Key Points
Burkitt-like lymphoma with 11q aberration harbors a mutational landscape distinct from sporadic BL.
Biallelic inactivation indicates a pathogenic role of the INO80 complex-associated NFRKB gene in mnBLL,11q.
Abstract
The new recently described provisional lymphoma category Burkitt-like lymphoma with 11q aberration comprises cases similar to Burkitt lymphoma (BL) on morphological, immunophenotypic and gene-expression levels but lacking the IG-MYC translocation. They are characterized by a peculiar imbalance pattern on chromosome 11, but the landscape of mutations is not yet described. Thus, we investigated 15 MYC-negative Burkitt-like lymphoma with 11q aberration (mnBLL,11q,) cases by copy-number analysis and whole-exome sequencing. We refined the regions of 11q imbalance and identified the INO80 complex-associated gene NFRKB as a positional candidate in 11q24.3. Next to recurrent gains in 12q13.11-q24.32 and 7q34-qter as well as losses in 13q32.3-q34, we identified 47 genes recurrently affected by protein-changing mutations (each ≥3 of 15 cases). Strikingly, we did not detect recurrent mutations in genes of the ID3-TCF3 axis or the SWI/SNF complex that are frequently altered in BL, or in genes frequently mutated in germinal center–derived B-cell lymphomas like KMT2D or CREBBP. An exception is GNA13, which was mutated in 7 of 15 cases. We conclude that the genomic landscape of mnBLL,11q, differs from that of BL both at the chromosomal and mutational levels. Our findings implicate that mnBLL,11q, is a lymphoma category distinct from BL at the molecular level.
Introduction
Recently, a subgroup of germinal center–derived B-cell (GCB) lymphomas has been described that resembles Burkitt lymphoma (BL) with regard to morphology, immunophenotype, and gene-expression profile but it lacks the IG-MYC translocation typical for BL.1-5 Instead, these cases are cytogenetically characterized by a peculiar pattern of an 11q aberration consisting of a gain in 11q23.2-23.3 followed by a telomeric loss in 11q24.1-qter. According to the revised 4th edition of the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue, these lymphomas have been described as a new provisional entity called “Burkitt-like lymphoma with 11q aberration” (abbreviated herein as “mnBLL,11q,”).6 In addition to the 11q aberration, mnBLL,11q, shows various secondary imbalances and harbors a more complex karyotype than BL.2
Recently, the mutational landscape of IG-MYC–translocated BL has been thoroughly investigated7-10 and MYC, ID3, TP53, SMARCA4, and GNA13, for example, have been identified as recurrently mutated genes. In contrast, mutational analyses of mnBLL,11q, have mainly focused on single genes, showing, for example, lack of ID3 mutations in 14 mnBLL,11q, cases.2 This, together with the lack of IG-MYC translocations and the different imbalance patterns, suggests that mnBLL,11q, shows a genetic makeup quite different from IG-MYC–translocated BL.
To more comprehensively delineate the profile of copy-number alterations (CNAs), single-nucleotide variants (SNVs) as well as small insertions and deletions (indels) of mnBLL,11q, we performed array-based imbalance mapping and whole-exome sequencing (WES) in 15 mnBLL,11q.
Study design
In this retrospective analysis, tumor samples of 15 patients were included that had been diagnosed as BL, atypical BL/BL-like or other aggressive B-cell lymphomas, and lacking an IG-MYC translocation. In all cases, the diagnosis of mnBLL,11q, had been considered (refer to supplemental Methods [available on the Blood Web site] for detailed description) and an 11q aberration pattern was detected by fluorescence in situ hybridization and imbalance profiling applying the OncoScan CNV FFPE assay (12 cases). For 3 of these cases, the clinical, immunophenotypical, and copy-number data have been published previously.1,2,5 DNA extracted from formalin-fixed paraffin-embedded tissue of all 15 patients was subjected to WES. Refer to supplemental Methods for details.
Results and discussion
We analyzed tumor samples of 15 patients retrospectively diagnosed with mnBLL,11q. The median age at diagnosis was 15.5 years (range, 4-52 years) and the male-to-female ratio was 2.75:1. An underlying immunodeficiency was clinically reported in 2 of the cases, both of which had no evidence for an Epstein-Barr virus infection. The characteristics of the cohort are summarized in supplemental Table 1.
As a part of the retrospective workup, we analyzed the CNAs in 12 cases not reported before (excluding the 3 cases reported in Salaverria et al2 ). The median number of CNAs in these 12 cases (gains, losses, and copy-number neutral losses of heterozygosity) was 6.5 (range, 3-38), not significantly differing from the findings reported by Salaverria et al using a different array platform (median, 12.2; range, 2-28).2 Considering the 12 novel CNA profiles and those of the 3 cases previously reported,2 the typical pattern of chromosome 11q gain/loss was observed in 13 of 15 mnBLL,11q, cases (supplemental Figure 1; supplemental Table 2). In the remaining 2 cases, in agreement with fluorescence in situ hybridization studies (supplemental Table 1), only a telomeric loss in 11q24.1-qter was detected without concomitant gain in 11q23. We consider such alterations, which have also recently been reported in 1 mnBLL,11q, case,2 as a variant 11q aberration. This might point to a more pronounced pathogenic role of the genes in the deleted rather than in the gained region. Besides the changes on chromosome 11, other recurrent imbalances included (partial) trisomy 12 (7 of 15 cases, minimally gained region in 12q13.11-q24.32), and gain in 7q34-qter and loss in 13q32.3-q34 (both 3 of 15 cases) (supplemental Figure 1).
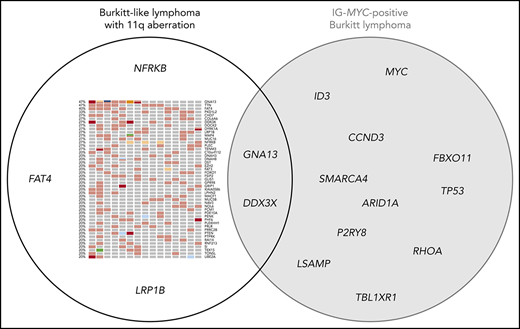
Next, we analyzed the mutational profile (SNVs and indels) of mnBLL,11q, using WES. We identified 47 genes showing potentially protein-changing SNVs in ≥3 of 15 mnBLL,11q, cases (Figure 1A; supplemental Table 3). Strikingly, the genes recurrently mutated in BL (>15% of BL patients [Cristina López, K.K., Sietse M. Aukema, Marius Rohde, Stephan H. Bernhart, D.H., R.W., Umut H. Toprak, F.R., Markus Kreuz, Sebastian M. Waszak, Zhiqin Huang, Lina Sieverling, Nagarajan Paramasivam, J.S., Stephanie Sungalee, R.B.R., Julia Bausinger, Helene Kretzmer, Ole Ammerpohl, Anke K. Bergmann, Hans Binder, Arndt Borkhardt, Benedikt Brors, Alexander Claviez, Gero Doose, Lars Feuerbach, Andrea Haake, Martin-Leo Hansmann, Jessica Hoell, Michael Hummel, Jan O. Korbel, Chris Lawerenz, Dido Lenze, Bernhard Radlwimmer, Julia Richter, Philip Rosenstiel, Andreas Rosenwald, Markus B. Schilhabel, Harald Stein, Stephan Stilgenbauer, Peter F. Stadler, Monika Szczepanowski, Marc A. Weniger, Marc Zapatka, Roland Eils, Peter Lichter, Markus Loeffler, Peter Möller, Lorenz Trümper, W.K., Steve Hoffmann, Ralf Küppers, B.B., M.S., and R.S., accepted for publication January 2019; Schmitz et al7 ; and Love et al9 ]) like MYC, ID3, TCF3, TP53, and SMARCA4 were not recurrently (>15%) or not at all mutated in any of the mnBLL,11q, cases (Figure 1B; supplemental Figures 2 and 3). The notable exceptions were GNA13 and DDX3X, mutated in 7 and 4 of the 15 mnBLL,11q, respectively. The almost complete absence of the BL-associated mutational pattern prompted us to compare the mutational profile of mnBLL,11q, to that of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) (www.icgc.org and Morin et al11 ). Again, we observed only a minor overlap in genes recurrently mutated in both mnBLL,11q, and DLBCL/FL (>15% of cases) including GNA13, TTN, and EZH2. Notably, the mutations in EZH2 affected, in all 3 cases, the Y641 mutational hotspot.12 In contrast, genes like KMT2D or CREBBP most frequently mutated in GCB-like non-BL, which share with mnBLL,11q, a GCB-like gene-expression signature, were not at all mutated in our case series (supplemental Figures 4 and 5). Accordingly, our findings show the mutational profile of mnBLL,11q, to be overall distinct from that of BL as well as FL and DLBCL with only a few exceptions.
Mutational landscape of mnBLL,11q, showing recurrent GNA13 mutations. (A) Depicted are the potentially protein-changing SNVs and indels. Columns encode samples and rows different genes. Different mutation types are color-coded in the oncoprint, where different types of mutation can coexist in 1 sample. *Mutations within these genes are considered as dubious hits as reported by Lawrence et al.15 (B) Overall, the mutational profile differs between the 2 lymphoma entities, and only a few genes are frequently mutated in both including GNA13 and DDX3X. Included are those genes that are mutated in ≥4 of 15 mnBLL,11q, cases and the 13 genes recurrently mutated in >6 of 39 (>15%) BL cases (median age at diagnosis 8 years [range, 2-18 years]) based on whole-genome sequencing data accessible at www.icgc.org.
Mutational landscape of mnBLL,11q, showing recurrent GNA13 mutations. (A) Depicted are the potentially protein-changing SNVs and indels. Columns encode samples and rows different genes. Different mutation types are color-coded in the oncoprint, where different types of mutation can coexist in 1 sample. *Mutations within these genes are considered as dubious hits as reported by Lawrence et al.15 (B) Overall, the mutational profile differs between the 2 lymphoma entities, and only a few genes are frequently mutated in both including GNA13 and DDX3X. Included are those genes that are mutated in ≥4 of 15 mnBLL,11q, cases and the 13 genes recurrently mutated in >6 of 39 (>15%) BL cases (median age at diagnosis 8 years [range, 2-18 years]) based on whole-genome sequencing data accessible at www.icgc.org.
Among the most frequently mutated genes in mnBLL,11q, were GNA13 and TTN, mutated in each 7 of 15 cases. GNA13 mutations, which frequently occur in GCB lymphomas like BL7,9 and GCB-DLBCL,13 lead to a loss of protein function.14 In line, 5 of 11 GNA13 mutations detected in 7 mnBLL,11q, were likely deleterious (frameshift, stop-gain, stop-loss, splice-site), whereas the remaining 6 mutations were nonsynonymous mutations located in functional domains of the protein (supplemental Figure 6A-B). We could verify 9 of 11 GNA13 mutations by Sanger sequencing. In line, in silico modeling of the GNA13 mutations confirmed the deleterious nature of the mutations leading to a loss of function or destabilizing the G-protein’s function (supplemental Table 3; supplemental Figure 6A-B). Extending the analysis to all genes belonging to the Gα13-signaling pathway, we identified, in 8 of 15 mnBLL,11q, cases, mutations in 1 of the pathway genes (supplemental Figure 6C) with GNA13 being the most frequently affected (7 of 8 cases). Most of the mutations in the other genes co-occurred with the GNA13 mutations (3 of 4 cases).
The role of TTN mutations in mnBLL,11q, is unclear. It needs to be considered that several other large and/or late replicating genes are contained within the list of recurrently mutated genes in mnBLL,11q, besides TTN (Figure 1A). Such genes are mutated in several cancer types (refer to supplemental Results and discussion) but their pathogenic role in tumor development is unclear.15 Modeling of the TTN mutations confirmed the mutations to be rather nononcogenic and, hence, constitute most likely passenger variants (supplemental Table 3).
Next, we asked whether genes in the minimal regions of gain and loss were targeted by mutations in addition to imbalances. Three of 45 genes (PCSK7, DSCAML1, TMEM25) mapping to the minimal region of gain were mutated in each only 1 mnBLL,11q, case. With regard to genes located in the minimally deleted region, we confirmed previous findings of recurrent mutations in ETS1 (2 of 15 cases, including 1 published mutation2 ) and detected recurrent mutations in NFRKB (4 of 15), which were verified by Sanger sequencing (supplemental Table 3). Interestingly, 3 of 4 NFRKB mutations were stop-gain mutations and, accordingly, NFRKB function is supposed to be biallelically lost in these 3 cases, that is, by deletion of 1 allele and mutation of the other. In silico modeling of the mutations further supported the deleterious character of the mutations (Figure 2; supplemental Table 3; supplemental Results and discussion). In line, NFRKB expression was described to be significantly lower in mnBLL,11q, compared with BL based on expression-array analysis (P < .007; supplemental Figure 7).2 NFRKB encodes a nuclear factor related to the κB-binding protein, belonging to the INO80 chromatin-remodeling complex,16 which plays a role in transcriptional regulation.17 Extending the analysis to all genes of the INO80 complex showed mutations in 5 of 15 mnBLL,11q, cases. Given the absence of recurrent mutations in genes of the SWI/SNF chromatin-remodeling complex in mnBLL,11q, it is intriguing to speculate that mutations in the INO80 complex in mnBLL,11q, have a comparable function as the mutations in the SWI/SNF complex in BL.
Modeling of NFRKB mutations.NFRKB mutations (red lollipops) annotated on protein primary sequence with additional information regarding posttranslational modifications (PTMs) and domain composition. Three-dimensional structural model showing the deubiquitinating enzyme-binding domain (Protein Data Bank identifier [PDB ID]: 4UF5), the winged domain (PDB ID: 3U21), and the transcriptional regulation domain (by homology; PDB ID: 3BY6). The 2 stop-gain mutations R290* and K322* are predicted to delete the protein region responsible for mediating interactions with transcriptional regulators and/or DNA whereas leaving the N-terminal region deputed to the interaction with deubiquitnating enzymes. The R174C mutation is predicted to perturb adjacent phosphosites, for example, S176, and is located in a motif predicted by the Eukaryotic Linear Motif (ELM; elm.eu.org) to be either the phosphorylation site of GSK3 and MAPK or the docking site for USP7 or the Pin1 WW domain. The C-terminal deletion by Q1103* is predicted to have detrimental consequences, as it is rich in phosphorylation sites (eg, CK1 and GSK3) or recognition sites for FHA and WDR5.
Modeling of NFRKB mutations.NFRKB mutations (red lollipops) annotated on protein primary sequence with additional information regarding posttranslational modifications (PTMs) and domain composition. Three-dimensional structural model showing the deubiquitinating enzyme-binding domain (Protein Data Bank identifier [PDB ID]: 4UF5), the winged domain (PDB ID: 3U21), and the transcriptional regulation domain (by homology; PDB ID: 3BY6). The 2 stop-gain mutations R290* and K322* are predicted to delete the protein region responsible for mediating interactions with transcriptional regulators and/or DNA whereas leaving the N-terminal region deputed to the interaction with deubiquitnating enzymes. The R174C mutation is predicted to perturb adjacent phosphosites, for example, S176, and is located in a motif predicted by the Eukaryotic Linear Motif (ELM; elm.eu.org) to be either the phosphorylation site of GSK3 and MAPK or the docking site for USP7 or the Pin1 WW domain. The C-terminal deletion by Q1103* is predicted to have detrimental consequences, as it is rich in phosphorylation sites (eg, CK1 and GSK3) or recognition sites for FHA and WDR5.
Finally, besides NFRKB, only MACF1, UBE2A, and DST, each mutated in 3 of 15 mnBLL,11q, have been reported to be differentially expressed in comparison with BL.2
Taken together, our data clearly show that besides the chromosomal translocation and imbalance patterns, the mutational profile of mnBLL,11q, is also strikingly different from BL and non-BL. Our findings suggest a role of GNA13 in the pathogenesis of mnBLL,11q, and identify the INO80 complex-associated gene NFRKB as a candidate gene in the deleted region in 11q24.3. Moreover, our findings support the recognition of mnBLL,11q, as an entity distinct from not only MYC-positive BL but also from other aggressive GCB lymphomas like DLBCL. Finally, the observations presented, particularly the lack of a BL-specific mutation pattern, combined with lack of an IG-MYC fusion, might aid in the differential diagnostic process distinguishing IG-MYC–translocated BL from mnBLL,11q.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the teams of the tumor genetic laboratories at the Institutes of Human Genetics in Kiel and Ulm for expert technical assistance. The authors thank the Omics IT and Data Management Core Facility of the German Cancer Research Center (DKFZ, Heidelberg) for excellent technical support.
This work was supported by the German Ministry of Science and Education (BMBF) in the framework of the MMML-MYC-SYS project (036166B). R.W., W.K., and R.S. are recipients of grant support from the KinderKrebsInitiative Buchholz/Holm-Seppensen. R.W. is the recipient of a fund by the Medical Faculty of Ulm University. B.B. is the recipient of a grant of the Deutsche Kinderkrebsstiftung (DKS 2016.24 A/B).
Authorship
Contribution: B.B., H.H., L.L., G.O., G.R., E.S.J., R.A.-Y., and W.K. provided tumor samples and clinical data; S.B. and I.N. performed cytogenetic analysis; J.A., H.T., and P.N. performed WES; J.S., K.K., D.H., and M.S. analyzed the WES data; C.W.K. performed expression analysis; F.R. and R.B.R. modeled the functional impact of the mutations; R.W. analyzed the OncoScan data and interpreted the WES data; R.S. designed the study and coordinated the project; R.W., J.S., and R.S. interpreted the data and wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: R.S. received a speaker’s honorary from Roche. The remaining authors declare no competing financial interests.
The current affiliation for D.H. is Division of Stem Cells and Cancer, DKFZ, Heidelberg, Germany and Heidelberg Institute for Stem Cell Technology and Experimental Medicine (HI-STEM gGmbH), Heidelberg, Germany.
Correspondence: Reiner Siebert, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: reiner.siebert@uni-ulm.de.
REFERENCES
Author notes
R.W. and J.S. share first authorship.

![Figure 1. Mutational landscape of mnBLL,11q, showing recurrent GNA13 mutations. (A) Depicted are the potentially protein-changing SNVs and indels. Columns encode samples and rows different genes. Different mutation types are color-coded in the oncoprint, where different types of mutation can coexist in 1 sample. *Mutations within these genes are considered as dubious hits as reported by Lawrence et al.15 (B) Overall, the mutational profile differs between the 2 lymphoma entities, and only a few genes are frequently mutated in both including GNA13 and DDX3X. Included are those genes that are mutated in ≥4 of 15 mnBLL,11q, cases and the 13 genes recurrently mutated in >6 of 39 (>15%) BL cases (median age at diagnosis 8 years [range, 2-18 years]) based on whole-genome sequencing data accessible at www.icgc.org.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/133/9/10.1182_blood-2018-07-864025/4/m_blood864025f1.png?Expires=1765889783&Signature=DPxYLYlzYPPmgqNsYBGxAtxFUu6PsvgxIUMvSzyjAAPnPyigFX~DmgznmqM4pVMB-lR5~kZza9nlgHdTNo4RFl32GA4Uk5JbptH1Vpr7oqC2t3ZUSYXb21NNT2Bv3qFRa4FHe~y7jzs7VlbivEKt6uEBPaCvi4zpFMSv7LV2bwNfKpIZS49wyfsCUFFjd~R19GnUhsYil7B58snolh3qPzBVYLTqC~TntWAJPMa88UwKKscqYlYR4nKsyzssHERxhnmmEBm6PtCsoZW1Z9zy73umiJRxCDv5~awp5FJvsB1c95sqfdJ2iK3hGedwDQagnlwRlpGRmJZ8h8tQ1gwe8w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Modeling of NFRKB mutations. NFRKB mutations (red lollipops) annotated on protein primary sequence with additional information regarding posttranslational modifications (PTMs) and domain composition. Three-dimensional structural model showing the deubiquitinating enzyme-binding domain (Protein Data Bank identifier [PDB ID]: 4UF5), the winged domain (PDB ID: 3U21), and the transcriptional regulation domain (by homology; PDB ID: 3BY6). The 2 stop-gain mutations R290* and K322* are predicted to delete the protein region responsible for mediating interactions with transcriptional regulators and/or DNA whereas leaving the N-terminal region deputed to the interaction with deubiquitnating enzymes. The R174C mutation is predicted to perturb adjacent phosphosites, for example, S176, and is located in a motif predicted by the Eukaryotic Linear Motif (ELM; elm.eu.org) to be either the phosphorylation site of GSK3 and MAPK or the docking site for USP7 or the Pin1 WW domain. The C-terminal deletion by Q1103* is predicted to have detrimental consequences, as it is rich in phosphorylation sites (eg, CK1 and GSK3) or recognition sites for FHA and WDR5.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/133/9/10.1182_blood-2018-07-864025/4/m_blood864025f2.png?Expires=1765889783&Signature=eTFwa8Co2n0WspmLZEHOuD6AUU2cNq3V5czRUubazS3HPWCXD9DP5KXTNh2sRX4fO8LY41aQ4ykWhB1ZybQ8MF-3gHwPusCJXbfqyGtG9atW5kpdsJo0XpPw7yUIbQQwjRH1QPNqD1~L1my4XV9Hb9ALj6PesvzPKI9~iZW5~vIe0VRpLV23mynk3lurcTSd5VIFzJ6amfTbmr~Vgo~z9lBJWeLQhsu0z8sFpCAgXg6riPAGbO9By2hfKRTWx~eNtfGIJSbaNREk--zYbQh9UaL8CVQMYrb8wQ8s2RDP1OdisXDjyd0drpQH4NJNrPlivtm7OVKvkv5EAZ~z-Nfz0g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal