Methylation profiling in myeloid malignancies such as Acute Myeloid Leukemia (AML) and Chronic Myelomonocytic Leukemia (CMML) has demonstrated the ability to define distinct biological and clinical subgroups, including predicting which patients will respond to therapy with a hypomethylating agent (HMA).
The Philadelphia-chromosome negative myeloproliferative neoplasms (MPNs) carry an inherent risk of progression to an accelerated-phase disease (AP; 10-19% blasts in the peripheral blood or bone marrow), as well as to blast phase disease (BP; ≥ 20% blasts in the peripheral blood or bone marrow), which is associated with a poor prognosis. It is unknown whether the methylation profiles of MPN-AP/BP cases may further help identify distinct biological, genomic, and clinical subgroups, including identifying patients more likely to respond to HMA.
We recently carried out a phase I/II study to test the safety and efficacy of combination therapy with the JAK1/2 inhibitor ruxolitinib (RUX) and the HMA Decitabine (DAC) in patients with MPN-AP/BP (MPD-RC 109 study; NCT02076191). A total of 46 patients were accrued to the phase I and II studies. 37 patients were evaluable for response. Complete response (CR) occurred in 10%, Complete Response with incomplete count recovery (CRi) in 24%, Partial Response in 24%. 42% of patients had no response to therapy.
Using samples available from the MPD-RC 109 study, we sought to assess whether the baseline global methylation profile predicts for response to this regimen. Further, we sought to utilize this dataset to determine if IDH2 mutations (amongst the most common mutations in MPN-AP/BP) are associated with a distinct methylation profile, as has been demonstrated in de novo AML.
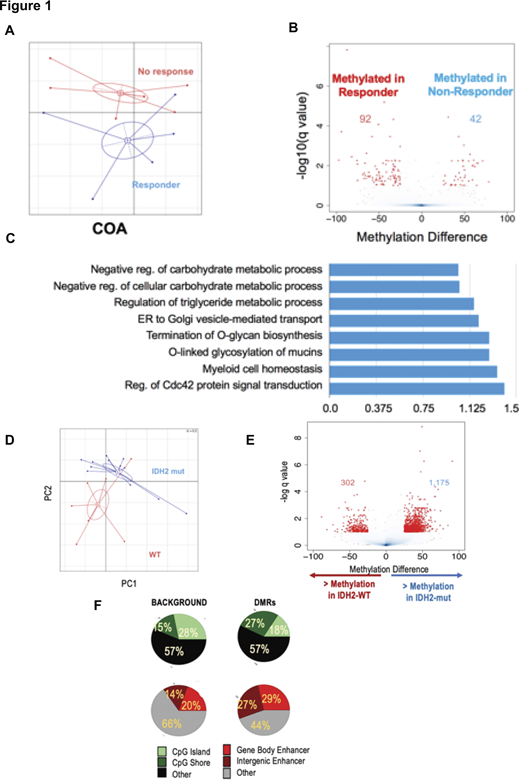
We carried out a pilot study of 11 MPN-AP/BP patients from the MPD-109 phase I/II trial and performed Enhanced Reduced Representation Bisulfite Sequencing (ERRBS) for DNA methylation quantification at ~3M CpG sites across the genome. Baseline DNA methylation profiles were compared between Responder (R) and Non-responder (NR) patients. Notably, unsupervised analysis using correspondence analysis (COA) demonstrated an almost complete separation of the two groups of patients (Fig 1A), while supervised analysis using a beta binomial model identified 134 differentially methylated regions (DMRs) (FDR<10% and absolute methylation difference >25%) between the two groups at diagnosis (Fig 1B). Similar to our prior observation in CMML, response-associated DMRs were depleted from promoter regions (p<0.001) and enriched at enhancers (p<0.001), and were annotated to genes in pathways related to myeloid biology and metabolic processes (Fig 1C).
We next carried out a pilot study to characterize the epigenetic abnormalities of IDH2-mutant MPN-AP/BP cases. For this purpose, we compared the genome-wide DNA methylation profiles of 12 IDH2-mutant to 7 IDH1/2 wild type MPN-AP/BP cases using ERRBS. Unsupervised analysis based on the DNA methylation profiles alone showed a strong trend to naturally segregate mutant from wild-type cases, indicating strong underlying epigenetic differences (Fig.1D). A supervised analysis using the beta binomial method identified 1,477 differentially methylated regions (DMRs) between the two groups (average absolute methylation difference ≥25% and FDR <10%) (Fig 1E). Eighty percent of these DMRs corresponded to sites that were hypermethylated in IDH2-mutant cases compared to wild type. These DMRs were strongly enriched in CpG shores and enhancer regions (p value < 0.001 for both) (Fig 1F).
Our data demonstrate that the methylation profile of MPN-AP/BP may predict for response to HMA-based therapy. Such data could be used to guide therapeutic decisions and select patient for whom HMA has the highest likelihood of procuring a response. As well, these findings indicate that IDH2-mutant MPN-AP/BP are epigenetically distinct, and given the preferred targeting of regulatory elements, these epigenetic differences may play a functional role in disease biology. Further validation of these observations is required. Updated data, including analysis of further cases, and RNA-sequencing analysis of gene-expression and pathway enrichment of genes differentially methylated between responders and non-responders, and IDH2 mutated and wildtype cases will be presented at the conference.
Rampal:Constellation: Research Funding; Pharmaessentia: Consultancy; CTI Biopharma: Consultancy; Promedior: Consultancy; Celgene: Consultancy; Incyte: Consultancy, Research Funding; Abbvie: Consultancy; Galecto: Consultancy; Jazz Pharmaceuticals: Consultancy; Blueprint: Consultancy; Stemline: Consultancy, Research Funding. Mascarenhas:Celgene, Prelude, Galecto, Promedior, Geron, Constellation, and Incyte: Consultancy; Incyte, Kartos, Roche, Promedior, Merck, Merus, Arog, CTI Biopharma, Janssen, and PharmaEssentia: Other: Research funding (institution). Levine:Morphosys: Consultancy; Prelude Therapeutics: Research Funding; Novartis: Consultancy; Amgen: Honoraria; Lilly: Consultancy, Honoraria; Janssen: Consultancy; Astellas: Consultancy; Roche: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Imago: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; C4 Therapeutics: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Isoplexis: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Loxo: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Qiagen: Current equity holder in publicly-traded company, Membership on an entity's Board of Directors or advisory committees; Gilead: Honoraria. Hoffman:Novartis: Membership on an entity's Board of Directors or advisory committees; Protagonist: Consultancy; Forbius: Consultancy; Dompe: Research Funding; Abbvie: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal