Key Points
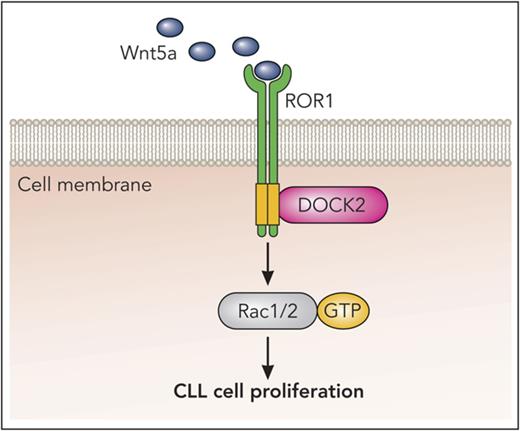
Wnt5a enhances activation of Rac1/2 by inducing ROR1 to interact with DOCK2.
ROR1-DOCK2 interaction contributes to Wnt5a-enhanced CLL cell proliferation.
Abstract
Receptor tyrosine kinase-like orphan receptor 1 (ROR1) is an oncoembryonic protein expressed on chronic lymphocytic leukemia (CLL) that can serve as a receptor for Wnt5a, which can promote leukemia cell migration, proliferation, and survival. We found Wnt5a could induce ROR1 to complex with DOCK2 (dedicator of cytokinesis 2) and induce activation of Rac1/2; these effects could be blocked by cirmtuzumab, a humanized anti-ROR1 monoclonal antibody. We find that silencing DOCK2 specifically impaired the capacity of Wnt5a to induce activation of Rac1/2 or enhance CLL cell proliferation. We generated truncated forms of ROR1 and found the cytoplasmic proline-rich domain (PRD) of ROR1 was required for Wnt5a to induce ROR1 to complex with DOCK2 and activate Rac1/2 in the CLL cell-line MEC1. We introduced single amino acid substitutions of proline (P) to alanine (A) in the ROR1-PRD at potential binding sites for the Src-homology 3 domain of DOCK2. In contrast to wild-type ROR1, or other ROR1 P→A variants, ROR1P808A was unable to recruit DOCK2 in response to Wnt5a. Moreover, unlike MEC1 cells transfected with wild-type ROR1 or ROR1 with P→A substitutions at positions 784, 826, or 841, MEC1 cells transfected to express ROR1P808A did not have a growth advantage over MEC1 cells that do not express ROR1. This study reveals that the recruitment of DOCK2 may be critical for the capacity of Wnt5a to enhance CLL proliferation, which may contribute to the observed increased tendency for disease progression in patients who have CLL cells that express high levels of ROR1.
Introduction
ROR1 (receptor tyrosine kinase-like orphan receptor 1) is an evolutionarily conserved, type I membrane protein that is expressed during embryogenesis, in which it plays a key role in skeletal and neural organogenesis.1-4 Expression of ROR1 attenuates during fetal development and, with few exceptions,5 is negligible on virtually all postpartum tissues.6 However, we and others have found that the leukemia cells of most patients with chronic lymphocytic leukemia (CLL) express ROR1,6-8 suggesting it may play a role in pathogenesis. Consistent with this notion are studies showing that expression of ROR1 can enhance disease progression in mouse models of this leukemia9 and in patients with CLL.10 However, the molecular mechanism or mechanisms for how ROR1 contributes disease progression in patients with CLL is largely unknown.
We found that ROR1 can serve as a receptor for Wnt5a,6 which prior studies showed could induce noncanonical Wnt signaling leading to enhanced leukemia cell migration and proliferation.11 Recently, we described that ROR1 could interact with hematopoietic lineage-specific protein 1 at P(841) in the proline-rich domain (PRD) of ROR1, and subsequently activate RhoA in CLL cells to enhance migration.12 We found the humanized monoclonal antibody (mAb) cirmtuzumab (UC-961)13 could inhibit these effects.11 However, the full mechanism or mechanisms whereby ROR1 promotes leukemia cell migration and proliferation is still not resolved.
DOCK2 (dedicator of cytokinesis 2) is a cytoplasmic protein member of the DOCK-A subfamily of guanine exchange factors (GEFs) specific for Rac1 and Rac2 (Rac1/2) that is expressed primarily in leukocytes.14,15 In structural analysis, DOCK proteins differ from other GEFs in that they do not possess the canonical tandem Dbl-homology and Pleckstrin-homology domains that elicit nucleotide exchange. Instead, DOCK2 possesses a DOCK homology region 2 domain, which mediates Rac activation by stabilizing Rac in its nucleotide-free state. Moreover, DOCK2 has a SH3 domain, which allows it to bind characteristic motifs (-P-X-X-P-) in the PRDs of other proteins.14-17 DOCK2 apparently plays a role in hematopoietic cell migration and proliferation.18-21 However, the role of DOCK2 in the pathogenesis of CLL is not known. Here, we report finding that DOCK2 is expressed in CLL B cells, in which it can associate with ROR1 in response to Wnt5a to activate Rac1/2 and promote CLL cell proliferation.
Materials and methods
Cell culture and CLL specimens
MEC1 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum and 1% penicillin/streptomycin and maintained at 37°C in a humidified atmosphere of 5% CO2. Media and supplements were purchased from Life Technologies (Carlsbad, CA). Blood samples were collected from patients with CLL at the Moores Cancer Center. Peripheral blood mononuclear cells were isolated by density centrifugation with Ficoll-Paque PLUS (GE Healthcare Life Sciences) and suspended in 90% fetal bovine serum (Omega Scientific) and 10% dimethyl sulfoxide (Sigma-Aldrich) for viable storage in liquid nitrogen.
Immunoprecipitation analysis
Immunoprecipitation analysis was performed as described.9,12 Cells were lysed in a buffer containing 1% Nonidet P-40, 10 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, and 1 mM EDTA with protease inhibitors (Roche). The lysates were cleared by centrifugation at 16 000g for 15 minutes. Immune precipitates were isolated using protein A agarose beads, followed by immunoblot or mass spectrometry analysis. Antibodies for immune precipitation were as follows: the anti-ROR1 antibodies (cirmtuzumab [UC-961] and 4A5) were generated in our laboratory; the anti-DOCK2 antibody was obtained from Santa Cruz Biotechnology.
Immunoblot analysis
Western blot analysis was performed as described.9,12 Equal amounts of total protein from each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membrane. Western blot analysis was performed using primary mAbs specific for ROR1 (Cell Signaling), DOCK2 (Santa Cruz), Rac1 (Cytoskeleton), Rac2 (Millipore), CrkL (Cell Signaling), or Vav (Abcam), which were detected using secondary antibodies conjugated with horseradish peroxidase (Cell Signaling Technology).
Nucleofection of siRNA
Human B Cell Nucleofector Kit for siRNA transfection was from Lonza. CLL cells or MEC1 cells (5 × 106) were suspended in 100 μL Nucleofector Solution with siRNA (Life Technologies) and transfected with the Nucleofector II device (program U-015). The transfected cells were cultured in 12-well plates in complete medium for 72 hours and then subjected to immunoblot analysis.
Cell proliferation assay
The CLL cell proliferation assay was performed as described.11,22 CLL cells were labeled by carboxyfluorescein diacetate succinimidyl ester (Life Technologies) and plated at 1.5 × 106/well/mL in a 24-well tray on a layer of irradiated HeLaCD154 cells (80 Gy) at a CLL/HeLaCD154 cell ratio of 15:1 in complete RPMI 1640 medium supplemented with 5 ng/mL recombinant human IL-4 (R&D Systems), and 15 ng/mL recombinant human IL-10 (R&D Systems). Wnt5a (200 ng/mL, R&D Systems) was added, as indicated in the text. Carboxyfluorescein diacetate succinimidyl ester-labeled CLL cells were analyzed by flow cytometry; Modfit LT software (version 3.0, Verity Software House) was used for analysis of cell proliferation.
The MEC1 cell proliferation assay also was performed as described.11 MEC1, MEC1-ROR1, or MEC1 transfected with truncated or mutant forms of ROR1 were plated in 96-well plates at 2 × 103 cells/well in complete RPMI 1640 medium. A total of 10 μL CCK-8 (Dojindo Molecular Technologies) was added to each well at different points, and the cells were cultured for an additional 3 hours. The absorbance at 450 nm was measured to calculate the numbers of viable cells in each well.
Cell migration assay
The cell migration assay was performed as described.12,23 Briefly, A total of 5 × 105 cells were washed twice, cultured overnight in serum-free medium, and then treated with or without Wnt5a (200 ng/mL) for 30 minutes. The cells then were placed into the top chamber of a Transwell culture polycarbonate insert with 6.5-mm diameter and 5-μm pore size (Corning). Cells were incubated for 2 hours in serum-free medium at 37°C and 5% CO2 and the migration toward chemokine (CXCL12, 200 ng/mL) was analyzed by flow cytometry. The percentage of migrating cells was calculated as the number of migrated cells in response to chemokine divided by the total number of input cells.
Rac1/2 activation assay
Rac1 or Rac2 activation assay reagents were purchased from Cytoskeleton or Millipore, respectively, and used per the manufacturer’s instructions. Briefly, guanosine triphosphate (GTP)-bound active Rac1/2 was pulled down with p21-activated kinase–p21-binding domain beads for 1 hour at 4°C, and then subjected to immunoblot analysis. Immunoblots of whole-cell lysates were used to assess for total Rac1/2. The integrated optical density (IOD) of bands was evaluated by densitometry and analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics).
RhoA activation assay
RhoA activation assay reagents were purchased from Cytoskeleton and used per the manufacturer’s instructions. Briefly, GTP-bound active RhoA was pulled down with Rhotekin-RBD beads for 1 hour at 4°C and then subjected to immunoblot analysis. Immunoblots of whole-cell lysates were used to assess for total RhoA. The IOD of bands was evaluated by densitometry and analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics).
Statistical analysis
Data are presented as mean ± SD. Differences between 2 groups were determined by unpaired 2-tailed Student t test. P values of less than .05 were considered significant. Analysis for significance was performed with GraphPad Prism 6.0 (GraphPad Software Inc.). Longitudinal mixed effects model (nlme in R) analysis was performed for the differences in growth rate among multiple groups of MEC1 cells, identifying experiment as a random effect and nesting day within experiment.
Study approval
Blood samples were collected from patients at the Moores Cancer Center who satisfied diagnostic criteria for CLL and who provided written, informed consent, in compliance with the Declaration of Helsinki and the Institutional Review Board of the University of California, San Diego (Institutional Review Board approval number 080918).
Results
Wnt5a induces ROR1 to associate with DOCK2 in primary CLL cells
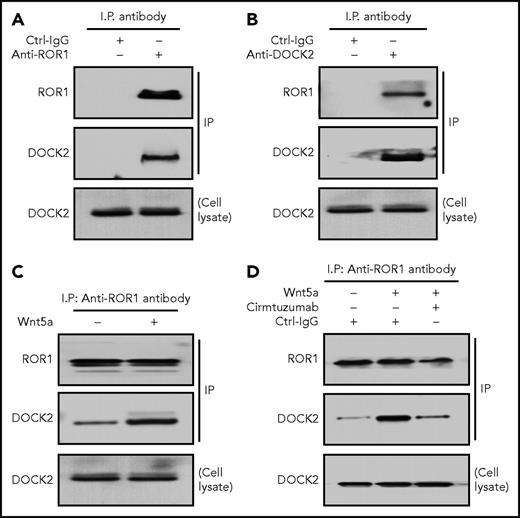
We performed mass spectrometry-based proteomic analysis on anti-ROR1 immune-precipitates from CLL cell lysates and detected DOCK2, in addition to ROR1 (supplemental Figure 1, available on the Blood Web site). Immunoblot analysis of anti-ROR1 or anti-DOCK2 immune precipitates confirmed that ROR1 associated with DOCK2 in freshly isolated primary CLL cells (Figure 1A-B). However, this association was less apparent in lysates of CLL cells cultured overnight in Wnt5a-deficient media unless the CLL cells were treated for 30 minutes with exogenous Wnt5a (Figure 1C). We also treated the CLL cells with cirmtuzumab, a humanized anti-ROR1 mAb that binds a different epitope of ROR1 than that recognized by 4A5, the anti-ROR1 mAb used for generating immune precipitates. Treatment of the CLL cells with cirmtuzumab, but not a control immunoglobulin G (IgG) of irrelevant specificity, inhibited the capacity of Wnt5a to induce ROR1 to associate with DOCK2 in freshly isolated CLL cells (Figure 1D).
Association of ROR1 with DOCK2 in primary CLL cells. (A) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip or control IgG (Ctrl-IgG) ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients); the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (B) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients); the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (C) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip, using lysates prepared from overnight, serum-starved primary CLL cells (representative of 3 patients) that subsequently were treated for 30 minutes without (−) or with (+) Wnt5a (100 ng/mL), as indicated on the top; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (D) Immunoblot analysis of anti-ROR1 (4A5) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients) that had been treated with cirmtuzumab (10 μg/mL), without (−) or with (+) Wnt5a (100 ng/mL), as indicated on the top; membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL treated with cirmtuzumab and probed with anti-DOCK2 mAb is provided in the bottom panel.
Association of ROR1 with DOCK2 in primary CLL cells. (A) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip or control IgG (Ctrl-IgG) ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients); the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (B) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients); the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (C) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip, using lysates prepared from overnight, serum-starved primary CLL cells (representative of 3 patients) that subsequently were treated for 30 minutes without (−) or with (+) Wnt5a (100 ng/mL), as indicated on the top; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL is provided in the bottom panel. (D) Immunoblot analysis of anti-ROR1 (4A5) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from freshly isolated primary CLL cells (representative of 3 patients) that had been treated with cirmtuzumab (10 μg/mL), without (−) or with (+) Wnt5a (100 ng/mL), as indicated on the top; membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL treated with cirmtuzumab and probed with anti-DOCK2 mAb is provided in the bottom panel.
DOCK2 functions in Wnt5a-enhanced CLL cell proliferation
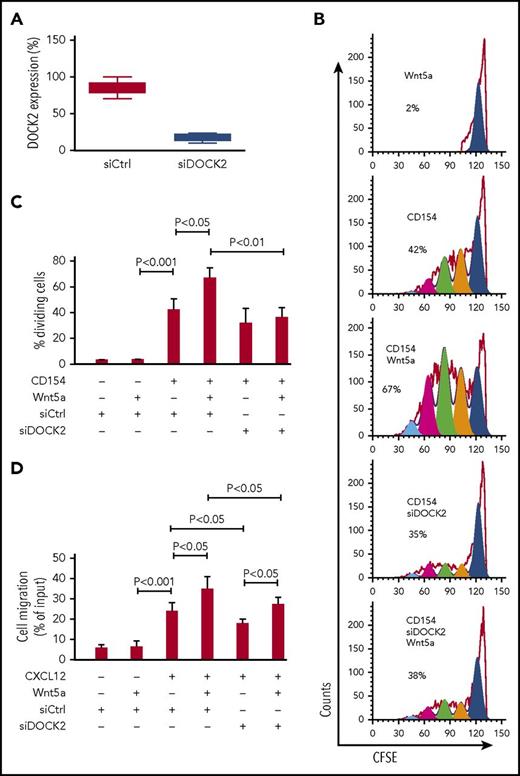
Prior studies demonstrated that Wnt5a could enhance proliferation of CLL cells cocultured with accessory cells (HeLa) that express CD154 in media supplemented with exogenous IL-4 and IL-10.11,12 Wnt5a also could enhance chemokine-directed CLL-cell migration.11,12 These effects of Wnt5a on CLL cells could be inhibited by cirmtuzumab.11,12 In this study, we performed siRNA-mediated silencing of DOCK2 (siDOCK2) in CLL cells that use either unmutated (U) or mutated (M) immunoglobulin heavy-chain variable region genes without acutely affecting cell viability relative to that of cells transfected with control siRNA (siCtrl) (Figure 2A; supplemental Figure 2). Transfection with siDOCK2 effectively reduced the amount of DOCK2 protein to ≤ 25% of that observed in siCtrl-treated CLL cells by immunoblot analyses (Figure 2A). Whereas Wnt5a could enhance proliferation of either U or M CLL cells transfected with siCtrl, Wnt5a did not enhance proliferation of CLL cells transfected with siDOCK2, (Figures 2B-C; supplemental Figure 3). Moreover, ROR1-positive (ROR1Pos) CLL cells silenced for DOCK2 behaved similarly to exogenous Wnt5a, as did ROR1-negative (ROR1Neg) CLL cells, which did not have enhanced proliferation or migration in response to Wnt5a and were unaffected by treatment with siDOCK2 (supplemental Figure 4).10 In contrast, Wnt5a could enhance chemokine-directed migration of ROR1Pos CLL cells treated with siDOCK2 (Figure 2D), indicating that silencing DOCK2 affected the capacity of Wnt5a to enhance proliferation of ROR1Pos CLL cells, but did not affect the capacity of Wnt5a to enhance chemokine-directed migration.
DOCK2 factors in the capacity of Wnt5a to enhance CLL proliferation. (A) CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL) transfected with control siRNA or siRNA targeting DOCK2 and examined 72 hours later by western blot. Data are shown as mean ± SD (n = 6). Cell viability was examined more than 80% both in control or DOCK2-siRNA transfected cells. (B) Fluorescence of CLL cells stained with carboxyfluorescein diacetate succinimidyl ester and treated with CD154, without (−) or with (+) Wnt5a. The percentage of dividing cells is indicated in each histogram. (C) Mean proportions of dividing CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL) under conditions indicated at the bottom. P < .05; P < .01; P < .001, as assessed by 2-tailed Student t test. (D) CLL cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2. Cell viability was higher than 80% both in control or DOCK2-siRNA transfected cells. CLL cell migration in response to CXCL12 (200 ng/mL) was assessed without (−) or with (+) addition of exogenous Wnt5a (200 ng/mL), as indicated at the bottom. Data are shown as mean ± SD from 3 independent experiments of CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL). P < .05; P < .001, as assessed by 2-tailed Student t test.
DOCK2 factors in the capacity of Wnt5a to enhance CLL proliferation. (A) CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL) transfected with control siRNA or siRNA targeting DOCK2 and examined 72 hours later by western blot. Data are shown as mean ± SD (n = 6). Cell viability was examined more than 80% both in control or DOCK2-siRNA transfected cells. (B) Fluorescence of CLL cells stained with carboxyfluorescein diacetate succinimidyl ester and treated with CD154, without (−) or with (+) Wnt5a. The percentage of dividing cells is indicated in each histogram. (C) Mean proportions of dividing CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL) under conditions indicated at the bottom. P < .05; P < .01; P < .001, as assessed by 2-tailed Student t test. (D) CLL cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2. Cell viability was higher than 80% both in control or DOCK2-siRNA transfected cells. CLL cell migration in response to CXCL12 (200 ng/mL) was assessed without (−) or with (+) addition of exogenous Wnt5a (200 ng/mL), as indicated at the bottom. Data are shown as mean ± SD from 3 independent experiments of CLL cells from each of 6 patients (3 U-CLL and 3 M-CLL). P < .05; P < .001, as assessed by 2-tailed Student t test.
ROR1 associates with DOCK2 in MEC1-ROR1 cells
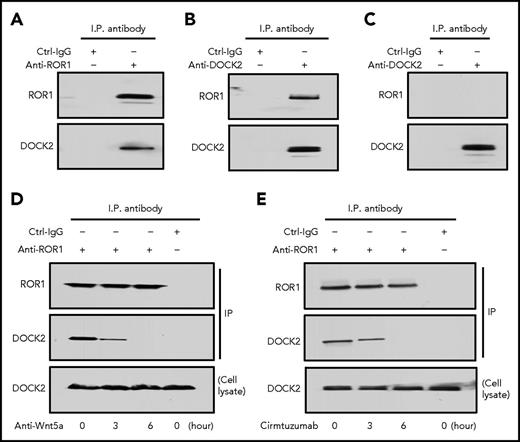
MEC1 cells are derived from CLL cells and have been used a model system for studying this leukemia.24 Prior studies found that MEC1 does not express ROR1, but does express high levels of Wnt5a, obviating the addition of exogenous Wnt5a to the culture media.11 Transfection of MEC1 cells with an expression vector encoding ROR1 generated MEC1-ROR1, which expressed high levels of surface ROR1. Immunoblot analysis of anti-ROR1 or anti-DOCK2 immune precipitates revealed that ROR1 associates with DOCK2 in MEC1-ROR1 cells (Figure 3A-B). As expected, ROR1 was not observed in anti-DOCK2 immune precipitates of lysates of MEC1 cells (Figure 3C). Treatment of MEC1-ROR1 cells for at least 3 hours with a neutralizing antibody specific for Wnt5a caused ROR1 to dissociate from DOCK2, which no longer associated with ROR1 after 6 hours of treatment (Figure 3D). Treatment of MEC1-ROR1 cells with cirmtuzmab for at least 3 hours also disrupted the ROR1-DOCK2 complex, which was disrupted entirely after 6 hours of treatment (Figure 3E). DOCK2 also could associate with other cytosolic proteins, such as CrkL or Vav, in leukemia cell lines.25 In agreement with these previous findings, we also found that DOCK2 associated with CrkL or Vav in the CLL cell line MEC1, which does not express ROR1 (supplemental Figure 5). However, the association of CrkL or Vav with DOCK2 in MEC1-ROR1 was less apparent than in MEC1 cells, suggesting that ROR1 may compete with CrkL or Vav for binding to DOCK2 (supplemental Figure 5).
ROR1 in MEC1-ROR1 cells associates with DOCK2. (A) Immunoblot analysis of anti-ROR1 (cirmtuzumab) immunoprecipitation (ip) or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (B) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells; membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (C) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from ROR1-negative cell line, MEC1; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (D) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells that had been treated with Wnt5a neutralizing antibody (2 μg/mL; R & D) for the times indicated at the bottom (in hours); membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the MEC1-ROR1 cells treated with Wnt5a neutralizing antibody and probed with anti-DOCK2 mAb is provided in the bottom panel. (E) Immunoblot analysis of anti-ROR1 (4A5) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells that had been treated with cirmtuzumab (10 μg/ml) for the times indicated at the bottom (in hours); membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL treated with cirmtuzumab and probed with anti-DOCK2 mAb is provided in the bottom panel.
ROR1 in MEC1-ROR1 cells associates with DOCK2. (A) Immunoblot analysis of anti-ROR1 (cirmtuzumab) immunoprecipitation (ip) or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (B) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells; membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (C) Immunoblot analysis of anti-DOCK2 ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from ROR1-negative cell line, MEC1; the membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (D) Immunoblot analysis of anti-ROR1 (cirmtuzumab) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells that had been treated with Wnt5a neutralizing antibody (2 μg/mL; R & D) for the times indicated at the bottom (in hours); membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the MEC1-ROR1 cells treated with Wnt5a neutralizing antibody and probed with anti-DOCK2 mAb is provided in the bottom panel. (E) Immunoblot analysis of anti-ROR1 (4A5) ip or Ctrl-IgG ip, as indicated at the top, using lysates prepared from MEC1-ROR1 cells that had been treated with cirmtuzumab (10 μg/ml) for the times indicated at the bottom (in hours); membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. An immunoblot of the whole-cell lysates of the CLL treated with cirmtuzumab and probed with anti-DOCK2 mAb is provided in the bottom panel.
DOCK2 contributes Wnt5a-induced activation of Rac1/2, but not RhoA
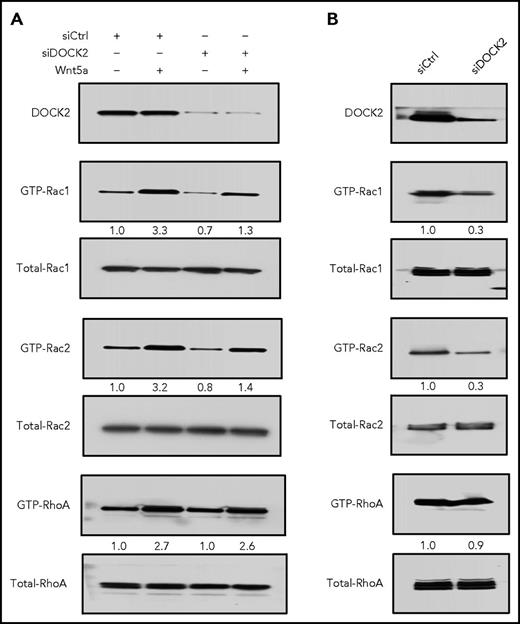
As noted in prior studies, Wnt5a enhanced activation of Rac1 and RhoA in primary CLL cells, as well as in MEC1-ROR1.11 We found that siDOCK2-silencing of DOCK2 in primary CLL cells (Figure 4A, upper panel) inhibited the capacity of Wnt5a to induce activation of Rac1/2 (Figure 4A, middle panels), but not RhoA (Figure 4A, lower panel). Moreover, we found that siDOCK2-silencing of DOCK2 in MEC1-ROR1 (Figure 4B, upper panel) reduced the level of activated Rac1/2 relative to that of MEC1-ROR1 treated with siCtrl (Figure 4B, middle panels), but did not reduce the level of activated RhoA (Figure 4B, lower panel), indicating that DOCK2 contributes to ROR1-dependent activation of Rac1/2, but not RhoA.
DOCK2 factors in Wnt5a-induced activation of Rac1/2, but not RhoA. (A) Immunoblot analysis of lysates prepared from freshly isolated primary CLL cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2, then treated without (−) or with (+) Wnt5a; expression of DOCK2, total Rac1/2, RhoA, and activated Rac1/2, RhoA was measured, as indicated on the left (representative of 3 patients). The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 or GTP-RhoA vs total Rac1/2 or RhoA, respectively. (B) Immunoblot analysis of lysates prepared from MEC1-ROR1 cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2, then treated without (−) or with (+) Wnt5a; expression of DOCK2, total Rac1/2, RhoA, and activated Rac1/2, RhoA was measured, as indicated on the left. The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 or GTP-RhoA vs total Rac1/2 or RhoA, respectively.
DOCK2 factors in Wnt5a-induced activation of Rac1/2, but not RhoA. (A) Immunoblot analysis of lysates prepared from freshly isolated primary CLL cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2, then treated without (−) or with (+) Wnt5a; expression of DOCK2, total Rac1/2, RhoA, and activated Rac1/2, RhoA was measured, as indicated on the left (representative of 3 patients). The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 or GTP-RhoA vs total Rac1/2 or RhoA, respectively. (B) Immunoblot analysis of lysates prepared from MEC1-ROR1 cells transfected 72 hours previously with control siRNA or siRNA targeting DOCK2, then treated without (−) or with (+) Wnt5a; expression of DOCK2, total Rac1/2, RhoA, and activated Rac1/2, RhoA was measured, as indicated on the left. The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 or GTP-RhoA vs total Rac1/2 or RhoA, respectively.
P-808 is necessary for ROR1 to recruit DOCK2 on stimulation with Wnt5a
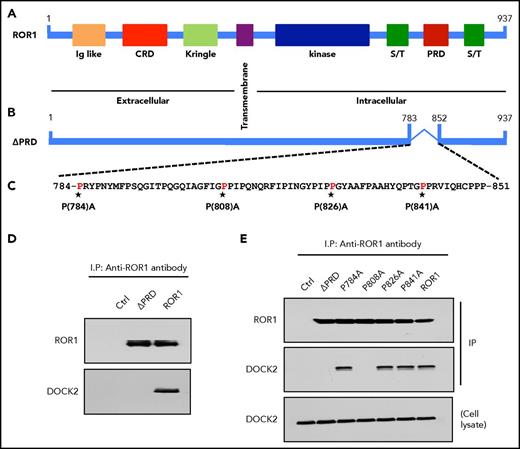
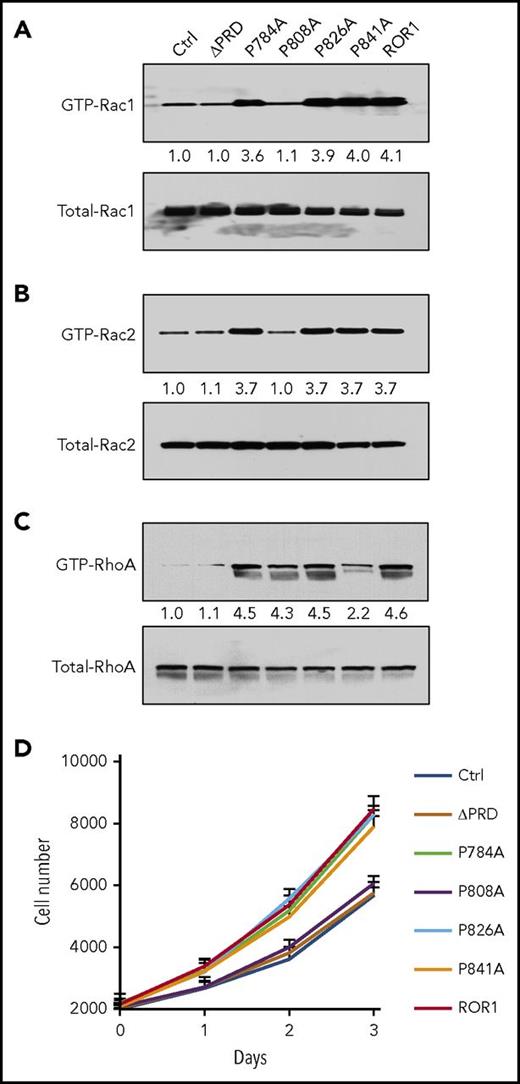
DOCK2 contains a SH3 domain, which can bind to -P-X-X-P- motifs found in the PRDs of many proteins, including ROR1.1,16,17,26,27 We examined the PRD of ROR1 for putative SH3-binding sites that could enable it to dock with DOCK2. We found that anti-ROR1 ip using lysates of MEC1-ROR1 contained DOCK2 (Figure 5D). However, anti-ROR1 ip using lysates of MEC1-∆PRD-ROR1 cells, which expressed a mutant ROR1 lacking its PRD, did not contain any detectable DOCK2, indicating the PRD of ROR1 was necessary for DOCK2 binding. We generated various mutant forms of ROR1 with proline (P) to alanine (A) substitutions at the -P-X-X-P- motifs identified in ROR1; namely, at positions 784, 808, 826, and 841 (Figure 5A-C). Comparable levels of wild-type (W/T) or truncated forms of ROR1 were expressed by each of the various transfectants (Figure 5E, upper panel). Moreover, comparable levels of DOCK2 were found in each of the various MEC1 transfectants and in the MEC1 parental cell line (Figure 5E, lower panel). We found that ROR1 with P→A substitutions at 784, 826, or 841 bound to DOCK2 as effectively as W/T ROR1 (Figure 5E, middle panel). MEC1 cells transfected with each of these forms of ROR1 had enhanced Rac1/2 activation and proliferation, as did MEC1-ROR1 cells expressing W/T ROR1 (Figures 6A, B and D). However, ROR1 with a P→A substitution at 808 (ROR1P808A) did not associate with DOCK2 in MEC1 transfected to express this mutant form of ROR1 (Figures 5E). Moreover, MEC1 cells expressing ROR1P808A did not have enhanced activation of Rac1/2 relative to that seen in MEC1 cells, but did have enhanced activation of RhoA (Figures 5E and 6A-C). Consistent with the notion that activation of Rac1/2 is responsible for the capacity of ROR1 to enhance cell proliferation, we found that MEC1 cells transfected with ROR1P808A did not have enhanced cell growth relative to that of MEC1 cells, in contrast to MEC1 cells transfected with W/T ROR1 or other mutant forms of ROR1 with P→A substitutions at 784, 826, or 841 (Figure 6D).
ROR1P808A is unable to associate with DOCK2, induce Rac1/2 activation, or enhance leukemia cell proliferation. (A) Schematic depicts the structure of ROR1 protein with different domains. (B) ΔPRD represents the truncated form of ROR1 without its P-rich region. (C) Amino acid sequences of the P-rich domain of ROR1. Asterisks indicate the P amino acid residues that had been substituted with A. (D) Immunoblot analysis of anti-ROR1 (cirmtuzumab) immune precipitates from lysates of MEC1-Ctrl, MEC1-ΔPRD, or MEC1-ROR1 cells, as indicated on the top. Membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (E) Interaction of ROR1 with DOCK2 was confirmed by immunoblot analysis of anti-ROR1 (cirmtuzumab) immune precipitates from lysates of MEC1, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells transfected with each of the various mutated forms of ROR1, as indicated on the top (upper panel). In the lower panel is an immunoblot of the whole-cell lysates of the MEC1-Ctrl, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells transfected with each of the various mutated forms of ROR1, as indicated on the top, and probed with anti-DOCK2 antibody.
ROR1P808A is unable to associate with DOCK2, induce Rac1/2 activation, or enhance leukemia cell proliferation. (A) Schematic depicts the structure of ROR1 protein with different domains. (B) ΔPRD represents the truncated form of ROR1 without its P-rich region. (C) Amino acid sequences of the P-rich domain of ROR1. Asterisks indicate the P amino acid residues that had been substituted with A. (D) Immunoblot analysis of anti-ROR1 (cirmtuzumab) immune precipitates from lysates of MEC1-Ctrl, MEC1-ΔPRD, or MEC1-ROR1 cells, as indicated on the top. Membranes were probed with anti-ROR1 or anti-DOCK2 antibody, as indicated on the left. (E) Interaction of ROR1 with DOCK2 was confirmed by immunoblot analysis of anti-ROR1 (cirmtuzumab) immune precipitates from lysates of MEC1, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells transfected with each of the various mutated forms of ROR1, as indicated on the top (upper panel). In the lower panel is an immunoblot of the whole-cell lysates of the MEC1-Ctrl, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells transfected with each of the various mutated forms of ROR1, as indicated on the top, and probed with anti-DOCK2 antibody.
ROR1P808A has impaired capacity to induce activation of Rac1/2 or to enhance leukemia cell proliferation. (A) Activated Rac1, Rac2 (B), and RhoA (C) were measured in MEC1-Ctrl, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells expressing any 1 of the mutated forms of ROR1, as indicated on the top. The numbers above each lane are ratios of band IOD of GTP-Rac1/2 vs total Rac1/2. The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 vs total Rac1/2, or GTP-RhoA vs total-RhoA. (D) Mean numbers of MEC1-Ctrl (blue), MEC1-ROR1 (red), or MEC1 cells expressing each of the mutated forms of ROR1 (colors indicated in the legend) at the days indicated below the graph. Data are shown as mean ± SD from 5 independent experiments. Longitudinal mixed effects model analysis was performed to get a global P value for the differences in proliferation rate compared with control. This model takes into account growth overtime (baseline is day 0 and then subsequent measurements were on days 1, 2, and 3), as well as cell type; this showed ROR1, P874A, P826A, and P841A each grew significantly more rapidly compared with Ctrl cells (P < .0001); ΔPRD and P808A cells did not differ from Ctrl cells (P = .60 and .26, respectively).
ROR1P808A has impaired capacity to induce activation of Rac1/2 or to enhance leukemia cell proliferation. (A) Activated Rac1, Rac2 (B), and RhoA (C) were measured in MEC1-Ctrl, MEC1-ΔPRD, MEC1-ROR1, or MEC1 cells expressing any 1 of the mutated forms of ROR1, as indicated on the top. The numbers above each lane are ratios of band IOD of GTP-Rac1/2 vs total Rac1/2. The numbers between 2 lanes are ratios of band IOD of GTP-Rac1/2 vs total Rac1/2, or GTP-RhoA vs total-RhoA. (D) Mean numbers of MEC1-Ctrl (blue), MEC1-ROR1 (red), or MEC1 cells expressing each of the mutated forms of ROR1 (colors indicated in the legend) at the days indicated below the graph. Data are shown as mean ± SD from 5 independent experiments. Longitudinal mixed effects model analysis was performed to get a global P value for the differences in proliferation rate compared with control. This model takes into account growth overtime (baseline is day 0 and then subsequent measurements were on days 1, 2, and 3), as well as cell type; this showed ROR1, P874A, P826A, and P841A each grew significantly more rapidly compared with Ctrl cells (P < .0001); ΔPRD and P808A cells did not differ from Ctrl cells (P = .60 and .26, respectively).
Discussion
Here we demonstrate that ROR1 interacts with DOCK2 in primary CLL cells that are stimulated with exogenous Wnt5a, and in MEC1-ROR1 cells, which constitutively express high levels of Wnt5a. We also found that ROR1/DOCK2 interaction is important for Wnt5a-enhanced production of activated Rac1/2, but not activated RhoA. Reducing expression of DOCK2 with specific siRNA inhibited Wnt5a-enhanced CLL cell proliferation. As such, these studies reveal that DOCK2 plays an important role in ROR1-dependent noncanonical Wnt5a signaling leading to enhanced CLL cell proliferation.
The cytoplasmic domain, and more specifically the PRD, of ROR1 was required for it to associate with DOCK2 and enhance proliferation in response to Wnt5a. DOCK2 contains a SH3 domain, which allows it to bind P residues within motifs that commonly are found in PRDs of other proteins.16,17,26,27 Substitution of the P residue at position 808 with A (ROR1P808A) precluded DOCK2 from complexing with ROR1 in MEC1 cells. Moreover, we did not observe enhanced activation of Rac1/2, or proliferation in MEC1 cells transfected with ROR1P808A, in contrast to MEC1 cells transfected with W/T ROR1 or ROR1 with A substitutions at any other P residue within the ROR1-PRD. As such, the P residue in ROR1 at 808 appears critical for binding DOCK2 and enhancing proliferation in MEC1 cells.
DOCK2 is a direct activator of Rac1/2 in hematopoietic cells.15,18 Rac1/2 have been implicated in a variety of cellular signaling processes, including those involved in B-cell development or neoplasia.28-30 Rac2−/− mice, which are deficient in Rac2, have peripheral B lymphocytosis and marked reductions in marginal zone B cells, peritoneal cavity B-1a B cells (CD5 B cells), and IgM-secreting plasma cells, indicating that Rac2 plays an important role in B-cell development.31 Moreover, when Rac2−/− mice are bred with mice housing a conditional inactivation of Rac1 in the B-cell lineage, the progeny that had progenitor B cells lacking Rac1 and Rac2 had marked reductions in mature marrow and lymph node B cells, as well as severe reductions in mature follicular B cells, and lacked peritoneal B-1a B cells.32 Moreover, there were no mature B cells in such animals that lacked both Rac1 and Rac2, which appeared required for B-cell receptor signaling leading to enhanced B-cell survival. Rac2 also appears to play an important role in initiating and sustaining p210–breakpoint cluster region protein–Abelson murine leukemia viral oncogene–induced leukemia stem cells in vivo.33 DOCK2 has been identified as a regulator of leukemia cell proliferation or tumor growth through the activation of Rac and extracellular signal-regulated kinase.21 In the present study, we found that reducing expression of DOCK2 with specific siRNA in CLL cells inhibited the capacity of Wnt5a to induce activation of Rac1/2 or enhance proliferation, indicating that DOCK2 is necessary for Wnt5a-induced activation of Rac1/2 in CLL cells.
Previously, we described that cytoplasmic GEFs such as ARHGEF1, ARHGEF2, and ARHGEF6 can be recruited to ROR1 in response to Wnt5a to induce activation of Rac1 or RhoA.11 In such cases, hematopoietic lineage-specific protein 1 acts an adopter protein for recruitment of ARHGEF1,12 and 14-3-3ζ for ARHGEF2 to ROR1.34 However, the molecular mechanism or mechanisms of how ROR1 could contribute to activation of Rac2 remained elusive. In the present study, we identified that DOCK2 can interact with ROR1 in response to Wnt5a, and specifically activate Rac1 and Rac2, but not RhoA. Knockdown studies reveal that DOCK2 was required for the Wnt5a-enhanced activation of Rac GTPases and proliferation of CLL cells. Overall, our results suggest that all these cellular events potentially contribute to the capacity of ROR1 to promote disease progression in patients with CLL.10
DOCK2 also may interact with proteins other than ROR1 to induce activation of Rac1 and Rac2 in hematopoietic cells.35,36 Upon stimulation, DOCK2 translocates from cytoplasm to the plasma membrane on generation of phosphatidylinositol 3,4,5-trisphosphate at the leading edge of cell migration. Subsequent accumulation of DOCK2 is promoted via phospholipase D-mediated synthesis of phosphatidic acid, which interacts with a polybasic amino acid cluster, resulting in increased local actin polymerization.20 As a consequence, DOCK2-deficient lymphocytes have reduced F-actin polymerization and impaired cell motility.19 Moreover, DOCK2-deficient neutrophils fail to form functional leading edges and have defective chemotaxis.20 In such cases, the SH3 domain of DOCK2 may associate with proteins other than ROR1, such as the engulfment and cell motility proteins 1 and 2, Crkl, or Vav, which appear to play a role in DOCK2-mediated Rac activation in lymphocytes or leukemia cell lines.25,35,36 However, our studies reveal that DOCK2 also can associate with ROR1 in CLL cells when stimulated with Wnt5a. Silencing DOCK2 inhibited the capacity of CLL cells to migrate in response to chemokines, such as CXCL12, but Wnt5a still could enhance chemokine-directed migration. Moreover, although CLL cells treated with siDOCK2 did not migrate as well as CLL cells treated with siCtrl in response to chemokine, siDOCK2-treated CLL cells still had enhanced migration when treated with Wnt5a, suggesting that the interaction of ROR1 with DOCK2 was not required for it to promote chemokine-directed migration, which appears dependent on the interaction of ROR1 with other cytosolic proteins. For example, ROR1P808A can associate with hematopoietic lineage-specific protein 1 in response to Wnt5a, thereby activating RhoA and enhancing chemokine-directed migration.12
In summary, the present study describes a previously unrecognized ROR1/DOCK2-dependent mechanism for activating Rac1/2 in response to Wnt5a. The findings reported here demonstrate the importance of DOCK2 in ROR1-dependent Wnt5a-induced signaling and highlight a pathway for potential drug development. To this end, we found that the capacity of Wnt5a to induce ROR1 to associate DOCK2 could be blocked by cirmtuzumab, a first-in-class humanized anti-ROR1 mAb, which is being evaluated in patients with CLL (https://clinicaltrials.gov/ct2/show/NCT02222688).13,37 Moreover, we found that cirmtuzumab could block the capacity of Wnt5a to induce recruitment of DOCK2 to ROR1, contributing to the noted capacity of cirmtuzumab to block ROR1-dependent, noncanonical Wnt5 signaling responsible for enhancing leukemia cell proliferation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the California Institute for Regenerative Medicine (grant DR3-06924) for supporting us in generating anti-ROR1 mAbs and cirmtuzumab. This work was supported by the University of California, San Diego Foundation Blood Cancer Research Fund (BCRF), a Specialized Center of Research grant (7005-14) from the Leukemia and Lymphoma Society, and a PO1 grant (5P01CA081534-14) from NIH for the CLL Research Consortium.
Authorship
Contribution: M.K.H. and T.J.K. designed research and conceived the project; M.K.H., J.Y., G.F.W., L.Z.R., and L.C. performed research; Z.S. and S.P.B. performed mass spectrometry analysis; D.S.N. performed longitudinal mixed effects model analysis; and M.K.H. and T.J.K. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas J. Kipps, Moores Cancer Center, University of California, San Diego, 3855 Health Sciences Dr, Rm 4307, La Jolla, CA 92093-082; e-mail: tkipps@ucsd.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal