Key Points
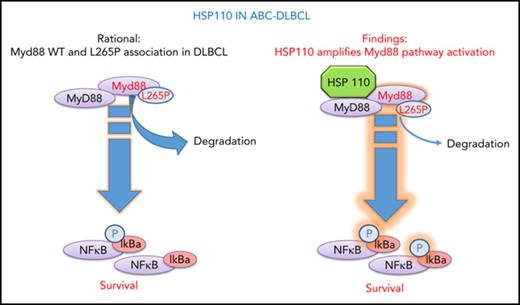
HSP110 sustains chronic NF-κB signaling in ABC-DLBCL through MyD88 stability.
HSP110 is highly expressed in cells of patients with ABC-DLBCL and correlates with MyD88 expression.
Abstract
Activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL) is an aggressive lymphoproliferative disorder involving chronic NF-κB activation. Several mutations in the BCR and MyD88 signaling pathway components, such as MyD88 L265P, are implicated in this aberrant activation. Among heat shock proteins, HSP110 has recently been identified as a prosurvival and/or proliferation factor in many cancers, but its role in ABC-DLBCL survival mechanisms remained to be established. We observed that short hairpin RNA–mediated HSP110 silencing decreased the survival of several ABC-DLBCL cell lines and decreased immunoglobulin M-MyD88 co-localization and subsequent NF-κB signaling. Conversely, overexpression of HSP110 in ABC-DLBCL or non-DLBCL cell lines increased NF-κB signaling, indicating a tight interplay between HSP110 and the NF-κB pathway. By using immunoprecipitation and proximity ligation assays, we identified an interaction between HSP110 and both wild-type MyD88 and MyD88 L265P. HSP110 stabilized both MyD88 forms with a stronger effect on MyD88 L265P, thus facilitating chronic NF-κB activation. Finally, HSP110 expression was higher in lymph node biopsies from patients with ABC-DLBCL than in normal reactive lymph nodes, and a strong correlation was found between the level of HSP110 and MyD88. In conclusion, we identified HSP110 as a regulator of NF-κB signaling through MyD88 stabilization in ABC-DLBCL. This finding reveals HSP110 as a new potential therapeutic target in ABC-DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of aggressive non-Hodgkin lymphoma (NHL) in adults. Although therapy with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) has led to an improvement in survival, a significant percentage of patients are still refractory to the treatment or they eventually relapse.1 Therefore, it is important to find new molecular targets for treating patients with DLBCL. Advanced genome sequencing studies have unveiled the complexity of DLBCL and revealed a heterogeneous group of tumors characterized by specific genetic alterations.2 One way to overcome the heterogeneity of this lymphoma is to identify and target mechanisms that drive or sustain oncogenic signaling and survival of lymphoma cells, regardless of their somatic mutations. Interestingly, a body of literature has highlighted the fact that tumor cells acquire a biological addiction to heat shock proteins (HSPs) and stress proteins during malignant transformation.3-5 HSPs are highly conserved molecular chaperone proteins whose expression is induced in response to multiple physiological and environmental stresses.6 In these contexts, they control a large array of cellular functions to rescue the cell from debilitating conditions. Cancer cells can be considered highly stressed cells because of their numerous mutations, translocation or polyploidy, and microenvironment.7,8 Therefore, HSPs are often overexpressed in cancer cells, where they contribute to cancer cell resistance, notably to apoptosis and to the clearance of cancer cells by immune cells.9,10 As a consequence, targeting HSPs in various types of cancer has been proposed. Several articles have shown that HSPs are also highly expressed in hematologic malignancies, in which they are involved in several key proliferative and survival functions.11 In particular, in lymphoma, HSP90 has been shown to have multiple roles in key DLBCL oncogenic pathways. Indeed, its capacity to enhance B-cell–like 6 (BCL6) oncogenesis4 and to interact with and stabilize key B-cell receptor (BCR) protein complexes (CD79A/B-SYK-BTK-PLCγ2)3 suggests that targeting HSP90 could represent an interesting additional option for treating DLBCL. Unfortunately, a recent clinical trial using the HSP90 inhibitor AUY922 in DLBCL (NCT01485536) was halted early because of limited response and adverse effects in 100% of patients, prompting the search for other potential targets.
Among HSPs, HSP110 is a high-molecular-weight chaperone with antiaggregation properties and, through its interaction with HSP70, participates in the correct folding of newly synthesized or misfolded proteins.12,13 We have recently demonstrated that HSP110 is a major factor in colon cancer growth through its intra- and extracellular functions.14,15 At the intracellular level, HSP110 binds directly to STAT3, facilitating its phosphorylation by JAK2 and contributing to STAT3-dependent tumor growth. We have demonstrated that colon cancer cells at the extracellular level secrete HSP110 to promote macrophage polarization toward a procancer phenotype (M2).15 Furthermore, we have identified a truncated form of HSP110 called HSP110DE9 in colorectal tumors with microsatellite instability that acts as an endogenous HSP110 inhibitor.16 The high expression of HSP110DE9 and/or the low expression of wild-type (WT) HSP110 in patients with colon cancer was associated with a highly effective response to chemotherapy.17 Recent studies have shown major new roles for HSP110 in human NHL. In particular, HSP110 was shown to be an immunogenic antigen in a xenograft model of NHL, and expression of HSP110 was shown to correlate with the aggressiveness and proliferation index of tumor cells from patients.18 A significant positive correlation between HSP110 and BCL6 expression was also found in germinal center (GC) BCL/BCL6+ DLBCL, which could be explained by the stabilization of BCL6.19
Given the emerging role of HSP110 in cancer and NHL in particular, we investigated the role of HSP110 in ABC-DLBCL, which relies on the NF-κB signaling pathway for growth and survival.20 Mutations affecting MyD88 L265P and the CD79/CARD11/MALT1/BCL10 signaling cascade are mostly responsible for the aberrant NF-κB activation.21,22 Here we show that HSP110 is an essential survival factor for these tumor cells. Furthermore, we demonstrate that HSP110 binds to and stabilizes wild-type and mutated MyD88 in cell lines and in patients’ samples and thereby amplifies the aberrant NF-κB signaling pathway. Thus, we have identified a new role for HSP110 in tumor cells and have confirmed that HSP110 is a potential target for the development of future therapies for NHL.
Methods
Primary tumors and cell lines
ABC-DLBCL cell lines were purchased from American Type Culture Collection (U2932/OCI-Ly3/OCI-Ly10/SUD-HL2) or kindly provided by D. Krappmann (Helmholtz Zentrum München, Munich, Germany) (HBL1/TMD8) and cultured in RPMI 1640 (Dominique Dutscher, Brumath, France) plus fetal bovine serum (Dominique Dutscher) at 10% (U2932/TMD8), 15% (OCI-Ly3), or 20% (HBL1/OCI-Ly10 and SUD-HL2). Tumors for immunoblots and immunohistochemistry were provided by the Ferdinand Cabanne Biological Resource Center (BB-0033-00044), Dijon, France, and by the Département de Biopathologie, Centre Léon Bérard, Lyon, France. Samples for messenger RNA (mRNA) analysis were from the Centre de Ressources Biologiques-Santé (BB-0033-00056) of Rennes Hospital. Patients with DLBCL were classified according the World Health Organization classification using the Hans immunohistochemical algorithm. The study using human biopsies was approved by each relevant institutional review board or ethics committee, and all human participants gave informed consent.
Transfection
ABC-DLBCL cell lines were transfected with the AMAXA Nucleofector 2b device (Lonza, Basel, Switzerland) and the Nucleofector kit T (HBL1, OCI-Ly3, OCI-Ly10) or V (U2932, SU-DHL2, TMD8). Short interfering RNA (siRNA) control (MISSION siRNA Universal Negative Control #1, Sigma-Aldrich, Lyon, France) or hsph1 targeting siRNA (Silencer Select Pre-Designed siRNAs, Life Technologies, Saint-Aubin, France) were used. Specific AMAXA programs were applied (U-15 for U2932; P005 for SU-DHL2, TMD8; G016 for HBL1, OCI-Ly3, OCI-Ly10). For other HSPs, Silencer Pre-Designed siRNA for HSP27 (ID: 122371-HSPB1) and HSP70 (IDs6966-HSPA1A) or the specific functional inhibitor of HSP90, 1 µM PU-H71 16 hours (Axon Medchem, Groningen, The Netherlands) were used. HEK293T transfections were performed using Genjet (Ver. II) reagent (TebuBio, Le Perray-en-Yvelines, France). Green fluorescent protein (GFP), HSP110-GFP, and HSP110DE9-mutant GFP were homemade plasmids. Myd88 WT and L265P plasmid were used as described,23 pGL4.32(luc2P/NF-κB-RE/Hygro) vector (Promega E849A) was kindly provided by P. Gaulard (Mondor Institute of Biomedical Research, Paris, France).
Retroviral particles
Human short hairpin RNA (shRNA) lentiviral vectors pseudotyped with measles virus glycoproteins or baboon retroviral glycoproteins have been described elsewhere.24-26 These lentiviral vectors were produced in 293T cells with GFP-expressing shRNA control (Sigma MISSION SHC016-1EA) or hsph1-targeting shRNA lentiviral constructs (Sigma MISSION shRNA SHCLND-NM_006644, clone TRCN0000275617). HBL1, Ly3, TMD8, and U2932 were incubated using viral supernatant at multiplicity of infection 2 for 24 hours before washing. At 48 hours after transduction, cells were subjected to puromycin selection for 96 hours. The viability of GFP+ cells over time was observed by using an LSRII Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ). For rescue experiments, the retroviral IKK2-EE vector was a gift from Stephan Ludwig (Zentrum für Molekularbiologie der Entzündung, Munster, Germany).27 This murine retroviral vector was produced by transfection in 293T cells together with murine leukemia virus gag-pol plasmid and the BAEV-encoding plasmid. HBL1 was infected 3 times every 12 hours, selected at 96 hours, and then transduced with the lentiviral vector–bearing shRNA control or HSP110 shRNA.
Immunoblot analysis
Primary antibodies used in this study were anti-Bcl-xL (2764S), c-Myc (9605S), cyclin D1 (2926S), p-STAT3 (9145S), STAT3 (9139S), and p-IκBα Ser32/36 (9246) from Cell Signaling Technologies (Danvers, MA); Mcl-1 (sc-819), HSC70 (sc-7298), HSP110 (sc-6241), GFP (sc-8334), BcL10 (331.3), and p-IRAK1 (sc-325147) from Santa Cruz Biotechnologies (Dallas, TX); anti-HSP110 (ab109624) and anti-MyD88 (ab119048) for IHC staining, anti-BcL10 (ab199011), immunoglobulin M (IgM; ab20054), IκBα (ab32518), p-IκBα (ab133462), Myd88 (ab119048), HSP90 (ab59459), HA (ab130275), HSP70 (ab5439), ubiquitin (linkage-specific K63) (ab179434) from Abcam (Paris, France); anti-IκBα (#4812), IRAK1 (#4504), Myd88 (#4283), c-Rel (#4727), TRAF6 (#8028), p65 (#8242), BcL2 (#2870), caspase-3 Asp175 (#9664), total caspase-3 (#9662), lamin A/C (#4777), and HSP60 (#12165) from Cell Signaling Technologies; anti-p50 (616702) from BioLegend (San Diego, CA); HSP27 (ADI-SPA-800) from Enzo Life (Villeurbanne, France), and monoclonal anti-β-actin-peroxidase antibody (A3854) from Sigma-Aldrich. Normal mouse serum IgG (sc-2025) and normal rabbit IgG (sc-2027) from Santa Cruz Biotechnologies were used as nonrelevant antibodies for control immunoprecipitation and Duolink assay. Cytoplasmic and nuclear extracts were obtained using the NE-PER Nuclear and Cytoplasmic Extraction reagent kit (Thermo Fisher Scientific, Waltham, MA). Immunoprecipitations were performed as previously described,21 and the HSP110 primary antibody (ab109624) was used.
NF-κB reporter assay
HEK293T cells were transfected using Genjet Ver. II reagent, 0.4 µg of pGL4.32 (luc2P/NF-κB-RE/Hygro) vector (Promega, Madison, WI), 0.4 µg Myd88 WT or L265P-hemagglutinin (HA) tagged, increasing amounts of HSP110-HA–tagged plasmid (0.1 to 0.7 µg), and 0.3 µg of GFP vector for normalization. Samples were prepared according to the Promega protocol (Luciferase Assay System E1500). Luminescence from equivalent amounts of lysate was read in triplicate on an EnVision Multilabel Plate Reader (PerkinElmer). All readings were normalized to the mean fluorescence intensity of GFP expression.
Immunofluorescence and Duolink proximity ligation assay (PLA)
Primary antibodies were anti-IgM (IM260), anti-IκBα (ab32518), anti-HSP105 (EPR4576), anti-HSP70 (ab5439), and anti-Myd88 (ab119048). Duolink experiments were performed according to the manufacturer’s protocol (Sigma-Aldrich, St Louis, MO). Microscopy images were taken on an Axio Imager 2 (Carl Zeiss Microscopy GmbH, Jena, Germany), and images were acquired by using an AxioCam MRm CCD camera (Carl Zeiss). The Spot detector plugin of Icy software was used to count the number of spots per cell. Statistical analysis was performed with GraphPad Prism 6 (unpaired Mann-Whitney U test; at least 100 cells counted for each condition).
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR for reactive lymph nodes and patients was performed using the TaqMan Universal Master Mix and specific TaqMan Gene Expression Assays (Life Technologies-Applied Biosystems): HSP110/hs-00971475-m1 and ABL/hs001104728-m1 (for internal standard gene). Statistical analyses were performed with Prism software (GraphPad Software Inc., San Diego, CA) using a Mann-Whitney nonparametric U test. For other RT-PCRs, PrimePCR SYBR Green Assay (RPS18, Human [Biorad]), MyD88 F (GTTGTCTCTGATGATTACCTG), MyD88 R (GGGGAACTCTTTCTTCATTG; Sigma-Aldrich), and the HSP110 primers used were described earlier.17
Results
HSP110 expression has an impact on B-cell lymphoma cell survival and NF-κB signaling
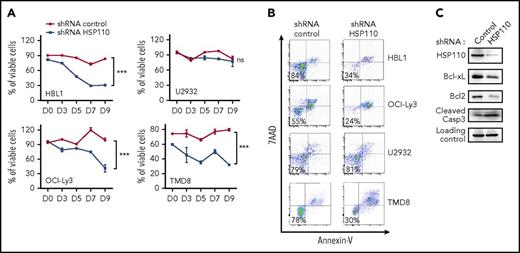
To explore the role of HSP110 in ABC-DLBCL cell lines, we transduced them with a lentivirus carrying HSP110 shRNA that was optimized to have high transduction capacities toward hard-to-transfect human B-cell lines.24,28 We observed a decreased survival in the HSP110 shRNA-infected cell lines we tested: mean percentage of viable cells in shRNA HSP110 vs shRNA control at day 9 for HBL1 was 30 ± 4 and 83 ± 1.5; for OCI-Ly3, 41 ± 13 and 83 ± 1.5; and for TMD8, 32 ± 0.7 and 80 ± 1.5; but no decrease for U2932 cells, 9.72 ± 16 and 76 ± 4 (Figure 1A-B). A decrease in the antiapoptotic proteins Bcl-xL and Bcl2, together with an increase in caspase-3 cleavage, suggested that depletion of HSP110 is deleterious in certain ABC-DLBCL cell lines and induced apoptotic cell death (Figure 1C). Because hyperactivation of the NF-κB signaling pathway is primarily responsible for ABC-DLBCL proliferation and survival, we studied the impact of HSP110 on this pathway in the following experiments. Interestingly, siRNA-mediated HSP110 knockdown induced a decrease in IκB and p65 phosphorylation in all ABC-DLBCL cell lines tested, with the exception of U2932 (Figure 2A-B). The silencing of HSP110 did not modulate AKT activation, another important signaling pathway in ABC-DLBCL (supplemental Figure 1A, available on the Blood Web site). Next, we compared the effect observed with HSP110 depletion with that observed by depleting or inhibiting other main HSPs (HSP27, HSP70, and HSP90). We used either an siRNA approach (for HSP27, HSP70, and HSP110 [specificity of HSP110 siRNA is shown in supplemental Figure 1A]) or a chemical inhibitor (PU-H71 for HSP90). As shown in supplemental Figure 1B, inhibition of HSP90, as already reported,3 induced a decrease in IκB phosphorylation similar to that obtained upon HSP110 depletion. In contrast, HSP27 or HSP70 knockdown did not affect p-IκB content. In keeping with the decrease in IκB phosphorylation induced by HSP110 knockdown, the nuclear localization of p65 and p50 were also decreased in cells depleted in HSP110 (Figure 2C; supplemental Figure 1C). Conversely, transfection of SU-DHL2 by increasing amounts of HSP110 induced an increase in IκB phosphorylation (Figure 2D). Finally, HBL1 cells were rescued from HSP110 shRNA–induced cell death by co-expression of the constitutively active IKK2-EE (Figure 2E). Taken together, these data suggest that HSP110 is involved in ABC-DLBCL survival through NF-κB signaling modulation.
HSP110 expression has an impact on ABC-DLBCL cell survival. (A) Kinetics of cell survival determined by annexin-V/7-aminoactinomycin D (7AAD) staining on shRNA-expressing cells, data are presented as the mean ± standard deviation (n = 3). (B) Representative annexin-V/7AAD staining on shRNA-expressing cells determined at day 9. (C) Immunoblot analysis of HSP110, Bcl-xL, Bcl2, and cleaved caspase-3 (Casp3) at 48 hours after transfection of HBL1 with an shRNA targeting HSP110 or a control shRNA (n = 3). β-actin served as a loading control. ns, not significant. ***P < .001.
HSP110 expression has an impact on ABC-DLBCL cell survival. (A) Kinetics of cell survival determined by annexin-V/7-aminoactinomycin D (7AAD) staining on shRNA-expressing cells, data are presented as the mean ± standard deviation (n = 3). (B) Representative annexin-V/7AAD staining on shRNA-expressing cells determined at day 9. (C) Immunoblot analysis of HSP110, Bcl-xL, Bcl2, and cleaved caspase-3 (Casp3) at 48 hours after transfection of HBL1 with an shRNA targeting HSP110 or a control shRNA (n = 3). β-actin served as a loading control. ns, not significant. ***P < .001.
HSP110 expression has an impact on NF-κB signaling. (A) Immunoblot analysis of HSP110, p-IκBα, p-p65, IκBα, and p65 at 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control. (B) p-IκB band intensity from immunoblots shown in (A) and determined from all cell lines (except U2932) relative to the loading control, data are presented as the mean ± standard deviation (n = 5; not U2932; P < .01). (C) Immunoblot analysis of HSP110, p65, and p50 content in the nucleus and cytosol of OCI-Ly3 and SU-DHL2 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control (n = 3). (D) Immunoblot analysis of HSP110, p-IkB, and IκB in SU-DHL2 cells 48 hours after transfection with plasmids coding for HSP110-GFP (n = 3). HSC70 served as a loading control. (E) Kinetics of cell survival determined by annexin-V/7AAD staining on HBL1 stably expressing either shRNA HSP110, shRNA control plus IKK2-EE, or shRNA HSP110 plus IKK2-EE, data are presented as the mean ± standard deviation (n = 3). **P < .01; ***P < .001.
HSP110 expression has an impact on NF-κB signaling. (A) Immunoblot analysis of HSP110, p-IκBα, p-p65, IκBα, and p65 at 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control. (B) p-IκB band intensity from immunoblots shown in (A) and determined from all cell lines (except U2932) relative to the loading control, data are presented as the mean ± standard deviation (n = 5; not U2932; P < .01). (C) Immunoblot analysis of HSP110, p65, and p50 content in the nucleus and cytosol of OCI-Ly3 and SU-DHL2 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control (n = 3). (D) Immunoblot analysis of HSP110, p-IkB, and IκB in SU-DHL2 cells 48 hours after transfection with plasmids coding for HSP110-GFP (n = 3). HSC70 served as a loading control. (E) Kinetics of cell survival determined by annexin-V/7AAD staining on HBL1 stably expressing either shRNA HSP110, shRNA control plus IKK2-EE, or shRNA HSP110 plus IKK2-EE, data are presented as the mean ± standard deviation (n = 3). **P < .01; ***P < .001.
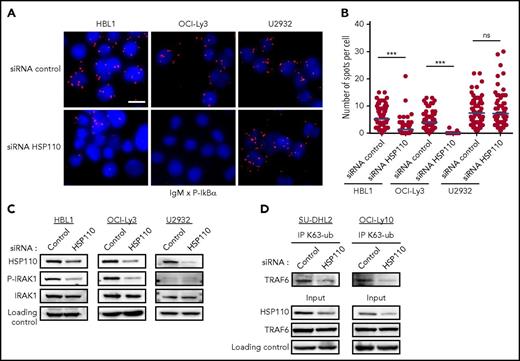
Recently, Louis M. Staudt pointed out that BCR components co-localized with MyD88, TLR9, and CARD11-BCL10-MALT1 (CBM) complex in endolysosomes, where they activate the NF-κB pathway (Louis M. Staudt, oral communication, American Association for Cancer Research Annual Meeting, 3 April 2017). To obtain more insights on the stage of the NF-κB pathway at which HSP110 interferes, we looked at the IgM-MyD88 complex formation when HSP110 is downregulated. By using PLA, we detected close proximity of IgM and p-IκB in the cytosol of ABC-DLBCL control cells (Figure 3A-B). Upon siRNA-mediated HSP110 down-expression, IgM and p-IκB proximity was strongly reduced in the cell lines bearing dual MyD88 and CBM mutations (HBL1 and OCI-Ly3), but not in the U2932 cell line, which is devoid of these mutations. This suggested that HSP110 could play a role in the IgM-MyD88 complex. We sought to confirm this hypothesis by studying the effect of HSP110 depletion in the IRAK1-TRAF6 complex formation immediately downstream of IgM and MyD88. Upon HSP110 downregulation, we found that IRAK1 was less phosphorylated in HBL1 and OCI-Ly3 (but not in U2932) (Figure 3C), and p-IRAK1 was less bound to TRAF6 (supplemental Figure 1D). Under normal IRAK1-TRAF6 complex formation, the auto-K63 ubiquitination of TRAF6 seems to enable the recruitment and activation of downstream signaling.29 Here, this TRAF6 K63 ubiquitination was also decreased (Figure 3D). These data suggest that HSP110 could intervene upstream of IRAK1-TRAF6 complex formation at an early stage of the NF-κB signaling pathway when IgM and MyD88 co-localize.
HSP110 intervenes upstream of the IRAK1-TRAF6 complex formation. (A) In cellulo interaction of IgM with p-IκBα was determined in HBL1, OCI-Ly3, and U2932 by Duolink technology after transfection with an siRNA targeting HSP110 or a control siRNA (n = 3; 1 representative image is shown). Scale bar represents 20 μm. (B) Quantitation of IgM–p-IκBα spots per cell determined by Duolink technology as in (A). (C) Immunoblot analysis of HSP110, p-IRAK1, and total IRAK1 in HBL1, OCI-Ly3, and U2932 cells at 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control (n = 3). (D) Immunoprecipitation of Lysine 63–linked polyubiquitin chain (K63-ub) in SU-DHL2 and OCI-Ly10 cells 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA, followed by immunoblot using a TRAF6 antibody. Immunoblot of total cell extract (input) shows HSP110 and TRAF6 analysis (n = 3). ***P < .001.
HSP110 intervenes upstream of the IRAK1-TRAF6 complex formation. (A) In cellulo interaction of IgM with p-IκBα was determined in HBL1, OCI-Ly3, and U2932 by Duolink technology after transfection with an siRNA targeting HSP110 or a control siRNA (n = 3; 1 representative image is shown). Scale bar represents 20 μm. (B) Quantitation of IgM–p-IκBα spots per cell determined by Duolink technology as in (A). (C) Immunoblot analysis of HSP110, p-IRAK1, and total IRAK1 in HBL1, OCI-Ly3, and U2932 cells at 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA. β-actin served as a loading control (n = 3). (D) Immunoprecipitation of Lysine 63–linked polyubiquitin chain (K63-ub) in SU-DHL2 and OCI-Ly10 cells 48 hours after transfection with an siRNA targeting HSP110 or a control siRNA, followed by immunoblot using a TRAF6 antibody. Immunoblot of total cell extract (input) shows HSP110 and TRAF6 analysis (n = 3). ***P < .001.
HSP110 enhances MyD88-mediated NF-κB signaling through direct interactions
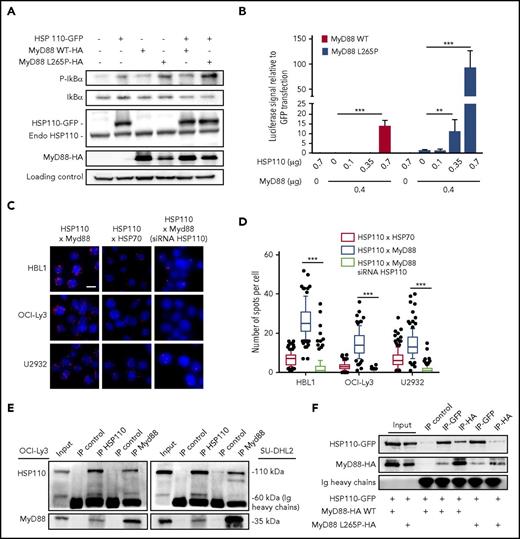
MyD88 activation upon BCR-TLR complex stimulation of B cells leads to NF-κB signaling and subsequent proliferation (Louis M. Staudt, oral communication, American Association for Cancer Research annual meeting, 3 April 2017). Furthermore, oncogenically active MyD88 in 30% of patients with ABC-DLBCL accounts for spontaneous assembling of the IRAK1-IRAK4 complex, leading to a strong and sustained NF-κB signal. Therefore, we hypothesized that HSP110 could enhance MyD88-mediated NF-κB signaling. We overexpressed HSP110 together with WT MyD88 or with the most common mutated form of MyD88 (MyD88 L265P) in a Burkitt lymphoma cell line (BJAB), that in contrast to ABC-DLBCL cells, is easy to transfect and does not bear either the MyD88 mutation or NF-κB hyperactivation. As shown in Figure 4A and supplemental Figure 1E, HSP110, MyD88, and MyD88 L265P alone enhanced IκB phosphorylation. As expected, MyD88 L265P promoted a stronger phosphorylation,23 which was enhanced by HSP110 overexpression.
HSP110 interacts with MyD88 to enhance NF-κB signaling. (A) Immunoblot analysis of p-IκB, IκB, HSP110, and MyD88 in Burkitt lymphoma (BJAB) cells 48 hours after transfection with plasmids coding for HSP110-GFP, and/or MyD88 WT, or MyD88 L265P. HSC70 served as a loading control (n = 3). (B) Luciferase reporter experiment in HEK293T cells. Cells co-transfected with GFP reporter (internal control) and NF-κB luciferase reporter plasmid with or without increasing concentrations of untagged HSP110 and with or without 0.4 μg of Myd88 L265P or Myd88 WT plasmids (n = 3). Shown are relative luciferase values (luciferase:GFP ratio). Data are presented as the mean ± standard deviation. (C) In cellulo interaction of HSP110 with MyD88 was determined in HBL1, OCI-Ly3, and U2932 by Duolink technology in the presence or not of siRNA HSP110. HSP70 was used as a positive control for HSP110 interaction (1 representative image is shown for each condition). Scale bar represents 20 μm. (D) Quantitation of HSP110-HSP70 and HSP110-MyD88 spots per cell determined by Duolink technology as in (A). Boxplots depict the mean (line), the 25-75 percentiles (box) and the 10-90 percentiles (whiskers). (E) Immunoprecipitation (IP) of HSP110 and MyD88 in SU-DHL2 and OCI-Ly3 cells followed by immunoblot using an anti-MyD88 and anti-HSP110 antibody. A nonrelevant antibody was used as a control (IP control) (n = 3). (F) Immunoprecipitation of HSP110-GFP (IP-GFP) and MyD88-HA (IP-HA) in HEK293 cells 48 hours after transfection with plasmids coding for HSP110-GFP with or without MyD88-HA L265P (0.7 µg) or MyD88-HA WT (0.7 µg) (n = 3). Endo, endogenous. **P < .01; ***P < .001.
HSP110 interacts with MyD88 to enhance NF-κB signaling. (A) Immunoblot analysis of p-IκB, IκB, HSP110, and MyD88 in Burkitt lymphoma (BJAB) cells 48 hours after transfection with plasmids coding for HSP110-GFP, and/or MyD88 WT, or MyD88 L265P. HSC70 served as a loading control (n = 3). (B) Luciferase reporter experiment in HEK293T cells. Cells co-transfected with GFP reporter (internal control) and NF-κB luciferase reporter plasmid with or without increasing concentrations of untagged HSP110 and with or without 0.4 μg of Myd88 L265P or Myd88 WT plasmids (n = 3). Shown are relative luciferase values (luciferase:GFP ratio). Data are presented as the mean ± standard deviation. (C) In cellulo interaction of HSP110 with MyD88 was determined in HBL1, OCI-Ly3, and U2932 by Duolink technology in the presence or not of siRNA HSP110. HSP70 was used as a positive control for HSP110 interaction (1 representative image is shown for each condition). Scale bar represents 20 μm. (D) Quantitation of HSP110-HSP70 and HSP110-MyD88 spots per cell determined by Duolink technology as in (A). Boxplots depict the mean (line), the 25-75 percentiles (box) and the 10-90 percentiles (whiskers). (E) Immunoprecipitation (IP) of HSP110 and MyD88 in SU-DHL2 and OCI-Ly3 cells followed by immunoblot using an anti-MyD88 and anti-HSP110 antibody. A nonrelevant antibody was used as a control (IP control) (n = 3). (F) Immunoprecipitation of HSP110-GFP (IP-GFP) and MyD88-HA (IP-HA) in HEK293 cells 48 hours after transfection with plasmids coding for HSP110-GFP with or without MyD88-HA L265P (0.7 µg) or MyD88-HA WT (0.7 µg) (n = 3). Endo, endogenous. **P < .01; ***P < .001.
To functionally test whether HSP110 was able to promote NF-κB target gene transcription in an MyD88-dependent manner, we used a luciferase reporter gene driven by an NF-κB promoter. HEK293 cells were co-transfected with increasing amounts of HSP110, with or without MyD88 WT or MyD88 L265P. We observed an HSP110 dose-dependent induction of luciferase activity (Figure 4B). Strikingly, whereas MyD88 L265P alone was able to promote this activity, the addition of HSP110 induced a 100-fold increase at the maximal concentration (Figure 4B). Overall, these data show that HSP110 enhances both MyD88 WT and mutated signaling. This suggests that HSP110 may interact directly with MyD88 to reinforce its action.
As shown in Figure 4C-D, HSP110 and MyD88 co-localized in ABC-DLBCL cells and associated in cellulo. Of note, in all 3 tested cell lines, HSP110-MyD88 binding spots were more abundant than HSP110-HSP70 binding spots, used here as a positive control because HSP70 is a well-known partner of HSP110. No close proximity of HSP70 and MyD88 was observed (data not shown). This interaction was confirmed by co-immunoprecipitation experiments in OCI-Ly3 and SU-DHL2 (Figure 4E). Because both cell lines expressed a mutated form of MyD88, we wondered whether HSP110 could also bind to the WT form. Co-immunoprecipitation experiments with HSP110 and MyD88 WT in HEK293 cells showed similar binding capacities (Figure 4F).
HSP110 stabilizes MyD88
Expression of increasing amounts of HSP110 in HEK293 cells induced a concomitant increase in the amount of MyD88 protein (Figure 5A-B; supplemental Figure 2A) for both L265P and WT MyD88. This phenomenon was not the result of transcriptional control of the MyD88 gene because MyD88 mRNA content did not increase (supplemental Figure 2B). Furthermore, expression of the HSP110 dominant negative mutant HSP110DE9 (known to inhibit HSP110 WT at a 1:1 ratio17 ) inhibited the expression of MyD88 (Figure 5A-B). Expression of an HSP110 mutant deficient in its nuclear localization signal also increased MyD88 protein expression, suggesting that HSP110 acts directly on MyD88 in the cytosol (Figure 5C). The study of MyD88 half-life in the presence of the protein synthesis inhibitor cycloheximide showed that HSP110 stabilizes MyD88 WT and MyD88 L265P in a similar way (Figure 5D). These observations were confirmed in HBL1 and OCI-Ly3 because HSP110 down expression by specific siRNA reduced the expression of MyD88 but not in U2932, which is in keeping with the previous results (Figure 5E). Finally, in the presence of the proteasome inhibitor MG132, the MyD88 level was restored, suggesting that HSP110 stabilized MyD88 by interfering with its proteasomal degradation (Figure 5F).
HSP110 stabilizes MyD88. Immunoblot analysis of HSP110-GFP, total HSP110, (A) MyD88 L265P, or (B) MyD88 WT 48 hours after transfection with plasmids coding for MyD88 L265P (0.7 µg) together with control GFP, HSP110 DE9GFP, or increasing concentrations of HSP110-GFP in HEK293 cells (n = 3). HSC70 served as a loading control. (C) Immunoblot analysis of HSP110-GFP and MyD88 L265P at 48 hours after transfection with plasmids coding for MyD88 L265P (0.5 μg) together with control GFP, increasing concentrations of the cytoplasmic-only mutant of HSP110 (HSP110 deficient in the nuclear localization signal [HSP110-NLS]) in HEK293 cells (n = 3). (D) Immunoblot analysis of HSP110-GFP, total HSP110, MyD88 L265P, or MyD88 WT in HEK293 cells treated with cycloheximide (CHX; 100 μg/mL) during the indicated times, 24 hours after transfection with control-GFP or HSP110-GFP plasmids. HSC70 served as a loading control. Lower panel shows the MyD88 immunoblot intensity from the above experiment. (E) Immunoblot analysis of HSP110 and MyD88 in HBL1, OCI-Ly3, and U2932 60 hours after transfection by a universal control siRNA or with an siRNA that targets HSP110 (n = 3). conc, concentration. (F) Immunoblot analysis of total HSP110 and MyD88 in HBL1 transfected as in (E) and treated or not with MG132 (10 μM) during the last 3 or 5 hours of the culture (n = 3).
HSP110 stabilizes MyD88. Immunoblot analysis of HSP110-GFP, total HSP110, (A) MyD88 L265P, or (B) MyD88 WT 48 hours after transfection with plasmids coding for MyD88 L265P (0.7 µg) together with control GFP, HSP110 DE9GFP, or increasing concentrations of HSP110-GFP in HEK293 cells (n = 3). HSC70 served as a loading control. (C) Immunoblot analysis of HSP110-GFP and MyD88 L265P at 48 hours after transfection with plasmids coding for MyD88 L265P (0.5 μg) together with control GFP, increasing concentrations of the cytoplasmic-only mutant of HSP110 (HSP110 deficient in the nuclear localization signal [HSP110-NLS]) in HEK293 cells (n = 3). (D) Immunoblot analysis of HSP110-GFP, total HSP110, MyD88 L265P, or MyD88 WT in HEK293 cells treated with cycloheximide (CHX; 100 μg/mL) during the indicated times, 24 hours after transfection with control-GFP or HSP110-GFP plasmids. HSC70 served as a loading control. Lower panel shows the MyD88 immunoblot intensity from the above experiment. (E) Immunoblot analysis of HSP110 and MyD88 in HBL1, OCI-Ly3, and U2932 60 hours after transfection by a universal control siRNA or with an siRNA that targets HSP110 (n = 3). conc, concentration. (F) Immunoblot analysis of total HSP110 and MyD88 in HBL1 transfected as in (E) and treated or not with MG132 (10 μM) during the last 3 or 5 hours of the culture (n = 3).
HSP110 is highly expressed in ABC-DLBCL patients and correlates with the amount of MyD88
Strong HSP110 expression has been found in samples from patients with different cancers, including those with GC-DLBCL. To get clinical insight on the role of HSP110 in ABC-DLBCL, we measured HSP110 mRNA levels in purified B cells from a cohort of patients with GC-DLBCL (n = 6) (as a positive control for HSP110), a cohort of patients with ABC-DLBCL (n = 9), and samples from nonmalignant reactive lymph nodes (n = 4). As for the GC-DLBCL samples, we observed stronger although heterogeneous HSP110 mRNA expression in ABC-DLBCL in comparison with reactive lymph node samples (a 21-fold increase; P = .009) (supplemental Figure 3A). This elevated HSP110 expression was then confirmed by IHC in biopsies from 35 patients with the ABC subtype (Figure 6A-B), as well as in biopsies from 7 patients with the GC subtype (provided for comparison) (supplemental Figure 3B). In all cases, more than 80% of cells were positive, with a score 3 to 4 for the staining intensity of cells from 57% of patients with the ABC subtype. Interestingly, although HSP110 was always present in the cytosol, frequent nuclear localization was also observed. HSP110 staining in reactive lymph nodes (n = 5) was limited to the GC, confirming its association with B-cell activation. We confirmed the HSP110 and MyD88 association in 2 ABC-DLBCL biopsies cells by PLA (Figure 6C). MyD88 expression was present in more than 80% of cells from this cohort with 3 levels of intensity (supplemental Figure 3C). Strong MyD88 expression was more frequent for biopsies with high HSP110 intensity (a score >2; 37%) compared with biopsies with low HSP110 intensity (score <2; 15%), but it was not statistically significant (supplemental Figure 3D). To get a more quantitative insight into MyD88-HSP110 expression, we used another cohort of patients with ABC-DLBCL; we were able to extract proteins from their tumor biopsies and we observed that the level of HSP110 correlated significantly with that of MyD88 (R2 = 0.7577 [n = 13]) (Figure 6D-E) and with that of p-IκBα (R2 = 0.5781 [n = 9]; supplemental Figure 3E). These patient data support our results showing that HSP110 plays a role in MyD88 protein expression in ABC-DLBCL.
High expression of HSP110 correlates with MyD88 expression in patients. (A) Representative images of reactive lymph nodes (n = 5) and ABC-DLBCL (n = 35) patients’ lymph node sections stained with HSP110 antibody by IHC. The 2 original magnifications are ×2.8 for A and C (scale bar represents 425 μm) and ×40 for B and D (scale bar represents 30 μm). Representative images of 4 HSP110 intensities are shown beside sections from patients with ABC-DLBCL (original magnification ×20; scale bar represents 60 μm). (B) Schematic representation of HSP110 IHC intensity in the cohort of patients analyzed (n = number of patients with each intensity). (C) Representative image of in cellulo interaction of HSP110 with MyD88 (determined by Duolink technology) in lymph node sections from 2 patients (scale bar represents 40 μm [upper panel] or 20 μm [lower panel]). (D) Immunoblot analysis of HSP110 and MyD88 in biopsies from 13 patients with ABC-DLBCL. HSC70 served as a loading control. (E) HSP110 immunoblot intensity relative to MyD88 from (C).
High expression of HSP110 correlates with MyD88 expression in patients. (A) Representative images of reactive lymph nodes (n = 5) and ABC-DLBCL (n = 35) patients’ lymph node sections stained with HSP110 antibody by IHC. The 2 original magnifications are ×2.8 for A and C (scale bar represents 425 μm) and ×40 for B and D (scale bar represents 30 μm). Representative images of 4 HSP110 intensities are shown beside sections from patients with ABC-DLBCL (original magnification ×20; scale bar represents 60 μm). (B) Schematic representation of HSP110 IHC intensity in the cohort of patients analyzed (n = number of patients with each intensity). (C) Representative image of in cellulo interaction of HSP110 with MyD88 (determined by Duolink technology) in lymph node sections from 2 patients (scale bar represents 40 μm [upper panel] or 20 μm [lower panel]). (D) Immunoblot analysis of HSP110 and MyD88 in biopsies from 13 patients with ABC-DLBCL. HSC70 served as a loading control. (E) HSP110 immunoblot intensity relative to MyD88 from (C).
Discussion
Genomic and functional investigations have shown that ABC-DLBCL tumors are driven by either chronic BCR and/or TLR9-MyD88 signaling in contrast to GC-DLBCL. Many downstream effectors are therefore activated, leading to sustained NF-κB activation, which ABC-DLBCL tumors rely on for survival. There is important genetic heterogeneity in ABC-DLBCL tumors that accounts for this chronic BCR and MyD88 signaling, because mutations in CD79a/b, MyD88, CARD11, and A20 have been detected in patients’ samples. In particular, 29% of tumors harbor a single nucleotide variant that changes a leucine residue at position 265 of the MyD88 coding region to a proline, leading to a mutant protein (MyD88 L265P) that spontaneously assembles with IRAK1 and IRAK4 into so-called myddosomes.23 We showed here that HSP110 directly binds to MyD88, independent of L265P mutation, and stabilizes both protein forms leading to the amplification of NF-κB signaling and to subsequent improved survival in ABC-DLBCL lines. We also observed a correlation between HSP110 and MyD88 levels in patients, thus confirming our in vitro results and suggesting that HSP110 could be a potential target for lymphoma therapy in the future. This effect seems specific for HSP110 because HSP27 and HSP70 do not alter NF-κB signaling. In contrast, HSP90 inhibition alters the BCR-dependent NF-κB pathway because HSP90 is a key component of the BCR signalosome.3 The involvement of HSP90 in MyD88 signaling, alone or in association with HSP110, deserves further investigation.
As an adaptor of Toll-like receptor signaling, MyD88 is regulated by endogenous mechanisms as well as pathogens. In macrophages, the E3 ubiquitin ligase Nrdp1 directly binds and polyubiquitinates MyD88, leading to its proteasomal degradation, thus limiting the overproduction of proinflammatory cytokines.30 In the same way, TGF-β1 drives MyD88 degradation through the Smad6-Smurf pathway.31 Pathogens have many ways to escape immune surveillance. For example, they can drive MyD88 through the Ub-proteasome pathway as shown for Kaposi’s sarcoma–associated herpesvirus.32 Because an adequate inflammatory response to pathogens must be balanced, molecular machinery that opposes MyD88 degradation must exist, and our work suggests that HSP110 could be involved because it promotes MyD88 stability.
HSPs have multiple roles in cancer cells that relate to their chaperone function; among these, stabilization of mutated or overexpressed proteins is common. In addition, proliferation- or survival-dependent activating mutations of signaling molecules show increased dependence on HSPs.3,33,34 In GC BCL/BCL6+ DLBCL, HSP90 interacts with the oncogene BC-6 and stabilizes BCL6 mRNA and protein.34 A significant positive correlation between HSP110 and BCL6 expression has also been found in GC-DLBCL cell lines and in patients with GC-DLBCL.18 In addition, in Burkitt lymphoma, HSP110 prevents degradation of the overexpressed oncogene c-Myc.19 Therefore, through their multiple oncogene targets in lymphomas, HSPs, and particularly HSP110, seem to be crucial to their proliferation and survival, thus confirming the overall addiction of tumors to HSPs. HSP110 plays an important role in GC-DLBCL, ABC-DLBCL, and Burkitt lymphoma, in which it acts on different oncogenes frequently associated with these particular diseases.
Beyond ABC-DLBCL, the missense mutation of MyD88 has been described in other diseases such as benign monoclonal IgM gammopathy of undetermined significance (47%), Waldenström macroglobulinemia (in almost 100% of patients), and chronic lymphocytic leukemia (less than 10%).35,36 It is very likely that HSP110 could also have the same MyD88 stabilizing function in these diseases, although this hypothesis needs to be investigated.
Tumor microenvironment may influence DLBCL progression because enriched expression of T-cell and natural killer cell activation molecules and CD4+ T-cell tumor infiltration are associated with good prognosis.37-39 The ability of extracellular recombinant HSP110 to improve antigen presentation and T-cell responses has been described.40-43 But the question remains of whether the cell-intrinsic antitumor effects of HSP110 targeting will be offset by reductions in T-cell responses.
Recent reports suggest that p-IκB and/or MyD88 expression can be used as prognosis factors.44,45 Given that HSP110 expression shows a good positive correlation with both markers, we can hypothesize that HSP110 is associated with more aggressive clinical behavior.
HSP110 protein level was found to be low in normal nonactivated B cells outside the GC (Figure 6A), and the currently available HSP110 knockout model shows no alteration of B-cell number in homeostatic conditions (not shown). This information could be of particular interest for the rationale of HSP110 inhibition in DLBCL because the current R-CHOP–based regimen targets normal and pathologic B cells. In contrast, HSP110 inhibition, because of its overexpression in NHL may selectively target lymphoma cells but not naive normal B cells. If new therapies use lymphoma oncogenic cell signaling to selectively kill DLBCL cells in preference to normal B cells, HSP110 inhibition could represent a promising new approach. Of course future therapies will need to combine strategies (BTK inhibitors) to completely eradicate tumor cells because some remaining cells persist despite HSP110 down expression. Once available, HSP110-specific inhibitors will probably play a preponderant role in the therapeutic arsenal for ABC-DLBCL and beyond.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the fonds européen de développement régional(FEDER) for their support; the Plateforme de Cytométrie Dijon and Cellimap Dijon for technical support; Gisèle Froment, Didier Nègre, and Caroline Costa from the lentivectors production facility SFR BioSciences Lyon (UMS3444/US8); and Thierry Defrance (INSERM U1111, Lyon, France) and Karen Leroy (Paris Descartes University, Paris, France) for helpful discussions and support.
This work was supported by grants from the Ligue Nationale Contre le Cancer (G.J. and C.G.), the Conseil Regional de Bourgogne, the French National Research Agency under the Investissements d’Avenir Program (ANR-11-LABX-0021-01-LipSTIC Labex), the German Research Foundation CRC685 Immunotherapy (A.W. and O.-O.W.), and the Juniorprofessoren-Programm of the Federal State of Baden-Württemberg, Germany (A.W. and O.-O.W.).
Authorship
Contribution: C.B., E.V., L.M., C.C.-C., T.F., A.N.R.W., and G.J. designed the study and experiments; C.B., E.V., L.S., R.M., and O.-O.W. performed the experiments; E.V., L.M., C.C.-C., C.P., T.F., S.R., F.J., and A.N.R.W. provided samples and expertise; C.B., E.V., L.M., C.C.-C., T.F., F.J., A.N.R.W., C.G., and G.J. analyzed the experiments; and G.J. directed the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gaetan Jego, INSERM, UMR 1231, Université Bourgogne Franche-Comté, 7, Boulevard Jeanne d’Arc, 21079 Dijon, France; e-mail: gaetan.jego@u-bourgogne.fr.


![Figure 5. HSP110 stabilizes MyD88. Immunoblot analysis of HSP110-GFP, total HSP110, (A) MyD88 L265P, or (B) MyD88 WT 48 hours after transfection with plasmids coding for MyD88 L265P (0.7 µg) together with control GFP, HSP110 DE9GFP, or increasing concentrations of HSP110-GFP in HEK293 cells (n = 3). HSC70 served as a loading control. (C) Immunoblot analysis of HSP110-GFP and MyD88 L265P at 48 hours after transfection with plasmids coding for MyD88 L265P (0.5 μg) together with control GFP, increasing concentrations of the cytoplasmic-only mutant of HSP110 (HSP110 deficient in the nuclear localization signal [HSP110-NLS]) in HEK293 cells (n = 3). (D) Immunoblot analysis of HSP110-GFP, total HSP110, MyD88 L265P, or MyD88 WT in HEK293 cells treated with cycloheximide (CHX; 100 μg/mL) during the indicated times, 24 hours after transfection with control-GFP or HSP110-GFP plasmids. HSC70 served as a loading control. Lower panel shows the MyD88 immunoblot intensity from the above experiment. (E) Immunoblot analysis of HSP110 and MyD88 in HBL1, OCI-Ly3, and U2932 60 hours after transfection by a universal control siRNA or with an siRNA that targets HSP110 (n = 3). conc, concentration. (F) Immunoblot analysis of total HSP110 and MyD88 in HBL1 transfected as in (E) and treated or not with MG132 (10 μM) during the last 3 or 5 hours of the culture (n = 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/5/10.1182_blood-2017-12-819706/4/m_blood819706f5.jpeg?Expires=1769078765&Signature=bc8E9JpZKsnoCrfQlJud-alhWAxgkQrJQFWlfuObtyAE4aEDKwLEMT1QJeqBK5wAsh~2FMmdXSgBAWDIL-YKfw80Vg~I42g0qUJaZbzt8JDxRNIhqIuKWJ8oedWCxEXedf8N6fRvHtMbwURVMXuEMOHGgKTG3U8OoLjkGehE7AhLDVWc0bWAkj~mcjQZGgUtRG4XIsGILWw36Rc4dmVU4Ck8iBMm433OywiEZAlNE9JweglIQvn5QyuoovVfV7ByHYbeK-XQorSx9jPtZi9aAHrIBvDfcPnyN58VfabQzkYx7l3-q71qBrOahfxrnv9TY-XBZ-~cAMoVkPrnYRM9xg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. High expression of HSP110 correlates with MyD88 expression in patients. (A) Representative images of reactive lymph nodes (n = 5) and ABC-DLBCL (n = 35) patients’ lymph node sections stained with HSP110 antibody by IHC. The 2 original magnifications are ×2.8 for A and C (scale bar represents 425 μm) and ×40 for B and D (scale bar represents 30 μm). Representative images of 4 HSP110 intensities are shown beside sections from patients with ABC-DLBCL (original magnification ×20; scale bar represents 60 μm). (B) Schematic representation of HSP110 IHC intensity in the cohort of patients analyzed (n = number of patients with each intensity). (C) Representative image of in cellulo interaction of HSP110 with MyD88 (determined by Duolink technology) in lymph node sections from 2 patients (scale bar represents 40 μm [upper panel] or 20 μm [lower panel]). (D) Immunoblot analysis of HSP110 and MyD88 in biopsies from 13 patients with ABC-DLBCL. HSC70 served as a loading control. (E) HSP110 immunoblot intensity relative to MyD88 from (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/5/10.1182_blood-2017-12-819706/4/m_blood819706f6.jpeg?Expires=1769078765&Signature=tH1AUm2XsVC-r7xgjFtBDYCd3jDb-7TdABlkkfpNEAntNIPzGXRKCuMq4MXbMvTo~VCCJgvD7XfUd1QP-o2GpbV2OuRq6r70d2SGznyBOzjHfm8zrJ3wHHuFUO6bXN44qgjVETnTga-WLroTnEMwmEYn7Zdij6fblwfRHQ6eQaAZU-W9Lignkd~9JK22dLOuDX4zlYARbeLjUodJo2481lczYQPULvWCT-w54jivjewKUHGJiUaWT3Wjxu2HaUyU03z5amk7sVXpU83~Jf6riA2y1pdLc~3vUvYLvOrf7OGXFVjtq05kCP~px0YcTW3YM-plJvlEbJiONFABFp6z3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal