Key points
ATRA, by activating RARγ-IRF1-OAS1-RNase L pathway and modulating cellular RNA degradation, enhances MM sensitivity to Cfz treatment.
RARγ agonists ATRA and BMS961 are highly promising and translatable drugs for (re)sensitizing MM to Cfz treatment.
Abstract
Proteasome inhibitors (PIs) such as bortezomib (Btz) and carfilzomib (Cfz) are highly efficacious for patients with multiple myeloma (MM). However, relapses are frequent, and acquired resistance to PI treatment emerges in most patients. Here, we performed a high-throughput screen of 1855 Food and Drug Administration (FDA)-approved drugs and identified all-trans retinoic acid (ATRA), which alone has no antimyeloma effect, as a potent drug that enhanced MM sensitivity to Cfz-induced cytotoxicity and resensitized Cfz-resistant MM cells to Cfz in vitro. ATRA activated retinoic acid receptor (RAR)γ and interferon-β response pathway, leading to upregulated expression of IRF1. IRF1 in turn initiated the transcription of OAS1, which synthesized 2-5A upon binding to double-stranded RNA (dsRNA) induced by Cfz and resulted in cellular RNA degradation by RNase L and cell death. Similar to ATRA, BMS961, a selective RARγ agonist, could also (re)sensitize MM cells to Cfz in vitro, and both ATRA and BMS961 significantly enhanced the therapeutic effects of Cfz in established MM in vivo. In support of these findings, analyses of large datasets of patients’ gene profiling showed a strong and positive correlation between RARγ and OAS1 expression and patient’s response to PI treatment. Thus, this study highlights the potential for RARγ agonists to sensitize and overcome MM resistance to Cfz treatment in patients.
Introduction
Multiple myeloma (MM) is a plasma cell cancer characterized by tumor cell accumulation and expansion in the bone marrow (BM)1 and accounts for ∼10% of all hematologic malignancies.2 Patients can develop morbidity due to hypercalcemia, renal insufficiency, anemia, and bony lesions.3 Novel chemotherapies of proteasome inhibitors (PIs), such as bortezomib (Btz) and carfilzomib (Cfz), immune-modifying drugs, such as thalidomide and lenalidomide, and monoclonal antibodies, such as daratumumab, have greatly improved the median overall survival (OS) of patients with MM.3-5
PIs kill MM cells by disrupting the degradation of misfolded proteins, presumably derived from high-level immunoglobulin production, which provides an appealing explanation for why MM cells are so uniquely sensitive to PIs.6-8 Endoplasmic reticulum (ER) stress is considered one of the major determinants of PI cytotoxicity.8,9 Btz, a reversible PI, was initially found to be effective in refractory MM and later approved for relapsed MM. Btz-based combination therapies were approved for relapsed or refractory and newly diagnosed patients with MM. More recently, Cfz, an irreversible PI and approved for relapsed/refractory and then newly diagnosed MM, is associated with clinically significant reduction in the risk of death compared with Btz.10-12
However, despite the demonstrated benefits of PIs, relapses are frequent, and acquired resistance to PI treatment eventually emerges in most, if not all, patients.6,13 Therefore, identifying novel and safe drugs overcoming PI resistance in MM will aid in chemo-(re)sensitization, reducing PI-induced side effects, and maximizing the outcomes of PI therapy. To date, there are genes proposed to regulate PI sensitivity in MM. They include, but are not limited to, TJP1 impacting proteasome capacity,14 NRF2 mediating antioxidant capacity15,16 and inhibiting autophagy,17 and GCN2 inducing integrated stress response when activated by amino acid deprivation.18,19 However, it will take much effort and time to develop specific antagonists or agonists targeting these genes to overcome PI resistance. In this study, we performed a high-throughput screening of 1855 Food and Drug Administration (FDA)-approved drugs20,21 to identify old drugs that can enhance or resensitize MM cells to PIs/Cfz. Interestingly, we discovered that tretinoin (all-trans retinoic acid [ATRA]), a classic pan-retinoic acid receptor (RAR) agonist used successfully to treat acute promyelocytic leukemia (APL) by inducing cell differentiation,22-24 was able to (re)sensitize human MM cells to Cfz treatment. As MM is terminally differentiated plasma cells,25 we hypothesized that ATRA-induced MM (re)sensitization to Cfz is independent of plasma cell redifferentiation but caused by unknown mechanisms. To elucidate the underlying mechanisms and explore the potential of using ATRA and other RAR agonists to sensitize MM to Cfz regimens, a series of in vitro and in vivo studies using human MM cell lines and primary MM cells and murine models was performed.
Materials and methods
Primary MM cells and cell lines
This study was approved by the Institutional Review Boards at the Lerner Research Institute, Cleveland Clinic and Houston Methodist Research Institute, Houston Methodist Hospital, and all patients provided written informed consent. BM aspirates were obtained from newly diagnosed patients with MM as part of their routine clinical workup, and patients were asked if they wished to provide informed consent for use of their samples for research purposes. BM CD138+ MM cells were isolated by EasySep Human Whole Blood and CD138 Positive Selection Kit II (STEMCELL Technologies, Cambridge, MA). Human BM-derived mesenchymal stem cells (BMSCs) were generated from BM aspirates of patients with MM as previously described.26 MM cell line ARP-1 was kindly provided by the Arkansas Cancer Research Center.27,28 LP-1, Cfz-resistant KMS-11/Cfz, and its wild-type (WT) KMS-11 cells were kindly provided by Robert G. Hawley.29,30 KMS-12-PE and KMS-12-BM cells were kind gifts from Frederic J. Reu of the Cleveland Clinic.31 Other cell lines were purchased from the American Type Culture Collection (Rockville, MD). All MM cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA).32,33
Cell viability and apoptosis assay
Cell viability was assessed using 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay after 48 hours of treatment. For apoptosis assay, MM cells (2.5 × 105/mL) were pretreated with chemotherapeutic drugs for 24 hours. The fraction of apoptotic cells was determined by staining with APC-conjugated Annexin V and analyzed by flow cytometry (BD LSRFortessa, BD Biosciences) as described.34,35 Combination index (CI) for assessment of drug synergy was calculated and constructed using Compusyn (ComboSyn, Inc, Paramus, NJ) according to the method of Chou and Talalay.36,37
Mouse MM models
NSG mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were maintained in American Association of Laboratory Animal Care–accredited facilities, and the studies were approved by the Institutional Animal Care and Use Committee of the Houston Methodist Research Institute and the Lerner Research Institute of Cleveland Clinic.
Statistical analysis
All data are shown as mean ± standard deviation. The Student 2-tailed t test was used to compare 2 experimental groups; when >2 groups were included in an analysis, a Bonferroni corrected significance level was used. A P value <.05 was considered significant. In some experiments, linear regression analyses were performed using GraphPad Prism 8, and slope and R values were calculated. Patient survival in RARγ and OAS1 expression groups was analyzed by log-rank (Mantel-Cox) test using R and GraphPad Prism 8 software. For patient stratification, the cutoff threshold was determined using maximally selected log-rank statistics as implemented in the maxstat R package (http://cran.r-project.org/web/packages/maxstat/index.html).
Results
Screen of FDA-approved drugs identifies ATRA in the top hits sensitizing MM to Cfz treatment
To identify potential candidates that can promote the cytotoxicity of Cfz on human MM cells, we performed a high-throughput screen of 1855 FDA-approved drugs using cell viability assay (supplemental Figure 1A-B, available on the Blood Web site). To find the most potent and safe compounds, positive hits were defined as compounds that alone showed low toxicity in MM.1S cells (>50% cell viability of dimethyl sulfoxide [DMSO]-treated cells) and exhibited > twofold toxic effects of Cfz-treated MM cells compared with Cfz treatment alone (Figure 1A). Interestingly, the drug library has PIs Cfz and ixazomib,38 and histone deacetylase inhibitor resminostat,39 which are novel anti-MM chemotherapeutic drugs. These drugs not only directly killed but also promoted Cfz-mediated killing of MM.1S cells and thus were used as positive controls for our screening (Figure 1A). Among 73 positive hits, ATRA, a vitamin A analog used as first-line treatment of APL with low side effects, emerged as 1 of top 3 hits that elicited remarkable inhibitory effect in cell viability of Cfz-treated MM.1S cells compared with Cfz or ATRA alone (Figure 1B; supplemental Figure 1C). We validated that ATRA, which alone did not inhibit MM growth, was synergistic with Cfz against MM cells starting from 4 µM (Figure 1C; supplemental Figure 2; supplemental Tables 1 and 2). Furthermore, ATRA promoted Cfz-induced apoptosis not only in human MM cell lines (Figure 1D; supplemental Figure 3A) but also in primary MM cells (Figure 1E; supplemental Figure 3B). In addition, ATRA not only induced more cell death in Cfz-treated WT KMS-11 cells but also resensitized KMS-11/Cfz cells to Cfz treatment (Figure 1F; supplemental Figure 3C). In addition, although BMSCs protected MM cells from Cfz-induced apoptosis and promoted MM cell proliferation, ATRA significantly increased Cfz-induced apoptosis (Figure 1G; supplemental Figure 3D) and enhanced the antiproliferation effect of Cfz in human MM cell lines in the presence or absence of BMSCs (supplemental Figure 3E). Taken together, these results strongly suggest that ATRA is a promising drug to increase Cfz cytotoxicity in MM cells.
High-throughput screening of drugs identifies that ATRA enhances Cfz-killing effect of human MM cells. FDA-approved compound libraries containing 1855 drugs were screened for enhancing the anticancer efficiency of Cfz in human MM.1S cells. In the screening, 1-hour pulse treatment of 30 nM Cfz or DMSO followed by 4 µM FDA-approved drug (X) treatment for 48 hours. Cell viability of MM.1S cells were detected by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay. (A) Dot plot showing the inhibitory effect of Cfz plus X on MM cell viability (−log2 [Cfz+X], y-axis) and relative cell viability of X-treated cells normalized to DMSO-treated cells (x-axis). Cfz, Resminostat (HDAC inhibitor), and Ixazomib in the library served as positive controls. Black line indicates 30% inhibition of MM cell viability in culture with Cfz alone; red line indicates the potential cutoff threshold (>65% inhibition by Cfz+X treatment); blue line is the cutoff line of low toxicity of X treatment (>50% cell viability by X treatment compared with DMSO treatment). (B) Dot plot showing relative inhibitory effect of Cfz + X treatment compared with X treatment alone (log2X − log2[Cfz+X], y-axis) and Cfz alone (−log2 [Cfz+X], x-axis). The potential drug X from the cutoff area in panel A and blue point indicates ATRA. (C) CI plot showing synergistic effect of Cfz with 10 µM ATRA in MM cells across a range of effective drug doses (MM.1S and MM.1R: Cfz 5-30 nM; ARP-1 and LP-1: Cfz 30 to 120 nM; KMS-12-PE and U266: Cfz 15 to 90 nM; primary MM: Cfz 10 to 80 nM). Detailed information is provided in supplemental Table 1. CI was calculated using the CompuSyn software. Each point represents the CI value plotted against the corresponding mean fraction affected (Fa) by the combination treatment relative to DMSO control in 3 independent experiments. CI < 1 represents synergism; CI = 1 represents additive effect; and CI > 1 represents antagonism. (D-E) Summarized results of apoptosis of MM cell lines (D) or primary MM cells (E) in cultures with the drugs (ATRA for 10 µM. For Cfz pulse treatment, MM.1S and MM.1R: 30 nM; ARP-1: 100 nM; KMS-12-PE: 60 nM; primary MM cells: 80 nM Cfz) for 24 hours. (F) Summary results of apoptosis of WT KMS-11 and KMS-11/Cfz MM cells with indicated treatment (200 nM Cfz pulse treatment and 10 µM ATRA) for 24 hours. (G) Summarized results of apoptosis of MM.1S and ARP-1 cells alone or in coculture with patient-derived BMSCs for 2 days followed by the indicated treatments for 24 hours. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
High-throughput screening of drugs identifies that ATRA enhances Cfz-killing effect of human MM cells. FDA-approved compound libraries containing 1855 drugs were screened for enhancing the anticancer efficiency of Cfz in human MM.1S cells. In the screening, 1-hour pulse treatment of 30 nM Cfz or DMSO followed by 4 µM FDA-approved drug (X) treatment for 48 hours. Cell viability of MM.1S cells were detected by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay. (A) Dot plot showing the inhibitory effect of Cfz plus X on MM cell viability (−log2 [Cfz+X], y-axis) and relative cell viability of X-treated cells normalized to DMSO-treated cells (x-axis). Cfz, Resminostat (HDAC inhibitor), and Ixazomib in the library served as positive controls. Black line indicates 30% inhibition of MM cell viability in culture with Cfz alone; red line indicates the potential cutoff threshold (>65% inhibition by Cfz+X treatment); blue line is the cutoff line of low toxicity of X treatment (>50% cell viability by X treatment compared with DMSO treatment). (B) Dot plot showing relative inhibitory effect of Cfz + X treatment compared with X treatment alone (log2X − log2[Cfz+X], y-axis) and Cfz alone (−log2 [Cfz+X], x-axis). The potential drug X from the cutoff area in panel A and blue point indicates ATRA. (C) CI plot showing synergistic effect of Cfz with 10 µM ATRA in MM cells across a range of effective drug doses (MM.1S and MM.1R: Cfz 5-30 nM; ARP-1 and LP-1: Cfz 30 to 120 nM; KMS-12-PE and U266: Cfz 15 to 90 nM; primary MM: Cfz 10 to 80 nM). Detailed information is provided in supplemental Table 1. CI was calculated using the CompuSyn software. Each point represents the CI value plotted against the corresponding mean fraction affected (Fa) by the combination treatment relative to DMSO control in 3 independent experiments. CI < 1 represents synergism; CI = 1 represents additive effect; and CI > 1 represents antagonism. (D-E) Summarized results of apoptosis of MM cell lines (D) or primary MM cells (E) in cultures with the drugs (ATRA for 10 µM. For Cfz pulse treatment, MM.1S and MM.1R: 30 nM; ARP-1: 100 nM; KMS-12-PE: 60 nM; primary MM cells: 80 nM Cfz) for 24 hours. (F) Summary results of apoptosis of WT KMS-11 and KMS-11/Cfz MM cells with indicated treatment (200 nM Cfz pulse treatment and 10 µM ATRA) for 24 hours. (G) Summarized results of apoptosis of MM.1S and ARP-1 cells alone or in coculture with patient-derived BMSCs for 2 days followed by the indicated treatments for 24 hours. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
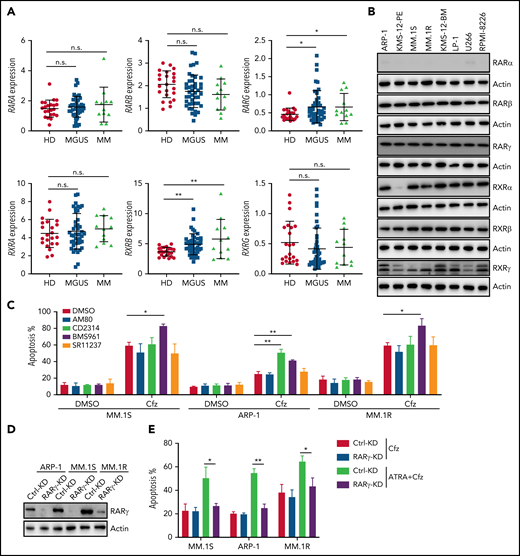
ATRA-induced RARγ activation is important for enhancing Cfz-induced cytotoxicity in MM cells
Because ATRA is a pan-RAR/RXR agonist, we sought to identify which RARs or RXRs are important for ATRA enhancement of Cfz-induced apoptosis in MM. First, we examined the expression of RARs and RXRs, which homodimerize or heterodimerize to regulate the transcription of target genes in ATRA response signaling pathways,40-42 in human MM cells. Based on analysis of Zhan et al MM-related datasets from the University of Arkansas for Medical Science,43 RARγ and RXRβ expressions were higher in MM cells from patients with monoclonal gammopathy of undetermined significance or MM than normal plasma cells from healthy donors (Figure 2A). Interestingly, all MM cell lines expressed high levels of RARγ, RXRβ, and RXRγ than normal plasma cells (Figure 2B; supplemental Figure 4A), whereas RARα, RARβ (actually RARβ2, a dominant isoform of RARβ44), and RXRα expressions in MM cells were similar to normal plasma cells or B cells. Furthermore, ATRA treatment only inhibited RXRα expression and did not affect the expression of other ATRA receptors in MM cells (supplemental Figure 4B). Second, by using selective RAR agonists and pan-RXR agonists, we found that BMS961, a selective RARγ agonist,45,46 but not RARα agonist (AM80),47 or pan-RXR agonist (SR11237),48 increased apoptosis in all 3 MM cells treated with Cfz, whereas RARβ agonist (CD2314)45,49 promoted Cfz-induced apoptosis in ARP-1 cells (Figure 2C; supplemental Figure 5A). Inhibiting RARγ by its antagonist MM1125350 abrogated ATRA’s ability to promote Cfz cytotoxicity in MM cells while inhibiting RARβ51 by its antagonist LE 135 (supplemental Figure 5B) or inhibiting RXRγ by its antagonist HX53152 (supplemental Figure 5C) did not. Third, knockdown (KD) of RARγ (Figure 2D) but not RARβ or RXRγ in MM cells decreased their apoptosis induced by ATRA+Cfz compared with control-KD (Ctrl-KD) MM cells (Figure 2E; supplemental Figure 5D-H), indicating that knocking down RARγ abrogated ATRA-enhanced apoptosis in Cfz-treated MM cells. In summary, these results demonstrate that RARγ activation is important for ATRA sensitizing MM cells to Cfz treatment.
RARγ plays an important role in ATRA sensitizing MM cells to Cfz treatment. (A) Gene-profiling data from Zhan et al’s patient database were analyzed. Dot plot showing ATRA receptors (RARs and RXRs) expression (median-centered intensity) in plasma cells from healthy donors (n = 22), Monoclonal gammopathy of undetermined significance (MGUS; n = 44) and MM (n = 12). (B) Western blot showing expression of ATRA receptors (RARs and RXRs) in human MM cell lines. (C) Summarized results of flow cytometry data showing the apoptosis of MM.1S, ARP-1, or MM.1R MM cells with indicated treatment (10 µM RARα agonist AM80, RARβ agonist CD2314, RARγ agonist BMS961, or pan-RXR agonist SR11237) for 24 hours. (D) Western blot showing RARγ expression in Ctrl-KD or RARγ-KD MM cell lines. (E) Summarized results showing apoptosis of Ctrl-KD or RARγ-KD MM cells pulsed with 1-hour Cfz treatment followed by ATRA treatment for 24 hours. Student t test was used to compare 2 samples. n.s., no significance. *P < .05; **P < .01.
RARγ plays an important role in ATRA sensitizing MM cells to Cfz treatment. (A) Gene-profiling data from Zhan et al’s patient database were analyzed. Dot plot showing ATRA receptors (RARs and RXRs) expression (median-centered intensity) in plasma cells from healthy donors (n = 22), Monoclonal gammopathy of undetermined significance (MGUS; n = 44) and MM (n = 12). (B) Western blot showing expression of ATRA receptors (RARs and RXRs) in human MM cell lines. (C) Summarized results of flow cytometry data showing the apoptosis of MM.1S, ARP-1, or MM.1R MM cells with indicated treatment (10 µM RARα agonist AM80, RARβ agonist CD2314, RARγ agonist BMS961, or pan-RXR agonist SR11237) for 24 hours. (D) Western blot showing RARγ expression in Ctrl-KD or RARγ-KD MM cell lines. (E) Summarized results showing apoptosis of Ctrl-KD or RARγ-KD MM cells pulsed with 1-hour Cfz treatment followed by ATRA treatment for 24 hours. Student t test was used to compare 2 samples. n.s., no significance. *P < .05; **P < .01.
ATRA-activated IFN-β responses are important for sensitizing MM to Cfz treatment
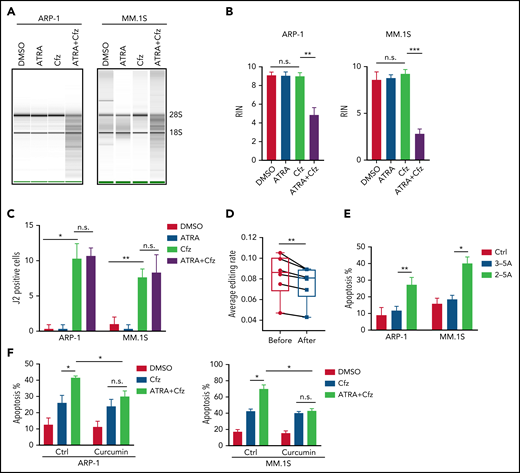
Consistent with the above findings, MM cells treated with ATRA+Cfz had enhanced expression of proapoptotic cleaved caspase 3 and PARP proteins compared with Cfz-treated MM cells, but prosurvival pAkt and pERK protein53 expression remained unchanged (supplemental Figure 6A-B). As PI cytotoxicity in MM cells has been linked to the induction of ER stress,9,54 we determined whether ATRA mediates its effect by affecting Cfz-induced ER stress in MM cells. We examined the expression of ER-stress proteins ATF4, ATF6, and XBP1s and found a similar expression pattern of these proteins in Cfz-treated and ATRA+Cfz–treated MM cells (supplemental Figure 6A), suggesting that ATRA sensitizes MM to Cfz independently of ER stress. To determine which signaling pathways are involved in ATRA+Cfz–treated MM cells, we performed gene set enrichment analysis (GSEA) of ATRA+Cfz– vs Cfz- or ATRA-treated MM cells. GSEA identified prominent enrichments of type 1 interferon (IFN) response, type 2 (IFN-γ) response, unfolded protein response, mTROC1 signaling, and xenobiotic metabolism gene sets in ATRA+Cfz–treated MM cells compared with Cfz-treated MM cells, whereas ATRA+Cfz–treated MM cells showed significant activation of apoptosis pathway compared with ATRA-treated MM cells (Figure 3A; supplemental Figure 6C-D). To determine which of the IFN response pathways are involved, we added IFN-β or IFN-γ with Cfz to treat MM cells. Similar to ATRA, IFN-β, which alone did not induce MM apoptosis, enhanced Cfz-induced MM cell apoptosis and displayed a synergistic effect with Cfz to kill MM.1S and ARP-1 cells, whereas IFN-γ exerted additive or slightly synergistic effect with Cfz against ARP-1 or MM.1S cells, respectively (Figure 3B-C; supplemental Figure 6E). By analyzing the expression of IFN-β response genes in microarray data, we found that ATRA induced the expression of OAS1, which is also 1 of top 20 upregulated genes in Cfz+ATRA–treated MM cells (supplemental Figure 6F), IRF1, OAS2, and OAS3 in Cfz-treated MM cells (Figure 3D). Using quantitative polymerase chain reaction and western blot, we confirmed that ATRA increased the expressions of IRF1, OAS1, OAS2, and OAS3 on both messenger RNA (mRNA; supplemental Figure 7A) and protein levels (Figure 3E). Similarly, IFN-β–treated MM cells displayed higher mRNA expression of IRF1, OAS1, and OAS3 except OAS2 (supplemental Figure 7B). Furthermore, halting ATRA-stimulated IFN-β response pathway by knocking down IRF1 in MM cells (Figure 3F) decreased their apoptosis induced by ATRA+Cfz compared with Ctrl-KD MM cells (Figure 3G; supplemental Figure 7C). Taken together, these results suggest that ATRA activates IFN-β response pathway, important for enhancing Cfz-induced apoptosis.
ATRA activates the expression of IFN-β response genes that are important for promoting Cfz-induced MM-killing effect. (A) Microarray data of MM.1S cells treated with ATRA+Cfz or Cfz for 12 hours were analyzed by GSEA. The top 6 representative Hallmark gene sets (with indicated colors) based on normalized enrichment score (NES) from data of ATRA+Cfz (positively correlated, red)- vs Cfz (negatively correlated, blue)-treated MM.1S cells are shown. (B) Summarized flow data showing apoptosis of MM.1S or ARP-1 cells pulsed with 1-hour Cfz treatment followed by 50 U/mL IFN-β (12.5 pM) or IFN-γ (150 pM) for 24 hours. (C) Histogram showing CI value for the combination of 50 U/mL IFN-β or IFN-γ with 30 nM Cfz in MM.1S or 100 nM ARP-1 cells. Each column represents the mean CI value obtained from 3 independent experiments plotted against the corresponding fraction affected (Fa) by the combination treatment in panel B. Apoptotic rates of MM cells treated with single-agent Cfz, IFN-β, or IFN-γ in serial dilution were obtained for calculating CI value of their combination therapy shown in supplemental Table 4. CI < 1 represents synergism; CI = 1 represents additive effect; and CI > 1 represents antagonism. (D) Heat map illustrating the log2-fold change of the genes involved in IFN-β response pathways. The color bar indicates gene expression value. (E) Western blot displaying protein expression of IRF1 and OAS1-3 in MM.1S or ARP-1 cells with indicated treatment for 14 hours. (F) Western blot showing IRF1 expression in Ctrl-KD or IRF1-KD MM cell lines with DMSO or ATRA treatment for 14 hours. (G) Summarized data showing apoptosis of Ctrl-KD or IRF1-KD MM cells with indicated treatment of 24 hours. Student t test was used to compare 2 samples, *P < .05; **P < .01.
ATRA activates the expression of IFN-β response genes that are important for promoting Cfz-induced MM-killing effect. (A) Microarray data of MM.1S cells treated with ATRA+Cfz or Cfz for 12 hours were analyzed by GSEA. The top 6 representative Hallmark gene sets (with indicated colors) based on normalized enrichment score (NES) from data of ATRA+Cfz (positively correlated, red)- vs Cfz (negatively correlated, blue)-treated MM.1S cells are shown. (B) Summarized flow data showing apoptosis of MM.1S or ARP-1 cells pulsed with 1-hour Cfz treatment followed by 50 U/mL IFN-β (12.5 pM) or IFN-γ (150 pM) for 24 hours. (C) Histogram showing CI value for the combination of 50 U/mL IFN-β or IFN-γ with 30 nM Cfz in MM.1S or 100 nM ARP-1 cells. Each column represents the mean CI value obtained from 3 independent experiments plotted against the corresponding fraction affected (Fa) by the combination treatment in panel B. Apoptotic rates of MM cells treated with single-agent Cfz, IFN-β, or IFN-γ in serial dilution were obtained for calculating CI value of their combination therapy shown in supplemental Table 4. CI < 1 represents synergism; CI = 1 represents additive effect; and CI > 1 represents antagonism. (D) Heat map illustrating the log2-fold change of the genes involved in IFN-β response pathways. The color bar indicates gene expression value. (E) Western blot displaying protein expression of IRF1 and OAS1-3 in MM.1S or ARP-1 cells with indicated treatment for 14 hours. (F) Western blot showing IRF1 expression in Ctrl-KD or IRF1-KD MM cell lines with DMSO or ATRA treatment for 14 hours. (G) Summarized data showing apoptosis of Ctrl-KD or IRF1-KD MM cells with indicated treatment of 24 hours. Student t test was used to compare 2 samples, *P < .05; **P < .01.
Activation of RARγ-IRF1-OAS1 pathway sensitizes MM cells to Cfz treatment
Based on the above results that ATRA activates type 1 IFN pathway and upregulates the expression of its downstream OASs in MM cells, and OAS1-3, which are pattern-recognition receptors for viral/cellular double-stranded RNA (dsRNA), synthesize 2-5A to activate RNase L, causing endonucleolytic cleavage of viral and cellular RNAs,55-57 we hypothesized that ATRA-activated expression of OASs induces degradation of cellular RNAs, leading to increased death of Cfz-treated MM cells. To test our hypothesis, we first determined cellular RNA degradation in MM cells. Degradation of cellular RNAs was exclusively observed in MM cells treated with ATRA+Cfz but not in ATRA or Cfz single-agent treated cells (Figure 4A-B; supplemental Figure 8A). Similar results were observed in MM cells treated with ATRA+Btz (supplemental Figure 8B). Next, we examined the expression of cellular dsRNAs that trigger activation of the OAS-RNase L system, in MM cells, which has never been explored before. Interestingly, Cfz induced dsRNA expression in MM cells, whereas ATRA did not (Figure 4C; supplemental Figure 9A). To confirm this novel finding, we examined the expression of adenosine deaminases acting on RNA 1 (ADAR1) in MM cells under ATRA, Cfz, or a combination treatment because ADAR1, unwinding dsRNA structure by its adenosine (A)-to-inosine (I) editing enzyme,58,59 has been reported to be highly expressed in human MM cells.60 Consistent with our findings, ATRA did not inhibit ADAR1 expression. However, Cfz did not affect ADAR1 expression either (supplemental Figure 9B-C). Therefore, we hypothesized that Cfz induces dsRNA expression by reducing A-to-I RNA editing activity in MM cells. We downloaded MM cell RNA-seq data from 6 CoMMpass patients before and after PI-based therapies and analyzed the global RNA editome by scanning A-to-I editing events at 15 681 872 reported editing sites of whole transcriptome in each sample. Interestingly, A-to-I editing level significantly decreased in MM cells of patients after PI-based treatment (Figure 4D), which is consistent with elevated dsRNA expression in Cfz-treated MM cells. Because protein kinase R (PKR, EIF2AK2) is known to induce cell apoptosis by inhibiting protein synthesis upon binding to dsRNA,61,62 we examined its expression by microarray and found that PKR expression was not affected by ATRA treatment (supplemental Figure 9B). By transfecting 2-5A into MM cells, we further demonstrated that expression of 2-5A directly induced MM cell apoptosis compared with nontransfected or 3-5A control-transfected cells (Figure 4E; supplemental Figure 9D). Moreover, using RNase L inhibitor curcumin,63,64 we observed that suppressing RNase L activity did not affect Cfz-induced apoptosis but completely abrogated ATRA ability to promote Cfz cytotoxicity (Figure 4F; supplemental Figure 9E). Thus, these data suggest that ATRA-induced OAS1-3s recognize cellular dsRNAs produced by Cfz treatment and in turn activate RNase L by synthesizing 2-5A, leading to enhanced cell apoptosis through degradation of cellular RNAs.
2-5A-RNase L activation mediates MM cell apoptosis in response to ATRA+Cfz. (A) RNA gel electrophoresis and (B) summarized results showing RNA integrity number (RIN) of ARP-1 (left panel) or MM.1S (right panel) cells with indicated treatment for 16 hours. The positions of 18S and 28S ribosomal RNA are indicated. (C) Summarized data of fluorescent confocal images showing expression of dsRNA, labeled by anti-dsRNA J2 antibody, in ARP-1 or MM.1S cells with indicated treatment for 8 hours. (D) Box plot showing the average level of A-to-I editing in MM cells from 6 CoMMpass patients before (red dots) or after (blue dots) PI-based therapy (the detailed clinical information is listed in supplemental Table 5) by analyzing global editing sites across the whole transcriptome in the RNA-seq data. (E) Summarized results displaying apoptosis of MM.1S or ARP-1 MM cells transfected with or without 2-5A for 24 hours. 3-5A served as a negative control. (F) Summarized flow data showing apoptosis of ARP-1 (left panel) or MM.1S (right panel) cells treated with Cfz or ATRA+Cfz in the presence or absence of 20 µM curcumin for 24 hours. For panel A, representative result of at least 3 independent experiments is shown. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
2-5A-RNase L activation mediates MM cell apoptosis in response to ATRA+Cfz. (A) RNA gel electrophoresis and (B) summarized results showing RNA integrity number (RIN) of ARP-1 (left panel) or MM.1S (right panel) cells with indicated treatment for 16 hours. The positions of 18S and 28S ribosomal RNA are indicated. (C) Summarized data of fluorescent confocal images showing expression of dsRNA, labeled by anti-dsRNA J2 antibody, in ARP-1 or MM.1S cells with indicated treatment for 8 hours. (D) Box plot showing the average level of A-to-I editing in MM cells from 6 CoMMpass patients before (red dots) or after (blue dots) PI-based therapy (the detailed clinical information is listed in supplemental Table 5) by analyzing global editing sites across the whole transcriptome in the RNA-seq data. (E) Summarized results displaying apoptosis of MM.1S or ARP-1 MM cells transfected with or without 2-5A for 24 hours. 3-5A served as a negative control. (F) Summarized flow data showing apoptosis of ARP-1 (left panel) or MM.1S (right panel) cells treated with Cfz or ATRA+Cfz in the presence or absence of 20 µM curcumin for 24 hours. For panel A, representative result of at least 3 independent experiments is shown. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
To confirm the importance of OAS1-3 in ATRA+Cfz–induced death in MM cells, we knocked down the expression of these genes in the cells under ATRA treatment (Figure 5A). Interestingly, OAS1-KD MM cells were resistant to ATRA+Cfz–mediated death compared with Ctrl-KD, OAS2-KD, or OAS3-KD MM cells (Figure 5B; supplemental Figure 10A). Furthermore, knocking in OAS1 in MM cells (Figure 5C) increased MM cell sensitivity to Cfz and promoted ATRA+Cfz–mediated cell death (Figure 5D; supplemental Figure 10B), indicating that OAS1 is essential for ATRA to sensitize MM cells to Cfz-induced apoptosis.
ATRA-induced OAS1 is responsible for MM cell death in response to Cfz. (A) Western blot showing the expression of OAS1-3 in Ctrl-KD or OAS1-3-KD ARP-1 and MM.1S cells under ATRA treatment of 14 hours. (B) Summarized results of apoptosis of Ctrl-KD or OAS1-3-KD ARP-1 and MM.1S cells with indicated treatment of 24 hours. (C) Western blot showing the expression of OAS1 in Ctrl-knockin (KI) or OAS1-KI ARP-1 or MM.1S cells. (D) Summarized results showing apoptosis of Ctrl-KI or OAS1-KI ARP-1 or MM.1S cells with indicated treatment for 24 hours. (E) ChIP analysis of IRF1 binding to OAS1 promoter under BMS961 treatment for 12 hours. The consensus IRF1 binding motif is listed as the colored DNA sequence at the top. The predicted IRF1 binding sequence in OAS1 promoter region is in the black box, and the binding positions are indicated at below. (F-G) Histogram showing mRNA expression of IRF1 (F) and OAS1 (G) in Ctrl-KD or RARγ-KD ARP-1 or MM.1S MM cells with ATRA treatment for 12 hours. The housekeeping gene GAPDH was used for normalization of quantitative reverse transcription polymerase chain reaction results. (H) Western blot of IRF1 and OAS1 expression in ATRA-treated Ctrl-KD or RARγ-KD ARP-1 or MM.1S MM cells with indicated treatment of 16 hours. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
ATRA-induced OAS1 is responsible for MM cell death in response to Cfz. (A) Western blot showing the expression of OAS1-3 in Ctrl-KD or OAS1-3-KD ARP-1 and MM.1S cells under ATRA treatment of 14 hours. (B) Summarized results of apoptosis of Ctrl-KD or OAS1-3-KD ARP-1 and MM.1S cells with indicated treatment of 24 hours. (C) Western blot showing the expression of OAS1 in Ctrl-knockin (KI) or OAS1-KI ARP-1 or MM.1S cells. (D) Summarized results showing apoptosis of Ctrl-KI or OAS1-KI ARP-1 or MM.1S cells with indicated treatment for 24 hours. (E) ChIP analysis of IRF1 binding to OAS1 promoter under BMS961 treatment for 12 hours. The consensus IRF1 binding motif is listed as the colored DNA sequence at the top. The predicted IRF1 binding sequence in OAS1 promoter region is in the black box, and the binding positions are indicated at below. (F-G) Histogram showing mRNA expression of IRF1 (F) and OAS1 (G) in Ctrl-KD or RARγ-KD ARP-1 or MM.1S MM cells with ATRA treatment for 12 hours. The housekeeping gene GAPDH was used for normalization of quantitative reverse transcription polymerase chain reaction results. (H) Western blot of IRF1 and OAS1 expression in ATRA-treated Ctrl-KD or RARγ-KD ARP-1 or MM.1S MM cells with indicated treatment of 16 hours. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.
To determine the mechanisms underlying ATRA-induced transcriptional regulation of OAS1 expression, we first examined the expression of STAT1/2, which are upregulated by type 1 IFN response65 and cooperate with IRF1 in mediating IFN response-downstream signaling.66 However, ATRA treatment did not elevate the expression of phosphorylated or total STAT1/2 in MM cells (supplemental Figure 10C). Knocking down IRF1 expression in MM cells abrogated the upregulated OAS1 mRNA (supplemental Figure 10D) or protein (supplemental Figure 10E) expressions induced by ATRA treatment, indicating that IRF1 plays an important role in ATRA-mediated transcriptional regulation of OAS1 expression. By analyzing public ChIP-seq data with Cistrome Data Browser, we found that IRF1 had a strong binding peak on OAS1 promoter region (supplemental Figure 10F). Using chromatin immunoprecipitation (ChIP) assay, we demonstrated that BMS961, a selective RARγ agonist, significantly increased the binding of IRF1 to OAS1 promoter in MM cells (Figure 5E). Microarray profiles from Zhan et al datasets67 showed that IRF1 expression positively correlated with OAS1 expression in MM patient’s tumor cells (supplemental Figure 10G). Therefore, we determined the activation of transcription factor RARγ on IRF1 and OAS1 expression. Consistently, expressions of IRF1 and OAS1 were significantly downregulated in ATRA-treated RARγ-KD MM cells as compared with Ctrl-KD cells (Figure 5F-H). Hence, these data indicate that ATRA-activated transcription regulation of RARγ-IRF1-OAS1 pathway contributes to the enhanced Cfz-induced MM cell apoptosis.
Clinical significance of RARγ activation in MM patient’s prognosis and chemoresponse
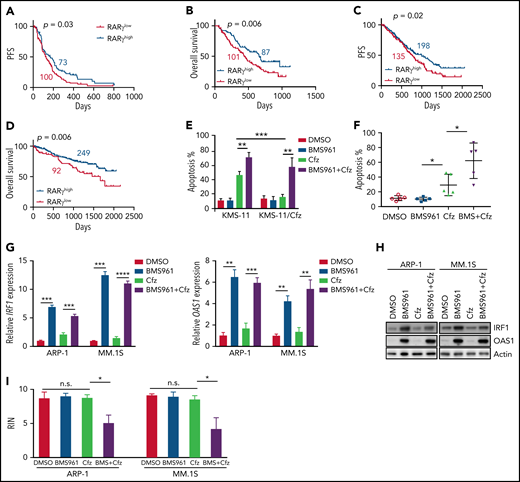
Based on our findings, we wanted to determine the impact and significance of RARγ and OAS1 in human MM pathogenesis and drug response. First, we downloaded the gene-profiling data of 264 patients with MM from Mulligan et al datasets.68 Patients with MM with low-RARγ expression had significantly shorter progression-free survival (PFS; Figure 6A) and OS (Figure 6B) than those with high-RARγ expression. Furthermore, we analyzed gene-profiling data of 1143 patients with MM from MMRF coMMpass study IA13. Patients treated with PI-based regimens with high-RARγ or OAS1 expression showed a superior OS and PFS compared with patients with low-RARγ or OAS1 expression (Figure 6C-D; supplemental Figure 11A-B). Hence, these data indicate that upregulating OAS1 expression by activating RARγ by ATRA may clinically benefit patients with MM in response to PI therapies.
Role of RARγ in MM cell sensitivity to Cfz treatment. Gene-profiling data from Mulligan et al study was analyzed. (A) PFS and (B) OS curves were evaluated in Btz-treated patients with MM based on high-RARγ (red, RARγhigh) or low-RARγ (black, RARγlow) expression in MM cells. The cutoff threshold was defined by the MaxStat package. (C-D) Gene-profiling data from MMRF coMMpass study were analyzed. PFS (C) and OS curves (D) were evaluated in newly diagnosed patients with MM treated with PI-based regimens as first-line treatment based on high-RARγ (blue, RARγhigh) or low-RARγ (red, RARγlow) expression in MM cells. (E) Summary results of apoptosis of WT KMS-11 and KMS-11/Cfz MM cells with indicated treatment for 24 hours. (F) Summary results of apoptosis of primary MM cells treated with DMSO, BMS961, Cfz, or their combinations (BMS+Cfz). (G) Histogram showing mRNA expression of IRF1 (left panel) and OAS1 (right panel) in ARP-1 or MM.1S cells treated with DMSO, BMS961, Cfz, or their combinations for 12 hours. (H) Western blot of IRF1 and OAS1 expression in ARP-1 or MM.1S cells treated with DMSO, BMS961, Cfz, or their combinations for 16 hours. (I) RNA integrity of ARP-1 (left panel) or MM.1S (right panel) cells treated with DMSO, BMS961, Cfz, or their combinations (BMS+Cfz) for 16 hours. The survival plots in panels A to D show Kaplan-Meier estimates of survival and comparisons using the log-rank test. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001; ***P < .0001.
Role of RARγ in MM cell sensitivity to Cfz treatment. Gene-profiling data from Mulligan et al study was analyzed. (A) PFS and (B) OS curves were evaluated in Btz-treated patients with MM based on high-RARγ (red, RARγhigh) or low-RARγ (black, RARγlow) expression in MM cells. The cutoff threshold was defined by the MaxStat package. (C-D) Gene-profiling data from MMRF coMMpass study were analyzed. PFS (C) and OS curves (D) were evaluated in newly diagnosed patients with MM treated with PI-based regimens as first-line treatment based on high-RARγ (blue, RARγhigh) or low-RARγ (red, RARγlow) expression in MM cells. (E) Summary results of apoptosis of WT KMS-11 and KMS-11/Cfz MM cells with indicated treatment for 24 hours. (F) Summary results of apoptosis of primary MM cells treated with DMSO, BMS961, Cfz, or their combinations (BMS+Cfz). (G) Histogram showing mRNA expression of IRF1 (left panel) and OAS1 (right panel) in ARP-1 or MM.1S cells treated with DMSO, BMS961, Cfz, or their combinations for 12 hours. (H) Western blot of IRF1 and OAS1 expression in ARP-1 or MM.1S cells treated with DMSO, BMS961, Cfz, or their combinations for 16 hours. (I) RNA integrity of ARP-1 (left panel) or MM.1S (right panel) cells treated with DMSO, BMS961, Cfz, or their combinations (BMS+Cfz) for 16 hours. The survival plots in panels A to D show Kaplan-Meier estimates of survival and comparisons using the log-rank test. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001; ***P < .0001.
RARγ agonists sensitize human MM cells to Cfz treatment in vitro and in vivo
As ATRA sensitizes MM cells to Cfz-induced apoptosis by activating RARγ pathway, we explored whether RARγ agonists may be used to treat MM. BMS961+Cfz treatment displayed similar apoptosis in MM.1S, MM.1R, and ARP-1 cells but induced more apoptosis in KMS-12-PE, U266, and LP-1 cells compared with treatment with ATRA+Cfz at the same agonist concentration (supplemental Figure 11C). BMS961 exerted stronger synergistic anti-MM effect with Cfz than ATRA at the same concentration in KMS-12-PE, U266, and LP-1 MM cells (supplemental Figure 11D; supplemental Table 3), suggesting that BMS961 is an equal or more potent drug than ATRA in sensitizing MM cells to Cfz treatment. BMS961 not only induced more cell death in Cfz-treated WT KMS-11 cells but also resensitized KMS-11/Cfz cells to Cfz (Figure 6E; supplemental Figure 11E). In addition, BMS961 promoted Cfz-induced apoptosis in primary MM cells (Figure 6F; supplemental Figure 11F). Similar as ATRA treatment, BMS961 upregulated IRF1 and OAS1 mRNA and protein expressions (Figure 6G-H). Furthermore, degradation of cellular RNAs was exclusively observed in MM cells treated with BMS961+Cfz (Figure 6I; supplemental Figure 11G). Taken together, these results strongly suggest that BMS961 also is a promising agent to enhance MM cell sensitivity to Cfz treatment.
Next, we determined whether ATRA and BMS961 can be used to enhance the therapeutic effects of Cfz in vivo. We established a human MM xenograft mouse model by injecting luciferase-expressing MM.1S-luc or ARP-1-luc cells into NSG mice via the tail vein.34,69 Although ATRA or BMS961 alone had no therapeutic effect in vivo, the combination of both significantly enhanced the therapeutic effects of Cfz in suppressing MM bioluminescence intensity, reducing tumor burden, and prolonging survival of MM-bearing mice (Figure 7A-D; supplemental Figure 12A-D). Furthermore, we examined the RNA integrity of MM cells from MM-bearing mice with indicated treatments ex vivo. Degradation of cellular RNAs was observed in MM cells from MM-bearing mice treated with ATRA or BMS961+Cfz but not from ATRA, BMS961, or Cfz single-agent treated mice (Figure 7E-F; supplemental Figure 12E-F). Finally, we determined the role of RARγ on the therapeutic effects of combination therapy in vivo. Interestingly, RARγ-KD MM-bearing mice showed higher tumor burden and poor survival than CTR-KD MM-bearing mice treated with Cfz or ATRA+Cfz. Combination therapy, compared with Cfz single-agent treatment, significantly reduced tumor burden and achieved better survival in CTR-KD but not in RARγ-KD MM-bearing mice (Figure 7G-H). Taken together, these results strongly suggest that RARγ agonists ATRA and BMS961 are potential therapeutic agents to sensitize human MM to Cfz treatment and to overcome Cfz resistance in human patients.
BMS961 enhances Cfz-killing effect of human MM cells in vivo. (A-F) NSG mice were injected IV with 2 × 106 MM.1S-luc MM cells. At day 16 after tumor establishment, DMSO, ATRA, or BMS961 (15 mg/kg for every 3 days), Cfz (3 mg/kg for 2 consecutive days weekly), ATRA+Cfz, or BMS961+Cfz were injected into MM-bearing mice (n ≥ 4 for each group). Blood samples were collected weekly starting at day 16. In vivo bioluminescent imaging (A) and quantification of bioluminescence intensity (B) showing tumor burden in MM-bearing mice treated with DMSO, ATRA, BMS961, Cfz, ATRA+Cfz, or BMS961+Cfz. Tumor burden (C), analyzed by enzyme-linked immunosorbent assay measuring human immunoglobulin light chain in mouse plasma which was normalized to control, and survival (D) in MM-bearing mice were calculated. (E-F) RNA gel electrophoresis showing ex vivo RNA integrity of CD138+ MM cells sorted out from MM.1S-bearing NSG mice received the indicated treatments (A+C: ATRA+Cfz, and B+C: BMS961+Cfz; n = 3 for each group) for 1 day. The positions of 18S and 28S ribosomal RNA are indicated. (G) Tumor burden and (H) survival of NSG mice bearing CTR-KD (dashed line) or RARγ-KD (solid line) ARP-1 cells receiving the indicated treatments (n = 5 for each group). Tumor bioluminescence intensity and burden were analyzed by 2-way analysis of variance. The survival plot showing Kaplan-Meier estimates of survival and comparisons using the log-rank test. For panel E, representative results of 3 are shown. Student t test was used to compare 2 samples. Veh., vehicle. *P < .05; **P < .01; ***P < .001; ****P < .0001.
BMS961 enhances Cfz-killing effect of human MM cells in vivo. (A-F) NSG mice were injected IV with 2 × 106 MM.1S-luc MM cells. At day 16 after tumor establishment, DMSO, ATRA, or BMS961 (15 mg/kg for every 3 days), Cfz (3 mg/kg for 2 consecutive days weekly), ATRA+Cfz, or BMS961+Cfz were injected into MM-bearing mice (n ≥ 4 for each group). Blood samples were collected weekly starting at day 16. In vivo bioluminescent imaging (A) and quantification of bioluminescence intensity (B) showing tumor burden in MM-bearing mice treated with DMSO, ATRA, BMS961, Cfz, ATRA+Cfz, or BMS961+Cfz. Tumor burden (C), analyzed by enzyme-linked immunosorbent assay measuring human immunoglobulin light chain in mouse plasma which was normalized to control, and survival (D) in MM-bearing mice were calculated. (E-F) RNA gel electrophoresis showing ex vivo RNA integrity of CD138+ MM cells sorted out from MM.1S-bearing NSG mice received the indicated treatments (A+C: ATRA+Cfz, and B+C: BMS961+Cfz; n = 3 for each group) for 1 day. The positions of 18S and 28S ribosomal RNA are indicated. (G) Tumor burden and (H) survival of NSG mice bearing CTR-KD (dashed line) or RARγ-KD (solid line) ARP-1 cells receiving the indicated treatments (n = 5 for each group). Tumor bioluminescence intensity and burden were analyzed by 2-way analysis of variance. The survival plot showing Kaplan-Meier estimates of survival and comparisons using the log-rank test. For panel E, representative results of 3 are shown. Student t test was used to compare 2 samples. Veh., vehicle. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Discussion
In this study, we used a high-throughput screen of 1855 FDA-approved drugs to identify potent drugs that can promote Cfz cytotoxicity in human MM cells. Our screen resulted in identifying ATRA, a first-line treatment of APL with low side effects, as a promising small molecule. We showed that ATRA promoted Cfz anti-MM effect in primary MM cells and MM cell lines and resensitized Cfz-resistant KMS-11/Cfz cells to Cfz treatment in vitro. Mechanistically, ATRA activated RARγ and IFN-β response pathways, elevated expression of IRF1, and initiated transcription of OAS1, which bound to dsRNA induced by Cfz and synthesized 2-5A to activate RNA cleavage activity of RNase L, leading to more MM cell death. We further showed that BMS961, a selective RARγ agonist, achieved a similar or even stronger effect than ATRA in promoting Cfz-induced MM killing in vitro. Both ATRA and BMS961 significantly enhanced the therapeutic efficacy of Cfz in vivo. Our findings provide strong evidence that activating RARγ by ATRA or BMS961 may be a promising strategy to enhance MM sensitivity to Cfz and to overcome Cfz resistance in patients. However, we cannot completely exclude the possibility that other RAR or RXR may also be involved in ATRA-mediated sensitization of MM cells to Cfz treatment, and future studies may be needed to investigate the possibility.
IFN-γ is known for overcoming Btz resistance in human hematological cancer cell lines, as it strongly induces the expression of β5i subunit of the immunoproteasome70 that is the target of Cfz, suggesting that IFN-γ should work for overcoming Cfz resistance.71 However, we obtained an interesting and novel finding that IFN-β had a stronger synergistic effect on Cfz-induced apoptosis in MM cells than IFN-γ, which depended on activation of the IRF1-OAS1-RNase L innate immune pathways. dsRNA directly activates 2 types of IFN-induced enzymes, protein kinase PKR that blocks translational initiation, and OAS1-3 that synthesizes 2-5A to activate RNase L.72 We observed that ATRA induced OAS1 but not PKR expression, and inhibition of RNase L activity or OAS1 expression rendered MM cells highly resistant to ATRA+Cfz–induced death. These findings demonstrate the importance of RNase L and OAS1 in ATRA-enhanced death of MM cells in response to Cfz.
The dependence of ATRA-induced sensitization of MM cells to Cfz on dsRNA-activated OAS-RNase L pathway raises a question as to what induces cellular dsRNA expression in MM cells. IFN-inducible ADAR1, which is highly expressed in MM cells,60,73 disrupts the normal A:U pairing in dsRNA by converting A to I upon binding to dsRNA and thus prevents the activation of OAS and RNase L.74 Based on the findings, we detected dsRNA expression in ATRA- and Cfz-treated MM cells. Neither ATRA nor Cfz altered ADAR1 expression in MM cells. However, we observed that Cfz was able to induce cellular dsRNA expression and reduce RNA editing activity in MM cells from treated patients, which is in line with findings that stress could rapidly upregulate dsRNA expression,75,76 suggesting that Cfz-induced stress may have caused ADAR1 loss of function in MM cells. Approximately one-third of patients with MM harbor amplification of chromosome 1q21,77 which contains ADAR1 loci and is a feature of high-risk MM associated with a poor prognosis.78 ADAR1 overexpression and high numbers of editing are strongly associated with disease progression and poor OS in patients with MM.73 Our findings that ATRA-induced OAS1 expression that synthesized 2-5A to activate RNase L and facilitated Cfz-induced cell death regardless of ADAR1 overexpression highlight the potential of combining ATRA and Cfz, or transfecting/delivering 2-5A to ADAR1-overexpressing MM cells to treat high-risk MM by overriding ADAR1 activation.
By analyzing clinical gene-profiling data, we noticed that patients with MM with high-RARγ expression had better response to Cfz treatment than those with low-RARγ expression, suggesting that MM cells with higher-RARγ expression might be activated by retinoic acid (RA) existing in the microenvironment. However, BM stroma creates an RA-low environment via CYP26, a P450 enzyme that degrades RA.79 Therefore, increasing intracellular concentrations of ATRA by developing CYP26-resistant RAs or combining CYP26 inhibitors such as talarozole80 may provide clinical benefit for improving therapeutic efficacy of Cfz in most, if not all, patients with MM. ATRA alone is not effective in treating MM but was well tolerated based on a previous phase 1/2 clinical study,81 indicating that ATRA could be used in patients to sensitize MM to Ctz and to overcome Cfz resistance. Furthermore, our results that the selective RARγ agonist BMS961 achieved similar or even stronger effects than ATRA in sensitizing MM cells to Cfz in vitro and effectively promoting Cfz-mediated anti-MM effect in vivo suggest that activating RARγ in MM cells by the agonists may be readily translatable in clinic.
Acknowledgments
The authors thank Xian Chen and the Cancer Genomics Center at The University of Texas for RNA-seq service.
This work was supported by Startup Support from Lerner Research Institute, Cleveland Clinic and Houston Methodist Research Institute, Houston Methodist Hospital, and Cancer Prevention & Research Institute of Texas (CPRIT) Recruitment of Established Investigator Award RR180044.
The Cancer Genomics Center in The University of Texas was supported by CPRIT grant RP180734. The authors thank Research Core services in the Lerner Research Institute of the Cleveland Clinic and in the Houston Methodist Research Institute for their support.
Authorship
Contribution: Q.Y., Q.W., and B.X. initiated the study; Q.Y., Q.W., and Z.L. designed the research and wrote the paper; Z.L., Q.W., Z.W., M.X., L.X., and P.S. performed experiments; Y.Z. and S.R.P. provided patient’s samples; and B.X., E.B., Y.-h.H., J.Q., L.L., X.M., M.Y., and W.X. provided critical suggestions; and L.Y. planned and performed the bioinformatics analysis of the clinical microarray and RNA-seq data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qing Yi, Center for Translational Research in Hematological Malignancies, Cancer Center, Houston Methodist Cancer Center/Houston Methodist Research Institute, Houston, TX 77030; e-mail: qyi@houstonmethodist.org; and Bing Xu, Department of Hematology, The First Affiliated Hospital of Xiamen University, Xiamen, Fujian, China; e-mail: xubingzhangjian@126.com.
The microarray data of Cfz- and ATRA+Cfz-treated MM.1S cells have been deposited into the Gene Expression Omnibus database with accession number GSE153196. The RNA-seq data of MM.1S cells treated with DMSO, ATRA, Cfz, or ATRA+Cfz has been deposited into the Gene Expression Omnibus database with accession number GSE178340.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
REFERENCES
Author notes
Q.W., Z.L., Z.W., and L.Y. contributed equally to this study.


![High-throughput screening of drugs identifies that ATRA enhances Cfz-killing effect of human MM cells. FDA-approved compound libraries containing 1855 drugs were screened for enhancing the anticancer efficiency of Cfz in human MM.1S cells. In the screening, 1-hour pulse treatment of 30 nM Cfz or DMSO followed by 4 µM FDA-approved drug (X) treatment for 48 hours. Cell viability of MM.1S cells were detected by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay. (A) Dot plot showing the inhibitory effect of Cfz plus X on MM cell viability (−log2 [Cfz+X], y-axis) and relative cell viability of X-treated cells normalized to DMSO-treated cells (x-axis). Cfz, Resminostat (HDAC inhibitor), and Ixazomib in the library served as positive controls. Black line indicates 30% inhibition of MM cell viability in culture with Cfz alone; red line indicates the potential cutoff threshold (>65% inhibition by Cfz+X treatment); blue line is the cutoff line of low toxicity of X treatment (>50% cell viability by X treatment compared with DMSO treatment). (B) Dot plot showing relative inhibitory effect of Cfz + X treatment compared with X treatment alone (log2X − log2[Cfz+X], y-axis) and Cfz alone (−log2 [Cfz+X], x-axis). The potential drug X from the cutoff area in panel A and blue point indicates ATRA. (C) CI plot showing synergistic effect of Cfz with 10 µM ATRA in MM cells across a range of effective drug doses (MM.1S and MM.1R: Cfz 5-30 nM; ARP-1 and LP-1: Cfz 30 to 120 nM; KMS-12-PE and U266: Cfz 15 to 90 nM; primary MM: Cfz 10 to 80 nM). Detailed information is provided in supplemental Table 1. CI was calculated using the CompuSyn software. Each point represents the CI value plotted against the corresponding mean fraction affected (Fa) by the combination treatment relative to DMSO control in 3 independent experiments. CI < 1 represents synergism; CI = 1 represents additive effect; and CI > 1 represents antagonism. (D-E) Summarized results of apoptosis of MM cell lines (D) or primary MM cells (E) in cultures with the drugs (ATRA for 10 µM. For Cfz pulse treatment, MM.1S and MM.1R: 30 nM; ARP-1: 100 nM; KMS-12-PE: 60 nM; primary MM cells: 80 nM Cfz) for 24 hours. (F) Summary results of apoptosis of WT KMS-11 and KMS-11/Cfz MM cells with indicated treatment (200 nM Cfz pulse treatment and 10 µM ATRA) for 24 hours. (G) Summarized results of apoptosis of MM.1S and ARP-1 cells alone or in coculture with patient-derived BMSCs for 2 days followed by the indicated treatments for 24 hours. Student t test was used to compare 2 samples. *P < .05; **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/139/1/10.1182_blood.2020009856/4/m_bloodbld2020009856f1.png?Expires=1767726514&Signature=GMWpLrAkR~ojXnw8qr4i~cV~S3YrHmQtM3yqpObOPlP~d0pGqcy1std1zCtwAuXelqEnukbfSKFS4DqMtrUFtnyhnzcSeQLkPF6v9e9sXZjljizO8p56BKYMII3Fv5GHQGHMkzDxH7HJq-nB8iRSLFC~VI1vKLH~uk1CC7w9J72giswRAR3Zy0BrIgj4SFLIse0CMyj9H9xH4m-lDiN49ak3S6SNCZnxdDnHtf9WvidLNmM~HrIAenWTO~OWQJqPyT2ArND1ghYeHvORX2qVyJt961jrDwgoPRLz3z-Y88tKMAaa04nXaTUKE4xWx~7P4kq430nu~lngbEaV8HtLag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal