Key Points
Human ADAMTS13 folds roughly in half to allow formation of an allosterically regulated complex of MDTCS domains, T7, T8, and CUB1.
Allosteric activation of ADAMTS13 by the substrate VWF depends upon binding of the VWF D4 domain to the T7 through CUB1 of ADAMTS13.
Abstract
Human ADAMTS13 is a multidomain protein with metalloprotease (M), disintegrin-like (D), thrombospondin-1 (T), Cys-rich (C), and spacer (S) domains, followed by 7 additional T domains and 2 CUB (complement components C1r and C1s, sea urchin protein Uegf, and bone morphogenetic protein-1) domains. ADAMTS13 inhibits the growth of von Willebrand factor (VWF)–platelet aggregates by cleaving the cryptic Tyr1605-Met1606 bond in the VWF A2 domain. ADAMTS13 is regulated by substrate-induced allosteric activation; without shear stress, the distal T8-CUB domains markedly inhibit VWF cleavage, and binding of VWF domain D4 or selected monoclonal antibodies (MAbs) to distal ADAMTS13 domains relieves this autoinhibition. By small angle X-ray scattering (SAXS), ADAMTS13 adopts a hairpin-like conformation with distal T7-CUB domains close to the proximal MDTCS domains and a hinge point between T4 and T5. The hairpin projects like a handle away from the core MDTCS and T7-CUB complex and contains distal T domains that are dispensable for allosteric regulation. Truncated constructs that lack the T8-CUB domains are not autoinhibited and cannot be activated by VWF D4 but retain the hairpin fold. Allosteric activation by VWF D4 requires T7, T8, and the 58–amino acid residue linker between T8 and CUB1. Deletion of T3 to T6 produced the smallest construct (delT3-6) examined that could be activated by MAbs and VWF D4. Columba livia (pigeon) ADAMTS13 (pADAMTS13) resembles human delT3-6, retains normal activation by VWF D4, and has a SAXS envelope consistent with amputation of the hairpin containing the dispensable T domains of human ADAMTS13. Our findings suggest that human delT3-6 and pADAMTS13 approach a “minimal” structure for allosterically regulated ADAMTS13.
Introduction
ADAMTS13 is a member of the ADAMTS type 1 repeats family of metalloproteases. It is a multidomain protein with Ca2+- and Zn2+-dependent metalloprotease (M), disintegrin-like (D), thrombospondin-1 repeat (T), Cys-rich (C), and spacer (S) domains, followed by 7 T domains and 2 CUB (complement components C1r and C1s, sea urchin protein Uegf, and bone morphogenetic protein-1) domains.1 ADAMTS13 limits the growth of von Willebrand factor (VWF)–platelet aggregates by cleaving a cryptic Tyr1605-Met1606 bond in the VWF A2 domain that is exposed under conditions of high fluid shear stress, thereby releasing adherent platelets.2-4 Congenital or acquired ADAMTS13 deficiency prevents the regulation of VWF-dependent platelet accumulation and causes microvascular thrombosis that characterizes thrombotic thrombocytopenic purpura.5
Interactions between VWF and the proximal MDTCS domains of ADAMTS13 have been studied extensively. When subjected to tensile stress in solution,6 bound to a surface,7 or on endothelial cell surfaces,8,9 VWF A2 domains unfold and expose several sites for binding to the proximal MDTCS domains of ADAMTS13.10-14 In addition, the distal T2-CUB2 domains of ADAMTS13 bind to domain D4 of native VWF.15-17 These multiple contacts enhance the highly specific interaction in vivo between ADAMTS13 and VWF, which are present at ∼1 μg/mL and ∼10 μg/mL, respectively, compared with the total plasma protein concentration ∼ 80 000 μg/mL. In addition, we have demonstrated that the distal T7-CUB domains inhibit the ADAMTS13 M domain and that this autoinhibition can be relieved by certain monoclonal antibodies (MAbs) against distal domains or by binding to VWF D4.18 There are 7 T domains between the proximal MDTCS domains and the 2 CUB domains. How these domains contribute to autoinhibition and allosteric regulation is not known.
To address this uncertainty, we used small angle X-ray scattering (SAXS) to characterize ADAMTS13 constructs with different complements of distal T and CUB domains. SAXS is a useful approach to obtain low-resolution (10-50 Å) structures for flexible proteins that are not amenable to crystallization or electron microscopy. Importantly, SAXS can be performed under physiological conditions of pH and ionic strength. We found that ADAMTS13 adopts a folded conformation with a hinge point between the T4 and T5 domains that allows distal T7-CUB domains to interact directly with the proximal MDTCS domains. This result is consistent with our earlier report that distal T-CUB domains interact with proximal MDTCS domains.18 Furthermore, autoinhibition depends on T8, the CUB domains, and the 58–amino acid residue linker between them. Allosteric activation by VWF D4 depends on the same motifs and, in addition, on T7, suggesting that other distal T domains are dispensable, and a minimal functional ADAMTS13 could be constructed without most distal T domains other than the essential T7 and T8. This conclusion was supported by the properties of ADAMTS13 with internal deletion of T3 through T6 (delT3-6) and Columba livia (pigeon) ADAMTS13 (pADAMTS13), which were allosterically regulated with only 3 distal T domains.
Methods
Recombinant ADAMTS13 and variants
Human ADAMTS13 (rADAMTS13) variants were constructed using a QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene) in tetracycline-inducible pcDNA4/TO (Invitrogen)19 or pTriEx-7 Ek/LIC (EMD Millipore), as previously reported.18 Constructs encode a mouse IgM signal peptide, Strep-Tag II, and enteropeptidase recognition site, followed by the ADAMTS13 sequence starting with the codon for Ala75. Construct MDTCS differs slightly by having the Strep-Tag II transposed to the C terminus after Ala685. ADAMTS13 with internal deletion of T2 through T7 (delT2-7; Trp686-Arg1075) or T2 through T8 (delT2-8L; Trp686-Ala1191), as well as delT3-6 (Trp746-Arg1015), was prepared by mutagenesis of wild-type ADAMTS13 in pTriEx-7 Ek/LIC (GENEWIZ, South Plainfield, NJ). Constructs were prepared in 2 versions: 1 with a wild-type M domain and a second with the active site mutation E225Q.16 A table of the constructs used and the ADAMTS13 variants that they encode are shown in supplemental Figure 1, available on the Blood Web site.
ADAMTS13 variants were expressed in stably transfected TRex 293 cells in roller bottles at 70% to 80% confluence in Freestyle serum-free media (Invitrogen) containing 0.2 mM AEBSF. For all variants, conditioned medium was centrifuged, filtered, supplemented with AEBSF to a final concentration of 0.2 mM, and stored at −20°C until purified, as described.18 After thawing, medium was diluted with 2 volumes of 25 mM Tris-HCl (pH 8.0) and applied to a 20-mL Q Sepharose FF column (GE Healthcare). After washing with the same buffer, bound rADAMTS13 was eluted with 25 mM Tris-HCl (pH 8.0) containing 1 M NaCl. Appropriate pooled fractions were desalted on a PD-10 column (GE Healthcare) equilibrated with 100 mM Tris-HCl (pH 8.0) containing 150 mM NaCl. Combined eluates were concentrated using a Centriprep YM-30 (Millipore) before adsorption on a StrepTrap HP 5-mL column (GE Healthcare). After washing with the same buffer, bound rADAMTS13 was eluted with 25 mM Tris-HCl (pH 8.0), containing 150 mM NaCl, and 2.5 mM desthiobiotin. Combined eluates were concentrated using an Amicon Ultra 10K device (Millipore) and further purified on a Superose 6 10/300 GL column (GE Healthcare) equilibrated with 50 mM HEPES (pH 7.4), 150 mM NaCl, 5 mM CaCl2, and 1 µM ZnCl2.
Proteolytically active ADAMTS13 variants were assayed for cleavage of FRETS-rVWF71 as described.20 Protein concentration was determined by the BCA method (Thermo Fisher).
SAXS
SAXS data were collected for ADAMTS13 variants on the SIBYLS Beamline 12.3.1 at the Advanced Light Source, a national user facility operated by the Lawrence Berkeley National Laboratory (Berkeley, CA) and supported by the US Department of Energy Integrated Diffraction Analysis (IDAT) under contract DE-AC02-05CH11231.21 Proteins were analyzed at 3 or 4 concentrations between 0.75 and 3 mg/mL following standard procedures. The scattering vector q (q = 4π sin θ/λ, where 2θ is the scattering angle) range was 0.01 to 0.32 Å−1. Data were collected for each sample and its matching buffer blank at 4 exposure times, ranging from 0.1 to 5.0 seconds. For each condition, scattering curves at different exposure times were merged with PRIMUS22 or SCÅTTER (SIBYLS Beamline at Lawrence Berkeley National Laboratory) and averaged after normalization for protein concentration. The average scattering curves were used for further calculations. The radius of gyration (Rg), pair distribution function [P(r)], and maximum interatomic distance (Dmax) were calculated with GNOM.23 Ab initio models were generated from scattering profiles using DAMMIN24 (P2 symmetry) and averaged (n = 15) using DAMAVER.25 PyMOL 2.2.0 (Schrodinger) was used for molecular rendering.
ADAMTS13 MAbs and VWF D4
Binding assays
Dissociation constants were determined by biolayer interferometry using an Octet Red96 (ForteBio). The VWF D4-CK construct was captured on Ni-NTA biosensors using its His tag. Binding to ADAMTS13 proteins was performed in 20 mM HEPES (pH 7.4), 150 mM NaCl, 2 mg/mL bovine serum albumin, and 0.02% Tween 20. A buffer-only reference was subtracted from all curves. Affinities were determined from global kinetic analysis for a 1:1 binding model using Octet RED software, version 8.0.
Results
SAXS of nested truncation constructs indicates that ADAMTS13 has a folded conformation
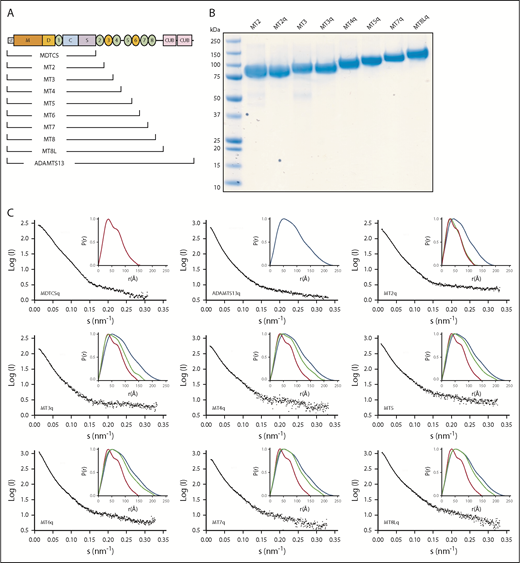
Based on enzymatic characterization and SAXS, we proposed that ADAMTS13 distal T8-CUB domains are adjacent to the proximal MDTCS domains.18 To characterize the disposition of the remaining distal domains, we prepared a series of ADAMTS13 variants truncated after each T domain from T2 through the linker between T8-CUB1 (named MT2 through MT8L) (Figure 1A). Although autoproteolysis is not a known physiological feature of ADAMTS13, we have observed degradation in some of the recombinant protein constructs during storage; however, degradation was markedly reduced by an inactivating mutation in the M domain (E225Q). From previous work,16 we know that the E225Q mutation abolishes catalytic activity without altering the tertiary structure of the M domain. Thus, to avoid degradation during SAXS data collection, the corresponding inactive E225Q mutants were prepared for MT2-8 (designated MT2q through MT8Lq) (Figure 1B) with the substitution E225Q in the M domain. The stability of purified inactive versions of those variants increased markedly.
SAXS data for ADAMTS13 and C-terminal truncations. (A) Schematic structure of ADAMTS13 and a nested series of C-terminal truncations. (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis of selected purified recombinant ADAMTS13 variants. Lane 1 is Precision Plus Protein All Blue Standards (Bio-Rad). (C) Scattering profiles (black) and P(r) plots (inset) for the indicated pure recombinant proteins. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included in each inset panel for comparison with other truncation constructs (green).
SAXS data for ADAMTS13 and C-terminal truncations. (A) Schematic structure of ADAMTS13 and a nested series of C-terminal truncations. (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis of selected purified recombinant ADAMTS13 variants. Lane 1 is Precision Plus Protein All Blue Standards (Bio-Rad). (C) Scattering profiles (black) and P(r) plots (inset) for the indicated pure recombinant proteins. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included in each inset panel for comparison with other truncation constructs (green).
SAXS data were collected for these ADAMTS13 variants (Figure 1C). Adding T2 through T5 to MDTCS shifted the P(r) toward that of full-length ADAMTS13, but adding T6 through T8 had little additional effect. As summarized in Table 1, the Rg increased ∼1.5-fold from MDTCS (∼45 Å) to MT6 (∼67 Å), which is consistent with a mass increase from 67 kilodaltons (kDa) to 103 kDa. In contrast, Rg was roughly constant for MT7 (∼64 Å) through full-length ADAMTS13 (∼67 Å), even though the protein mass increased ∼40%. Dmax exhibited a similar pattern, increasing ∼1.6-fold from MDTCS (∼148 Å) to MT6 (∼234 Å) and remained roughly constant for MT7 (∼210 Å) through ADAMTS13 (∼229 Å). The ∼40% increase in ADAMTS13 mass with no consistent change in dimensions suggests that constructs larger than MT6 adopt a folded conformation, with the more distal T domains close to the center of mass.
Molecular mass, Rg, Dmax, and activity assay at pH 6.0 and pH 7.4 for ADAMTS13 truncation mutants
| Variant . | Mass (kDa) . | Rg (Å), mean ± SD . | Dmax (Å), mean ± SD . | Enzymatic rate, pH 6.0 (min−1) (mean) . | Enzymatic rate, pH 7.4 (min−1) (mean) . |
|---|---|---|---|---|---|
| MDTCS | 67 | 45 ± 1 | 148 ± 2 | 9.9 | 10.0 |
| MT2 | 74 | 48 ± 3 | 155 ± 6 | 9.2 | 10.2 |
| MT3 | 81 | 54 ± 5 | 180 ± 10 | 9.7 | 9.7 |
| MT4 | 87 | 59 ± 4 | 193 ± 16 | 7.8 | 9.8 |
| MT5 | 96 | 63 ± 3 | 210 ± 9 | 7.2 | 9.4 |
| MT6 | 103 | 67 ± 5 | 234 ± 15 | 8.9 | 9.3 |
| MT7 | 109 | 64 ± 2 | 210 ± 11 | 8.3 | 8.9 |
| MT8 | 115 | 63 ± 1 | 215 ± 9 | 3.5 | 4.0 |
| MT8L | 121 | 65 ± 3 | 206 ± 2 | 5.3 | 4.7 |
| ADAMTS13 | 147 | 67 ± 2 | 229 ± 6 | 8.0 | 2.2 |
| Variant . | Mass (kDa) . | Rg (Å), mean ± SD . | Dmax (Å), mean ± SD . | Enzymatic rate, pH 6.0 (min−1) (mean) . | Enzymatic rate, pH 7.4 (min−1) (mean) . |
|---|---|---|---|---|---|
| MDTCS | 67 | 45 ± 1 | 148 ± 2 | 9.9 | 10.0 |
| MT2 | 74 | 48 ± 3 | 155 ± 6 | 9.2 | 10.2 |
| MT3 | 81 | 54 ± 5 | 180 ± 10 | 9.7 | 9.7 |
| MT4 | 87 | 59 ± 4 | 193 ± 16 | 7.8 | 9.8 |
| MT5 | 96 | 63 ± 3 | 210 ± 9 | 7.2 | 9.4 |
| MT6 | 103 | 67 ± 5 | 234 ± 15 | 8.9 | 9.3 |
| MT7 | 109 | 64 ± 2 | 210 ± 11 | 8.3 | 8.9 |
| MT8 | 115 | 63 ± 1 | 215 ± 9 | 3.5 | 4.0 |
| MT8L | 121 | 65 ± 3 | 206 ± 2 | 5.3 | 4.7 |
| ADAMTS13 | 147 | 67 ± 2 | 229 ± 6 | 8.0 | 2.2 |
SAXS was performed with E225Q variants. Rg was calculated from Guinier plots, and Dmax was calculated from P(r) profiles. Standard error for all enzymatic rates is <10%.
SD, standard deviation.
Ab initio envelopes generated from the scattering profiles (Figure 2) support this interpretation. The envelopes increased in length with each additional T domain for constructs MDTCS through MT5. Fitting a molecular model of ADAMTS1318 indicated that the envelopes can accommodate the stepwise addition of domains T2 through T4 in a linear arrangement, extending away from the proximal domains. Beginning with MT5, the envelopes stopped increasing in length, and placement of additional distal T domains required a reversal of the direction of packing. The linker connecting T4 and T5 has 27 extra amino acid residues and could serve as a flexible hinge to facilitate the change in packing direction. Starting with MT6, adding additional T domains increased the volume around the proximal MDTCS domains, providing space for the distal T8-CUB domains to approach and possibly regulate the proteolytic activity of the proximal domains.
Ab initio envelopes and molecular models of ADAMTS13 C-terminal truncations. Envelopes calculated from SAXS data are rendered as gray surfaces. ADAMTS13 domains are colored similarly to Figure 1A: M (orange), D (yellow), T1 (green), C (sky blue), S (lavender), T3 and T6 (yellow), T2, T4, T5, T7, and T8 (green), T4-T5 linker (gray), T8-CUB1 linker (gray), CUB1 (red), CUB2 (brick red). All constructs have the active site mutation E225Q.
Ab initio envelopes and molecular models of ADAMTS13 C-terminal truncations. Envelopes calculated from SAXS data are rendered as gray surfaces. ADAMTS13 domains are colored similarly to Figure 1A: M (orange), D (yellow), T1 (green), C (sky blue), S (lavender), T3 and T6 (yellow), T2, T4, T5, T7, and T8 (green), T4-T5 linker (gray), T8-CUB1 linker (gray), CUB1 (red), CUB2 (brick red). All constructs have the active site mutation E225Q.
Selected enzymatic properties of these C-terminal truncated ADAMTS13 variants are summarized in Table 1. In agreement with our previous report,18 ADAMTS13 was autoinhibited at pH 7.4 and cleaved substrate VWF71 ∼20% as rapidly as MDTCS, which is constitutively activated. ADAMTS13 was activated (or disinhibited) at pH 6 and cleaved VWF71 almost as rapidly as MDTCS, which was not further activated by low pH. MT2 through MT7 also were constitutively activated and cleaved VWF71 at rates similar to MDTCS. In contrast, MT8 and MT8L cleaved VWF71 at an intermediate rate that is consistent with partial autoinhibition. In addition, low pH did not significantly activate MT8 or MT8L, which suggests that the CUB domains mediate the allosteric activation of ADAMTS13 by protons. In addition, MT8 could be activated ∼1.2-fold by MAbs and ∼1.7-fold by VWF D4, whereas MT7 could not.28 These results support the proposed role for T8 and CUB domains in the regulation of ADAMTS13 activity18,29 and suggest that some T domains between T2 and T7 may be dispensable.
Autoinhibition and allosteric activation of ADAMTS13 depend on a few distal domains
To assess the role of other distal domains in ADAMTS13 allosteric regulation and conformation, variants were constructed with internal deletions of ≥1 T domains (Table 2). All of these constructs retained the CUB domains. All cleaved VWF71 at a rate (2.4 to 4.3 min−1) comparable to that of full-length ADAMTS13 (2.2 min–1) and slower than MDTCS (10 min−1), which is consistent with autoinhibition. All could be allosterically activated (1.4-fold to 3-fold) by MAbs against the T7-CUB domains, which indicates that these distal domains are close to the proximal MDTCS domains.
Molecular mass, Rg, Dmax, and allosteric activation for ADAMTS13 internal deletions
| Variant . | Mass (kDa) . | Rg (Å), mean ± SD . | Dmax (Å), mean ± SD . | Enzymatic rate, pH 7.4 (min−1) (mean) . | Activation by MAbs (fold) (mean) . | Activation by D4 (fold) (mean) . |
|---|---|---|---|---|---|---|
| delT4L | 144 | 64 ± 3 | 217 ± 15 | 2.4 | 2.0 | 1.3 |
| delT4pL | 138 | 75 ± 5 | 268 ± 5 | 3.1 | 1.8 | 1.7 |
| delT7 | 141 | 76 ± 4 | 258 ± 14 | 2.7 | 2.6 | 1.0 |
| delT8 | 141 | 77 ± 4 | 267 ± 16 | 4.1 | 2.0 | 1.0 |
| delT8L | 141 | 75 ± 4 | 256 ± 18 | 3.6 | 2.6 | 1.0 |
| delT8pL | 135 | IM | IM | 4.3 | 2.0 | 1.0 |
| delT2-7 | 109 | 57 ± 2 | 189 ± 14 | 3.6 | 3.0 | 1.0 |
| delT2-8L | 103 | AiS | AiS | n/a | 1.4 | 1.0 |
| delT3-6 | 117 | 70 ± 3 | 244 ± 8 | 2.6 | 2.5 | 1.5 |
| pADAMTS13 | 116 | 56 ± 1 | 190 ± 10 | 0.2 | 1.0 | 4.0 |
| ADAMTS13 | 147 | 67 ± 2 | 229 ± 6 | 2.2 | 3.8 | 1.8 |
| Variant . | Mass (kDa) . | Rg (Å), mean ± SD . | Dmax (Å), mean ± SD . | Enzymatic rate, pH 7.4 (min−1) (mean) . | Activation by MAbs (fold) (mean) . | Activation by D4 (fold) (mean) . |
|---|---|---|---|---|---|---|
| delT4L | 144 | 64 ± 3 | 217 ± 15 | 2.4 | 2.0 | 1.3 |
| delT4pL | 138 | 75 ± 5 | 268 ± 5 | 3.1 | 1.8 | 1.7 |
| delT7 | 141 | 76 ± 4 | 258 ± 14 | 2.7 | 2.6 | 1.0 |
| delT8 | 141 | 77 ± 4 | 267 ± 16 | 4.1 | 2.0 | 1.0 |
| delT8L | 141 | 75 ± 4 | 256 ± 18 | 3.6 | 2.6 | 1.0 |
| delT8pL | 135 | IM | IM | 4.3 | 2.0 | 1.0 |
| delT2-7 | 109 | 57 ± 2 | 189 ± 14 | 3.6 | 3.0 | 1.0 |
| delT2-8L | 103 | AiS | AiS | n/a | 1.4 | 1.0 |
| delT3-6 | 117 | 70 ± 3 | 244 ± 8 | 2.6 | 2.5 | 1.5 |
| pADAMTS13 | 116 | 56 ± 1 | 190 ± 10 | 0.2 | 1.0 | 4.0 |
| ADAMTS13 | 147 | 67 ± 2 | 229 ± 6 | 2.2 | 3.8 | 1.8 |
Rg data were calculated using the program AutoRg, and Dmax data were calculated using the program DATGNOM. Standard error for enzymatic rates and activation is <10%.
AiS, aggregation in solution; IM, insufficient material for SAXS; n/a, not available.
SAXS analysis supports this interpretation. Deletion of a single T4, T7, or T8 domain, or even deletion of 4 (delT3-6) or 6 (delT2-7) T domains, led to modest changes in Rg and Dmax (Table 2), but ab initio envelopes indicated that these deletion constructs still had a folded conformation that juxtaposes the distal and proximal domains (Figure 3).
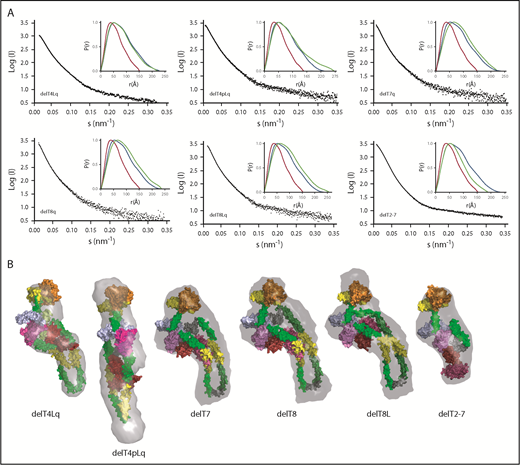
SAXS data, ab initio envelopes, and molecular models of ADAMTS13 internal deletions. (A) Scattering profiles and P(r) plots (inset) for the indicated pure recombinant ADAMTS13 variants. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included in each inset panel for comparison with other deletion constructs (green). (B) Molecular models were fitted into ab initio envelopes (gray surface) calculated from SAXS profiles for the indicated ADAMTS13 constructs.
SAXS data, ab initio envelopes, and molecular models of ADAMTS13 internal deletions. (A) Scattering profiles and P(r) plots (inset) for the indicated pure recombinant ADAMTS13 variants. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included in each inset panel for comparison with other deletion constructs (green). (B) Molecular models were fitted into ab initio envelopes (gray surface) calculated from SAXS profiles for the indicated ADAMTS13 constructs.
The analysis of sequential domain deletions (Figure 2) suggested that the folded structure of ADAMTS13 was achieved by bending the 29-residue linker between T4 and T5. However, deletion of T4L (delT4Lq) and deletion of T4 and T4L (delT4pLq) lack this linker and still retained a folded conformation. Construct delT4pLq had a larger Rg and Dmax and a longer solution envelope than ADAMTS13 (Figure 3), which suggests that other flexible T domains can rearrange to preserve a functional allosteric mechanism in the absence of T4 and the linker to T5.
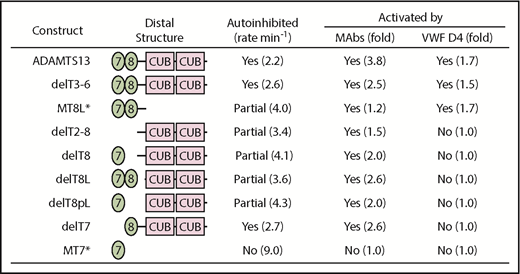
The enzymatic properties of ADAMTS13 deletion mutants indicate that autoinhibition and allosteric activation by MAbs or by VWF D4 depend on different combinations of ADAMTS13 domains. Because ADAMTS13 and delT3-6 had essentially similar properties (Table 2),28 T3 through T6 are dispensable for regulation. Therefore, we focused on the shared distal domains: T7, T8, CUBs, and the 58–amino acid residue linker between T8 and CUB1 (T8L) (Figure 4).
Summary of allosteric properties dependent on ADAMTS13 T7-CUB domains. Data from Table 2 except for constructs MT7 and MT8L designated with an asterisk (*).28
The extent of autoinhibition depended on T8, T8L, and CUBs but not on T7. The protease was fully autoinhibited like wild-type ADAMTS13 if all 3 segments were present, whereas partial inhibition resulted if CUBs, T8L, or T8 was deleted. It is notable that CUBs alone were sufficient to partially inhibit protease activity.
Activation of ADAMTS13 by MAbs against T7-CUB domains had similar requirements. CUB domains were sufficient to enable 1.5-fold activation, and addition of T8 and T8L further enhanced activation by MAbs. In contrast, addition of T7 had little or no effect.
Allosteric activation by VWF D4 had some requirements that were different from activation by MAbs. Deletion of the CUB domains had no effect. However, deletion of T7, T8, or T8L abolished activation by VWF D4 without affecting activation by MAbs (Figure 4). Deletion of T7 or T8 abolished binding to VWF D4-CK (supplemental Figure 2), explaining the loss of activation. By contrast, deletion of T8L did not abolish binding to VWF D4-CK but did cause a loss of activation by VWF D4. The striking loss of activation by VWF D4-CK upon deleting the linker segment between T8 and CUB1 (T8L) was unexpected and suggests that the length of T8L is critical to prevent CUB1 from interfering with allosteric activation by VWF D4.
Changes in the interface for binding VWF D4 affected the overall shape of ADAMTS13, as shown by Rg and Dmax values for delT7, delT8, and delT8L of 75 to 77 Å and 256 to 267 Å, respectively, which are ∼13% greater than for wild-type ADAMTS13 (Table 2). Ab initio envelopes for these variants (Figure 3B) show increased space where the proximal MDTCS and distal T8-CUB domains interact. These structural changes were associated with partial activation, increasing the rate of VWF71 cleavage from 2.2 min−1 for wild-type ADAMTS13 to 2.7 min−1 (delT7), 4.1 min−1 (delT8), and 3.6 min−1 (delT8L). Thus, deletion of key distal domains appears to prevent ADAMTS13 from achieving a fully inhibited compact conformation, and it disrupts allosteric activation by VWF D4.
pADAMTS13 and delT3-6 approach a “minimal” structure for allosterically regulated ADAMTS13
Through studies of ADAMTS13 in other species,28 we found that allosteric regulation of ADAMTS13 is evolutionarily conserved despite substantial variation in the number of distal T domains from as many as 8 to as few as 3. This finding led us to hypothesize that a “minimal” human ADAMTS13 with normal allosteric regulation could be designed based upon the structural variation observed among other vertebrates. For example, pADAMTS13 has 3 distal T domains and was activated potently by human VWF D4 (Table 2). Human delT3-6 was engineered with the same complement of distal T domains and, like human ADAMTS13, was fully autoinhibited and allosterically regulated by VWF D4. The further removal of T7 (construct delT2-7) rendered ADAMTS13 unresponsive to VWF D4, although delT2-7 could be activated by MAbs (Table 2). Thus, T7, T8, and the linker between T8 and CUB1 appear to be required for allosteric activation of ADAMTS13 by VWF D4.
At least 1 CUB domain also makes a significant contribution to allostery. For example, construct MT8L lacks both CUB domains and was partially autoinhibited, cleaving VWF71 at a rate (4.0 min−1) intermediate between uninhibited MT7 (9 min−1) and fully inhibited ADAMTS13 (2.2 min−1) (Figure 4). MT8L was allosterically activated ∼1.7-fold by VWF D428 (Figure 4), but MT8L bound to VWF D4-CK relatively weakly, with a dissociation constant ∼ 620 nM compared with ∼360 nM for ADAMTS13.18 A recent study reported that deletion of CUB2 had almost no effect on the allosteric properties of ADAMTS13 but that CUB1 could bind and inhibit ADAMTS13.30 Thus, the CUB domains, primarily CUB1, contribute to autoinhibition and to the affinity of binding VWF D4. Whether allosteric regulation would be preserved in a construct like delT3-6/CUB2 has not been assessed.
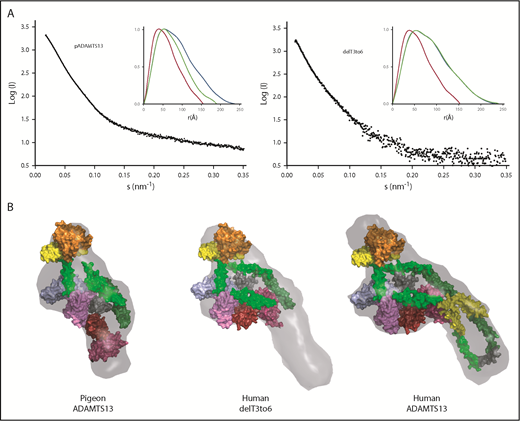
Comparison of the ab initio envelopes for rADAMTS13, pADAMTS13, and delT3-6 indicates that the distal T3 to T6 domains of rADAMTS13 extend away from the MDTCS and T7-CUB2 domains that are critical for autoinhibition and allosteric activation. The absence of 4 distal T domains in pADAMTS13 or delT3-6 appears to have a minimal effect on the association of MDTCS and T7-CUB domains (Figure 5), which is consistent with the preservation of allosteric regulation in these structural variants (Table 2).
Toward a minimal allosterically regulated ADAMTS13. (A) Scattering profiles (black) and P(r) plots (inset) for pADAMTS13, delT3-6, and human ADAMTS13. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included for comparison with those for other constructs (green). (B) Ab initio models (gray surface) calculated from scattering profiles for pADAMTS13, human delT3-6, and human ADAMTS13 superimposed on atomic models of MDTCS (orange = M domain; yellow = D domain; green = T1 domain; sky blue = C domain; lavender = S domain), Ts (surface), and CUBs. T2, green; T3, yellow; T4, green; T5, green; T6, yellow; T7, dark green; T8, green; linker between T8 and CUBs, gray; CUB domains, red and brick red.
Toward a minimal allosterically regulated ADAMTS13. (A) Scattering profiles (black) and P(r) plots (inset) for pADAMTS13, delT3-6, and human ADAMTS13. P(r) plots for MDTCSq (red) and ADAMTS13 (FLq: blue) are included for comparison with those for other constructs (green). (B) Ab initio models (gray surface) calculated from scattering profiles for pADAMTS13, human delT3-6, and human ADAMTS13 superimposed on atomic models of MDTCS (orange = M domain; yellow = D domain; green = T1 domain; sky blue = C domain; lavender = S domain), Ts (surface), and CUBs. T2, green; T3, yellow; T4, green; T5, green; T6, yellow; T7, dark green; T8, green; linker between T8 and CUBs, gray; CUB domains, red and brick red.
Discussion
ADAMTS13 is a multidomain protein with an M domain near the N terminus that is autoinhibited by association with T and CUB domains near the C terminus. These interacting domains are separated by ∼500 Å in an extended molecular model of human ADAMTS13, and bringing them together requires folding ADAMTS13 roughly in half.18 The packing of domains to achieve this folded structure is illustrated nicely by fitting progressively elongated ADAMTS13 constructs into ab initio envelopes determined by SAXS (Figure 2). The intervening nonessential T domains extend like a hairpin away from the core MDTCS and T7-CUB domains with a hinge between T4 and T5 that allows close packing of T domains (Figure 2).
The association of MDTCS and T7-CUB domains is surprisingly stable, persisting despite the deletion of the CUB domains (Figure 2; MT8L) or internal deletion of all distal T domains except T8 (Figure 3; delT2-7). MT8L and delT2-7 constructs were activated by selected MAbs, confirming the functional association between MDTCS and the few distal domains that remain in each construct. In addition, the T4-T5 linker may be a convenient hinge to allow folding of distal T domains in rADAMTS13, but if the T4-T5 linker is removed, the remaining T domains are sufficiently flexible to preserve allosteric regulation by adapting and maintaining the functional juxtaposition of MDTCS and T7-CUB.
In other comparative evolutionary studies, we found tremendous variability in the number of ADAMTS13 distal T domains, from a low of 3 in pigeon to a high of 8 in several reptiles, amphibians, and mammals.28 However, the current data show that human T7 and T8 are required for allosteric activation by VWF D4 (Figure 4), and these domains are invariably present in other species. Looking at the SAXS envelope and molecular model for full-length ADAMTS13 (Figure 2), the T7 and T8 domains appear closely associated with the proximal MDTCS domains. In contrast, the T3 to T6 domains extend away from the core domains required for normal allosteric regulation, are not positioned to contact the substrate VWF, and can be deleted from rADAMTS13 with little effect on substrate cleavage or allosteric regulation. These properties suggest that all distal T domains, with the exception of T7 and T8, are dispensable, and an allosterically regulated ADAMTS13 can be constructed without them.
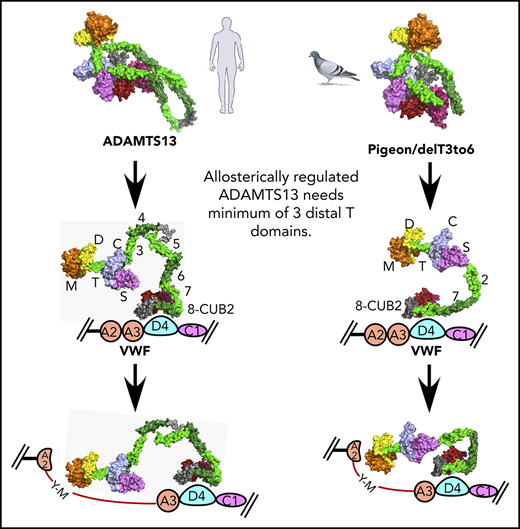
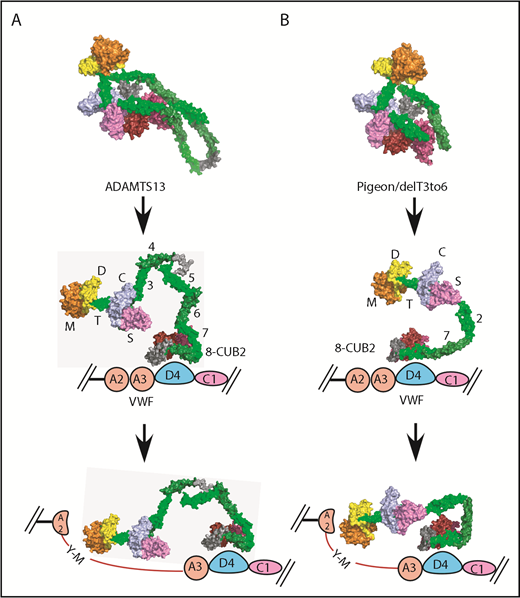
pADAMTS13 is an example of such a minimal ADAMTS13, with just 1 T domain in addition to the invariant T7 and T8. Indeed, when rADAMTS13 was engineered to resemble pADAMTS13, the result, delT3-6, was autoinhibited and allosterically activated by VWF D4–like pADAMTS13 and native rADAMTS13 (Table 2). The SAXS envelopes for these 3 proteases (Figure 5) indicate that all of them share a highly conserved core composed of proximal MDTCS and distal T7-CUB domains. Our current working model for ADAMTS13 allosteric activation is shown in Figure 6. The distal T7-CUB2 domains of ADAMTS13 (Figure 6A), as well as their counterparts in pigeon (or the similar rADAMTS13 construct; ie, delT3-6, Figure 6B), first bind to the D4 domain of VWF, leading to the activation of ADAMTS13. Under shear force, the A2 domain of VWF unfolds. This further stretches ADAMTS13, and the proximal exosites now are fully engaged with the A2 domain, resulting in cleavage of VWF.
Proposed allosteric activation of full-length human ADAMTS13, delT3-6, and pADAMTS13. Human ADAMTS13 (A) and pADAMTS13/delT3-6 (B). MDTCS (orange = M domain; yellow = D domain; green = T1 domain; sky blue = C domain; lavender = S domain), T2, T4, and T8, dark green; T3, T5, and T7, green; linker between T4 and T5 and between T8 and CUBs, gray; CUB1, red; and CUB2, brick red. VWF: A2 and A3, pink; D4, light blue; and C1, lavender.
Proposed allosteric activation of full-length human ADAMTS13, delT3-6, and pADAMTS13. Human ADAMTS13 (A) and pADAMTS13/delT3-6 (B). MDTCS (orange = M domain; yellow = D domain; green = T1 domain; sky blue = C domain; lavender = S domain), T2, T4, and T8, dark green; T3, T5, and T7, green; linker between T4 and T5 and between T8 and CUBs, gray; CUB1, red; and CUB2, brick red. VWF: A2 and A3, pink; D4, light blue; and C1, lavender.
With the exception of T7 and T8, most other distal domains are dispensable in vitro, and pigeons do perfectly well without them in vivo. In the vast majority of vertebrates, ADAMTS13 has between 2 and 5 more than the apparent minimum complement of 3 distal T domains.28 The mismatch between the apparent functional minimum and the evolutionary mean number of T domains suggests that at least some distal T domains perform a distinct selectable function that remains undiscovered.
For original data, please contact Linda J. Pike (pike@wustl.edu).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Ginsburg and Linda Pike for critical reading of the manuscript. They also thank Bruce Linders (Washington University) for tissue culture to prepare ADAMTS13 variants and Kevin Dyer, Kathryn Burnett, and Gregory Hura (Lawrence Berkeley Laboratory) for assistance with SAXS data collection.
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grants R01 HL72917, R01 HL89746, U54 HL112303, and T32 HL007088 and by American Heart Association Scientist Development grant 16SDG30740011 (J.M.). N.H.T. is supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: J.Z., J.M., G.G., N.H.T., and J.E.S. designed the research and analyzed data; J.Z., J.M., G.G., and L.A.W. performed research; K.V. provided essential materials; and J.Z. and J.E.S. wrote the manuscript. All authors revised the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jian Zhu, Department of Medicine, Washington University School of Medicine, 660 S. Euclid Ave, Box 8125, St. Louis, MO 63110; e-mail: zhujian@wustl.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal