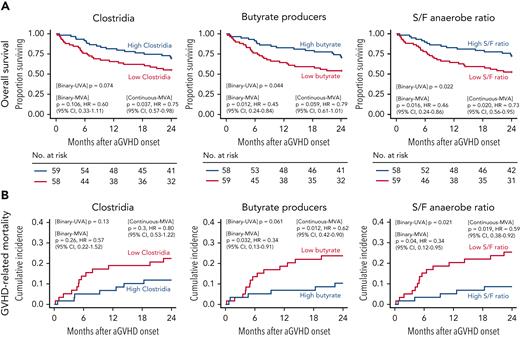
Microbial biomarkers in survival outcomes
Lastly, we investigated the association of microbial features measured in the 20 days prior to GVHD onset with clinical outcomes after GVHD onset. We analyzed the association between Clostridia, predicted butyrate producers, and S/F anaerobe ratio prior to GVHD onset with OS, GVHD-related mortality, TRM, and relapse. Lower mortality risk in the first 2 years of GVHD onset was associated with high S/F anaerobe ratio (hazard ratio [HR], 0.46; 95% confidence interval [CI], 0.24-0.86; P = .016) and high relative abundance of predicted butyrate producers (HR, 0.45; 95% CI, 0.24-0.84; P = .012) (Figure 6A; supplemental Table 16). Although not significant as a binary variable (P = .106), the higher relative abundance of Clostridia was associated with a reduced risk of death when analyzed as a continuous predictor (HR, 0.75; 95% CI, 0.57-0.98; P = .037). Higher abundance of predicted butyrate producers and S/F anaerobe ratio were both associated with lower risks of GVHD-related mortality (HR, 0.34; 95% CI, 0.13-0.91; P = .032 and HR, 0.34; 95% CI, 0.12-0.95; P = .04, respectively) (Figure 6B). Additionally, 2-year TRM was significantly lower in patients with high abundance of butyrate producers (P = .013) and S/F anaerobe ratio (P = .035) (supplemental Figure 9A). Moreover, older age, female sex, and nonablative conditioning (vs reduced intensity) were also associated with reduced survival in multivariate comparisons (supplemental Table 17), whereas no associations were observed between these summary microbial markers and incidence of relapse (supplemental Figure 9B). In summary, these data suggest that global biomarkers of microbial health prior to onset may be useful tools in predicting outcomes in patients with GVHD.
Preonset GVHD clostridia abundance, butyrate producers, and S/F anaerobe ratio predicts survival in allo-HCT patients. OS (A) and GVHD-related mortality (B). Patients were stratified according to the median relative abundances of class Clostridia, butyrate producers, and S/F anaerobe ratio prior to GVHD onset. Figure shows univariable (UVA) and multivariable (MVA) tests of associations for microbial determinants as binary or continuous variables. The proportional hazards multivariable regression models (OS) and the Fine and Gray multivariable regression models (GVHD-related mortality) were adjusted for sex, age, and conditioning regimen. Data corroborates the association of higher relative abundance of butyrate-producing bacteria and S/F ratio to increased OS and reduced GVHD-related mortality.
Preonset GVHD clostridia abundance, butyrate producers, and S/F anaerobe ratio predicts survival in allo-HCT patients. OS (A) and GVHD-related mortality (B). Patients were stratified according to the median relative abundances of class Clostridia, butyrate producers, and S/F anaerobe ratio prior to GVHD onset. Figure shows univariable (UVA) and multivariable (MVA) tests of associations for microbial determinants as binary or continuous variables. The proportional hazards multivariable regression models (OS) and the Fine and Gray multivariable regression models (GVHD-related mortality) were adjusted for sex, age, and conditioning regimen. Data corroborates the association of higher relative abundance of butyrate-producing bacteria and S/F ratio to increased OS and reduced GVHD-related mortality.
Discussion
The intestinal microbiome is a crucial component of gut health, and its relationship with the host includes exchange of factors and nutrients that promote gut homeostasis and modulate host immunity. Various studies in mice and humans have demonstrated that the intestinal microbiome composition is particularly relevant for allo-HCT outcomes, including GVHD.12-19 This study focused on the fecal microbiome of adult allo-HCT recipients with or without aGVHD, according to organ involvement. LGI GVHD patients had worse microbial injury than patients from other groups. Low abundance of Clostridia members in fecal samples of allo-HCT patients has been described previously in the peri-engraftment period.12,19 Here we extend this finding to the 20 days prior to onset of GVHD, specifically for Clostridia members such as Ruminococcus, Anaerostipes, Eubacterium, and Blautia. In postonset samples, we only observed this pattern of dysbiosis in those with GI GVHD. Increased abundance of Blautia has been associated with improved OS and reduced GVHD after HCT,15 and Blautia was reduced significantly in GI GVHD patients. Furthermore, a study evaluating the immunologic consequences of monocolonization of germ-free mice with Ruminococcus gnavus, a species suggested to belong to the Blautia genus,59 reported increased colon frequencies of regulatory T cells and innate lymphoid cells.60 These cells are of particular interest in GVHD, as regulatory cells may reduce GVHD in patients and preclinical models61,62 and innate lymphoid cells can promote intestinal regeneration through secretion of interleukin-22 and ameliorate aGVHD.63,64
The heterogeneity of microbial compositions across populations makes identifying associations between specific microbes and HCT outcomes complex. We therefore investigated whether broad markers of intestinal dysbiosis could be used as surrogates to characterize organ involvement in GVHD. We first evaluated microbiome diversity, which was decreased in patients with GI GVHD preonset, and continued postonset for the LGI cohort. These findings corroborate studies by our group and others demonstrating that lower diversity pretransplant and peri-engraftment is associated with GVHD and survival outcomes.13-15 Moreover, we found an association between exposure to antibiotics with high potential to disrupt the flora at preonset and dysbiosis in the gut, which is in agreement with other studies.14,16 The observational design of this study prevents definitively dissociating potential effects of antibiotics themselves on the microbiome from potential effects of the underlying condition the antibiotics were used to treat. Antibiotic-associated dysbiosis metrics were observed comparably in the overall cohort and in subsets excluding those with bloodstream infections, pointing to a prominent role for antibiotics in mediating microbiome injury in this population.
A diverse microbial community is important for the production of microbial metabolites, with beneficial effects on the host. Metabolites such as SCFAs, bile acids, vitamins, and tryptophan-derived molecules are important for intestinal gut health and immune regulation.63,65-67 The SCFA butyrate has an important role in maintaining the epithelial barrier68 and regulating local intestinal immunity by inducing differentiation of colonic T-regs.69 Importantly, in preclinical models, butyrate administration could mitigate acute and chronic GVHD.22,23 We found that the relative abundance of predicted butyrate producers was lower prior to onset in GI GVHD patients when compared with no-GVHD controls. Intestinal outgrowth of facultative anaerobes is also a significant risk factor for the development of aGVHD and worse survival after allo-HCT.14,25,55 All GVHD groups had a significantly lower S/F anaerobe ratio preonset when compared with no-GVHD cases, which persisted postonset in GI GVHD patients. Our findings extend previous reports that intestinal domination by facultative anaerobes is associated with worse aGVHD.14,25,55
In order to uncover microbial metabolites with potential to modulate organ involvement in GVHD, we further investigated metabolic pathway abundances in the fecal samples via PICRUSt and shotgun metagenomics. Patients without GVHD showed increased predicted abundance of pathways associated with carbohydrate degradation, amino acid biosynthesis, and fermentation, including butyrate-specific pathways, whereas antibiotic resistance and nucleotide biosynthesis pathways were increased in GI GVHD patients both pre and postonset. Notably, we observed a reduction in pathways responsible for fermentation to butyrate in patients with GI GVHD, particularly the LGI, which is consistent with our findings on reduced predicted butyrate producers within these cohorts. These findings are in agreement with preclinical studies, which have suggested a protective effect of butyrate in GVHD.23 However this protective effect may depend on the degree of intestinal damage and butyrate concentration available.26,27
Finally, we also demonstrate the clinical importance of these markers of microbial health after allo-HCT, in which higher relative abundance of predicted butyrate producers and S/F anaerobe ratio were associated with significant improvement in transplant outcomes, including overall survival, GVHD-related mortality, and TRM. In conclusion, we found significant associations between microbial diversity, the abundance of butyrate producing taxa, and the S/F anaerobe ratio with GI GVHD, particularly LGI GVHD. Our findings have practical implications for the development of microbiome-derived biomarkers and prophylactic and therapeutic interventions in aGVHD.
Acknowledgments
This work was supported in part by the MSK Internal Diversity Enhancement Award (IDEA); National Institutes of Health, National Cancer Institute award numbers R01-CA228358, R01-CA228308, P30 CA008748 MSK Cancer Center Core Grant and P01-CA023766; National Heart, Lung, and Blood Institute award numbers R01-HL123340, R01-HL147584, and K08HL143189; National Institute of Aging award number P01-AG052359; Starr Cancer Consortium; and Tri Institutional Stem Cell Initiative. Additional funding was received from the Lymphoma Foundation, The Susan and Peter Solomon Divisional Genomics Program, Cycle for Survival, MSKCC Cancer Systems Immunology Pilot Grant, Empire Clinical Research Investigator Program, the Society for MSK, the Staff Foundation, the American Society for Transplantation and Cellular Therapy, MSKCC Leukemia SPORE Career Enhancement Program, and the Parker Institute for Cancer Immunotherapy.
Authorship
Contribution: D.M.P., M.B.d.S., J.U.P., and M.R.M.v.d.B. designed the study, analyzed the data, and wrote the manuscript; M.B.d.S., A.D., S.M.D., A.L.C.G., and T.F. performed statistical analysis; G.M., A.C., J.S., and G.K.A. collected the data; G.K.A., E.F., L.A.A., and R.J.W. performed research; and R.S., S.D., E.F., L.A.A., R.J.W., H.A, O.M., M.-A.P., S.D., and Y.T. wrote the manuscript.
Conflict-of-interest disclosure: D.M.P. has served as advisory board member for Evive Biotechnology (Shanghai) Ltd (formerly Generon [Shanghai] Corporation Ltd); has consulted, received honorarium from, or particpated in advisory boards for Kadmon Corporation/Sanofi, CareDx, Ceramedix, and Incyte; has received research support from Incyte. A.L.C.G. reports equity, salary and is employed by Xbiome Co. R.S. received consultancy fees from Medexus and MyBiotics. M.-A.P. reports honoraria from Adicet, Allovir, Caribou Biosciences, Celgene, Bristol-Myers Squibb, Equilium, Exevir, Incyte, Karyopharm, Kite/Gilead, Merck, Miltenyi Biotec, MorphoSys, Nektar Therapeutics, Novartis, Omeros, OrcaBio, Syncopation, VectivBio AG, and Vor Biopharma. He serves on DSMBs for Cidara Therapeutics, Medigene, and Sellas Life Sciences, and the scientific advisory board of NexImmune. He has ownership interests in NexImmune and Omeros. He has received institutional research support for clinical trials from Incyte, Kite/Gilead, Miltenyi Biotec, Nektar Therapeutics, and Novartis. J.U.P. reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics and consulting fees from DaVolterra, CSL Behring, and MaaT Pharma. He serves on an advisory board of and holds equity in Postbiotics Plus Research. He has filed intellectual property applications related to the microbiome (reference numbers 62/843,849, 62/977,908, and 15/756,845). M.R.M.v.d.B. has received research support and stock options from Seres Therapeutics and stock options from Notch Therapeutics and Pluto Therapeutics; he has received royalties from Wolters Kluwer; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, Vor Biopharma, Rheos Medicines, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Ceramedix, Lygenesis, Pluto Therapeutics, GlaxoSmithKline, Da Volterra, Thymofox, Garuda, Novartis (Spouse), Synthekine (Spouse), Beigene (Spouse), Kite (Spouse); he has IP Licensing with Seres Therapeutics and Juno Therapeutics; and holds a fiduciary role on the Foundation Board of DKMS (a nonprofit organization). Memorial Sloan Kettering Cancer Center (MSK) has financial interests relative to Seres Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Doris M. Ponce and Marcel R. M. van den Brink, Memorial Sloan Kettering, 545 East 73rd St, New York, NY 10021; e-mail: vandenbm@mskcc.org.
References
Author notes
∗M.B.d.S., D.M.P., J.U.P., and M.R.M.v.d.B. contributed equally to this work.
Sequencing data have been deposited in National Center for Biotechnology Information databases; accession identifiers are tabulated in supplemental Table 2.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Allo-HCT patients with GVHD developed microbial disruption preonset GVHD. (A-C) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of preonset GVHD (day −20 to −1 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean relative abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). No-GVHD patients had higher abundance of commensals belonging to the class Clostridia when compared with non-GI, UGI, and LGI cohort. p, phylum; o, order; f, family; g, genus; Proteo, Proteobacteria.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/22/10.1182_blood.2021015352/5/m_blood_bld-2021-015352-gr1.jpeg?Expires=1769107837&Signature=e~flTzEz~PfmK2uDeHlgXVOdyWsJdCX~VGGbSZm5bXynFffNCPUB~dqEOXxCdLn0ZT-lxIQIi~I4nS8V8fcYGhJ0aFetXI8fZ5Hxu4Y~GSQnM0zjJ3vtvj0nVymygsbqd907Q-uGgeAGaVr2VAQGx6kzKnb9uf5521yhM5gSEB9FFobW0LIawN3xi6OOOtAZgSNwg5kNBfB2-Z9ceOW4WxLyFhNy8pD3lUuP6XZbmCSLpb2fnrDAQArg6ttUxY6euROgjiCiEEYKTV4LOazu0OCpnbllpo16xq7sPO~1Pj4KM67CGVwNJ9T8sbMqp1dIjQRlvJJQ98MlINtOiya2aQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Allo-HCT patients with GI GVHD developed microbial disruption postonset GVHD. (A-D) Volcano plot [−log10(FDR P value) vs log2(fold change)] representation of microbial dysbiosis of postonset GVHD (day 0 to +20 relative to onset) and no-GVHD (day −3 to +102 relative to allo-HCT) fecal samples. Plot includes taxa with mean abundance within samples >0.1% and highlights the 20 genera with strongest FDR corrected P-value significance (≤0.05). (A-B) No-GVHD compared with UGI and LGI patients had increased abundance of class Clostridia commensals Coprococcus and Blautia, respectively, post-GVHD onset. (C-D) Non-GI and UGI patients also had increased relative abundance of Blautia and Erysipelatoclostridium when compared with LGI patients. f, family; g, genus; p, phylum; Proteo, Proteobacteria.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/22/10.1182_blood.2021015352/5/m_blood_bld-2021-015352-gr2.jpeg?Expires=1769107837&Signature=i7WIrV2j8STXIcxhhFdCFMu266wCVRlmVGAEFPaY3k2Q7j8~nSY6o7eS9~i90IoK4jhKsamuaKRzOUXi3jh7YSEX2ce~XaTM0rKknBa0uqKpMQkZox~2VeCJoHvjBhq~ICffBfTgEw0X5p2VIge6336ZkDulgsBr~m5FoCmt-nRy2Jtfclu1Zjb3ZyZt0BSRFnvr51034bWmptBEd3fEbOVcNNth4vzdeiQz8jfzl-EDynmszH84WB6h6ceWRsDptuKihJN~hxqE2EnQR-soLKVO7jTY6hiUiV4n1iEulZLYRkkgCeCpTnmAcWhA4FdZFMboS7LF7BjBQjE0SjJnwA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal