Abstract
Age-related alterations in the human blood system occur in B cells, T cells, cells of the innate system, as well as hematopoietic stem and progenitor cells (HSPCs). Interestingly, age-related, reduced genetic diversity can be identified at the stem cell level and also independently in B cells and T cells. This reduced diversity is most probably related to somatic mutations or to changes in the microenvironmental niche. Either process can select for specific clones or cause repeated evolutionary bottlenecks. This review discusses the age-related clonal expansions in the human HSPC pool, which was termed in the past age-related clonal hematopoiesis (ARCH). ARCH is defined as the gradual, clonal expansion of HSPCs carrying specific, disruptive, and recurrent genetic variants, in individuals without clear diagnosis of hematological malignancies. ARCH is associated not just with chronological aging but also with several other, age-related pathological conditions, including inflammation, vascular diseases, cancer mortality, and high risk for hematological malignancies. Although it remains unclear whether ARCH is a marker of aging or plays an active role in these various pathophysiologies, it is suggested here that treating or even preventing ARCH may prove to be beneficial for human health. This review also describes a decision tree for the diagnosis and follow-up for ARCH in a research setting.
ARCH definition and history
It is established that mammals maintain hematopoiesis by the activity of thousands of hematopoietic stem and progenitor cells (HSPCs).1 During human aging, the expansion of 1 or more HSPCs will result in clones that will sustainably contribute more than others to the production of mature blood cells (Figure 1). Accordingly, age-related clonal hematopoiesis (ARCH) is defined as the expansion of HSPC clones, harboring specific, disruptive, and recurrent genetic variants, in individuals without clear diagnosis of hematological malignancies. Although the definition of recurrent variants is still rapidly evolving, it is important to suggest a list of recurrently mutated genomic regions that can define ARCH to avoid misdiagnosis. This list should be continuously updated and in the future include copy number variation (CNV). Moreover, new studies suggest that ARCH can also be attributed to nonrecurrent genetic variation, possibly the result of genetic drift.2 However, the current, agreed upon definition of ARCH considers recurrently mutated genes only.
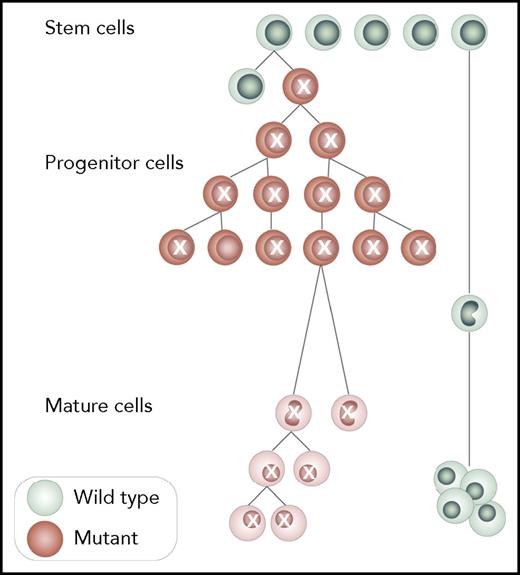
ARCH. ARCH is the clonal expansion of at least single HSPCs capable of multilineage differentiation. All cells belonging to the expanding clone share genetic variants, and at least some of them are recurrent and belong to ARCH-defining variants as in Table 1. A minimum of 1 somatic disrupting variant is needed for the diagnosis of ARCH, and wild-type cells that do not carry the mutation must be present. Although somatic mutations should be present both in hematopoietic stem and progenitor cells (HSPCs) and in mature cells, this concept is not mandatory for diagnosis because cell sorting is currently less feasible. Professional illustration by Somersault18:24.
ARCH. ARCH is the clonal expansion of at least single HSPCs capable of multilineage differentiation. All cells belonging to the expanding clone share genetic variants, and at least some of them are recurrent and belong to ARCH-defining variants as in Table 1. A minimum of 1 somatic disrupting variant is needed for the diagnosis of ARCH, and wild-type cells that do not carry the mutation must be present. Although somatic mutations should be present both in hematopoietic stem and progenitor cells (HSPCs) and in mature cells, this concept is not mandatory for diagnosis because cell sorting is currently less feasible. Professional illustration by Somersault18:24.
The first studies that described somatic clonal expansions identified them without realization of their mutational background. Initially X inactivation skewing (XIS) was used to identify clonal expansions. Studies on chronic myeloid leukemia3 found that all cells in the tumor expressed the same allele (the most extreme form of XIS), suggesting that all chronic myeloid leukemia cells were derived from a single cell and were therefore clonal. Contrastingly, if the observed frequency of either allele was ∼50%, it was interpreted that the cellular pool was evenly produced from a genetically diverse population of cells. Soon it became clear that XIS between 50% and 100% was more prevalent than expected by chance, or due to inaccuracies in the assay itself. Based on the relative protein amounts of the X-linked enzyme glucose-6-phosphate dehydrogenase (G6PD), Fialkow demonstrated that the level of XIS was highly similar in different tissues from the same individual, albeit different between individuals.4 Accordingly, he suggested that the different tissues, blood, skin, and lymph, were all derived from the same, relatively small progenitor pool. If the effective size of the founding population was small enough, it could explain the deviation from the expected 50% allele frequency.4 These early studies identified clonal hematopoiesis (CH), which is not age related and can be the result of early somatic genetic variation, that occurred in a subset of cells that generate the fetal hematopoietic system. This type of CH is not associated with recurrent genetic mutations and is termed here early development mosaic CH.
The first data to support age related somatic selection emerged from the clonal analysis of blood cells from a large cohort of South African women and their daughters around the 1980s. Hitzeroth et al noticed that the frequency of the heterozygous G6PD AB phenotype was lower than expected based on the Hardy-Weinberg equilibrium, whereas the homozygotes AA and BB were in excess of what was expected.5 This puzzling phenomenon could not be observed in the daughters of the women from the same cohort.6 The authors concluded correctly that the heterozygous phenotype undergoes negative selection, and they were able to demonstrate that the reduced heterozygosity in blood cells was increasing with age.6 Although this could have been the first evidence for ARCH, the authors favored the hypothesis that a specific G6PD allele is being selected because of its functional consequences; however, they could not identify specific mutations to support their hypothesis. Much later, in an attempt to develop a diagnostic test that could potentially differentiate between malignant clonal expansion and reactive polyclonal expansion, Fey et al noticed that a large proportion (∼20%) of healthy individuals had extreme XIS. This pattern of XIS was more frequent among the elderly (57 to 96 years) than among the younger (20 to 58 years) women tested. The authors used a different marker for XIS (M27beta) and not the G6PD to avoid the possible germline effects on the different G6PD alleles. The authors concluded that XIS cannot reliably indicate neoplastic states, because the observed XIS in leukemia was comparable to XIS in normal leukocytes and in many healthy individuals.7 Therefore, it was suggested that with aging a small number of HSPCs maintain normal hematopoiesis by gaining a selective advantage because of selection of 1 of their X chromosomes.7 Around the same time, other groups also reported high incidence of XIS in blood samples of healthy individuals (20% to 30%), however, with no clear correlation with aging.8,9
The notion that the observed XIS could be the result of other processes, and not solely due to fitness-determining elements on the X chromosome, was first suggested in 1996 by Busque et al.10 They argued that “blood cells are one of the most mitotically active tissues of the human body, and may therefore be particularly susceptible to mild selection pressures, or to other phenomenon such as stem cell depletion or development of clonal hematopoiesis. Each of these acquired processes could result in highly skewed X-inactivation ratios and should be associated with an increased incidence of skewing with age.”10 The major conceptual breakthrough made by Busque et al was the realization that the increase of XIS with age can be explained by other processes: genetic bottlenecks (stem cell depletion) and direct selection. Based on these conclusions, Busque et al also raised the hypothesis that ARCH might be correlated with hematological malignancies.10 In contrast to the predictions put forth by Busque et al, in 1998 Abkowitz et al identified ARCH from studies in cats, suggesting that a specific X chromosome allele is being selected over time with no contribution to clonal malignancy.11 This phenomenon was termed hemizygous selection. To conclude this part, these landmark studies demonstrated that, alongside with ARCH, early, developmental mosaic CH, which is not age related and is not associated with recurrent mutations, can be observed in healthy individuals.
For the next 15 years (1996-2011) no major advances were made regarding our understanding of ARCH. Studies in monozygous twins identified strong heritability, suggesting that ARCH has a germline genetic contribution to the phenotype and support the hemizygous selection.12 Other reports have identified increased frequency of blood XIS among individuals with breast and cervical carcinoma regardless of age.13,14 The degree of XIS proved to be stable over time (18 months) and to be present in both T cells and granulocytes.15 These data solidified the assumption that XIS is associated with clonality of long-lived HSPCs differentiation.
The next paradigm shift in our understanding of ARCH materialized in 2012. Both CNV analysis16,17 and next-generation sequencing18 studies collectively provided evidence that ARCH is associated with specific, leukemia-related somatic genetic lesions. The clear evidence that ARCH can be a preleukemic condition was provided by the identification of preleukemic stem cells (pre-L-HSPCs). These cells carried mutations that were also observed among healthy individuals and in mature, non–leukemic cells.19-21 At the same time (2014), 3 studies reported age-related accumulation of mutations in leukemia-related genes (DNMT3A, TET2, ASXL1, and several others) with variant allele frequency (VAF) >2% and a median of ∼12%. Moreover, by the age of 70, ∼10% of individuals with nonhematological malignancies carried specific, leukemia-related, somatic mutations.22-24 An important observation from these studies was the enrichment of disruptive somatic lesions (nonsynonymous, nonsense, frameshift, or splice-site disruption) in specific genes (DNMT3A, TET2, and ASXL1) among individuals with ARCH.22 The ability to accurately characterize ARCH by specific mutations transformed the field and cleared some of the ambiguities introduced by the previous XIS assays.
Common methods for detecting ARCH
Indirect methods
XIS
The major advantage of the XIS assays is its nonspecificity, meaning that XIS will demonstrate reduced diversity regardless of the driver mutation. The human androgen receptor gene (HUMARA) assay (prototype of the XIS assays) is based on indirect determination of the methylation state of 2 CCGG sites in the HUMARA. Using the HUMARA assay CH was identified among 30% of healthy females older than 60 who had an allelic ratio greater than 1:3.6 ,21 When a threshold ratio >1:10 was imposed, the incidence of CH was ∼22%.10 Such high ratios in the HUMARA assay imply that for these healthy individuals the >90% of mature blood cells originate from a single cell. This might point to the fact that XIS assays tend to overestimate the true proportions of clones. Alternately, XIS is probably affected by other, aging-associated parameters such as hemizygous selection.11
Single-cell analysis
The clonal structure of a tissue could be studied by single-cell phylogenetic26 or epigenetic approaches.27 Such analysis of human cells was performed for acute myeloid leukemia (AML),28 normal brain tissue,29 and other cases (reviewed by Shapiro et al26 ). Although single-cell analysis can potentially identify ARCH, these assays are still expensive.
Direct methods
CNV
Two large-scale studies16,17 were the first to associate ARCH with specific genetic lesions, identifying increased frequency of large (>2 Mb) indels, and neutral loss of heterogeneity (LOH) with VAF ≳10% among individuals with different diseases. Younger than the age of 50, <0.5% of the sampled population carried somatic CNV, compared with only ∼2% to 3% of older individuals. Specifically, somatic aneuploidies included 8+, 9+, 12+, 14+, 15+, 18+, 19+, 21−, and 22+, and whole-chromosome LOH was found on chromosomes 2, 3, 13, 14, and 15.16 There was a significant excess of subjects with multiple clonal events.16 Indels and LOH identified among the healthy individuals frequently overlapped with regions of CNV or LOH that are commonly identified in hematological malignancies: 20q− (myelodysplastic syndrome, MDS), 13q− (chronic lymphocytic leukemia, CLL), 11q− (CLL, MDS, AML), 17p– (MDS, AML, CLL), 12+, and 8+ (MDS, AML). Furthermore, they concluded that ARCH is associated with a 10-fold increased risk for developing any blood cancer16 and was more frequent in individuals with solid tumors.17 These studies could not detect CNVs in the sex chromosome due to technical limitations of the comparative genomic hybridization array technology; however, cytogenetic studies have previously suggested that the entire loss of chromosome Y (LOY) or deletions in chromosome Y become more frequent with aging.30 Pierre and Hoagland demonstrated in 1972 that ∼30% of healthy men have clones with LOY; these clones were stable over time and occurred more in the bone marrow (BM) compared with peripheral blood (PB).30 The frequency of LOY in the 802 unselected MPN, MDS, and AML cases was 7.7%, 10.7%, and 3.7%, respectively.31 Accordingly, LOY answers the criteria of an ARCH defining lesion in men. One of the main limitations of detecting ARCH based on CNV is the low sensitivity in detecting small clones. Furthermore, current next-generation sequencing (NGS) technologies demonstrate inaccuracies in estimating clone size for intermediate size indels (100 bp to 2 Mb). Future studies will need to further estimate indels’ role in ARCH once technology will allow their detection and accurate variant calling.
Single nucleotide variants and short indels
The first report to correlate single nucleotide variants with ARCH was focused on TET2 mutations.18 The authors first sequenced PB and buccal epithelial cells from 3 elderly individuals with XIS. In 1 subject, the authors identified somatic mutations in TET2, DNMT3A, SLC39A12, ERCC6, and KIAA1919.18 These specific genes were resequenced by Sanger sequencing in a larger cohort of individuals with and without XIS. Only somatic TET2 mutations were identified in 5.6% of elderly individuals with ARCH. Blood counts of TET2 mutation-carriers were comparable to controls. One of the TET2 carriers later developed JAK2-positive essential thrombocytosis.18 Because TET2 mutations were commonly found in myeloid malignancies,32,33 the authors concluded that specific ARCH mutations can lead to leukemia. The next conceptual advancement was to search for other leukemia-related somatic mutations in cohorts of healthy individuals who were already sequenced for other reasons. By this method, DNMT3A mutations were found among healthy individuals.21 Others have identified DNMT3A and mutations in epigenetic modifiers in mature blood cells of AML patients,20 and the term preleukemic mutations was coined.20,21 Preleukemic mutations can be found in both leukemic cells and normal mature cells in the same individuals. Accordingly, preleukemic mutations were part of the ARCH clone, which preceded leukemia. Hence, every leukemic patient with preleukemic mutations must evolve from ARCH. The next major breakthrough was reported in 3 large studies of individuals free from hematological cancers for whom exome sequencing data were used to identify somatic mutations in PB samples.22-24 The major common finding of these studies was that most somatic mutations identified in the PB among the elderly were located in genes that were reported to be recurrently mutated in different types of leukemia, and they accumulated with age. By the age of 70, ∼10% of the population carries specific somatic mutations related to leukemia, and the most frequently mutated gene is DNMT3A. The term ARCH was defined in 1 of these studies.23 It is also important to note that although the majority of mutations found were in leukemia-related genes, ARCH was also found to be associated with nonleukemic somatic variation in 1 of these studies22 and also later in a whole genome sequencing (WGS) study on blood samples from Iceland.2 According to this study, the majority of ARCH cases were due to non-leukemia-related mutations, and by the age of 70, only 5% of the population was defined as ARCH arising from known leukemia drivers, whereas >20% could be detected with CH by nondriver variation.2 Another important observation from this study was the high correlation between CH and LOY, suggesting that an important, non-leukemia-related lesion might be LOY itself.2 McKerrell et al used amplicon-based NGS of a limited target set of loci and found that the prevalence of specific DNMT3A and JAK2 mutations increased after the age of 40, whereas the mutational hotspots in SRSF2 and SF3B1 appeared at older age (∼70).34 Young et al performed a longitudinal study over a time period of 10 years, utilizing error-corrected sequencing (ECS) and the Illumina TruSight Myeloid Sequencing Panel.35 Their methods were sensitive enough to identify DNMT3a and TET2 mutations at low VAFs (as low as 0.0001) in 95% of the cases. Clonal kinetics suggested that not all clones answer the definition of ARCH, because some of the clones were very small and were either stable or decreased in their size over long periods of time (>10 years).
To conclude this section, ARCH can be identified by different methods, each identifying different lesions. In practice, WGS is well suited for detecting most lesion types; however, it is still too expensive to be conducted in a sensitive way, and currently, most probably introduce noise. Furthermore, it is not clear whether ARCH that is defined by specific recurrent lesions shares features with ARCH due to non-leukemia-related lesions. It is also not clear whether in the cases of ARCH identified by nondriver variants the clones also carry an unidentified recurrent variant like CNVs, translocations, noncoding region variants, which were not studied. Last, as sequencing technology improves and small clones can be detected, it is not clear whether any clone is clinically relevant.
Epidemiology
Prevalence
The prevalence of CH among different age groups is highly dependent on the methods used to detect it, and here, we will not provide an exhaustive description of all reported prevalence. Alternatively, we think it would be more useful to describe ARCH prevalence based on the current, most advanced technology and understanding. The most recent study describing ARCH performed ECS of a large cohort of 2000 individuals, and the following prevalence was reported (prevalence, age group [years]): 2.5%, 20-29; 3.2%, 30-39; 8.2%, 40-49; 13.2%, 50-59; 20.6%, 60-69.36 The median VAF of somatic mutations was 0.025 (2.5%),36 which was lower than previously reported (∼10%).22-24 This discrepancy is most probably associated with the different sequencing and analytical methods, because ECS can eliminate false-positive variants, resulting from errors that occurred during sample preparation or the sequencing reaction itself.
Sex differences
Sex differences were noted above the age of 60, with men having increased risk for ARCH in 1 of the studies (odds ratio 1.3).23 However, no such differences were noted in the recent ECS-ARCH study across all age groups without binning.36 Men were reported to have an increased risk for various hematological malignancies, including AML and MDS with an elevated risk over the age of 60.37
Heritability
Heritability of ARCH was determined by a twin study demonstrating that ARCH was significantly more concordant between monozygous twins compared with nonmonozygous twins, indicating a genetic effect on XIS pattern with heritability of 0.63 in young and 0.58 in elderly twins.12 More recently, it was reported that somatic TET2 mutations showed familial aggregation,38 and another study concluded that germline variant in the TERT gene was associated with ARCH based on WGS analysis.2 Taken together, current findings strongly suggest a germline effect on ARCH and certainly should be better studied.
Etiology and pathogenesis
ARCH arises from the clonal expansion of multipotential stem cells still capable of differentiation, such that the progeny is likely to harbor common mutations. These mutations will be ultimately found in the expanded, ARCH clones. The role of the various ARCH-related genetic alterations was studied mainly in animal models43 and in the leukemic phase of human diseases.40 Young et al genotyped ARCH-related mutations in B cells, T cells, and myeloid cells and provided further evidence that the same mutation was present in all blood lineages among healthy individuals,41 indicating a capacity for differentiation. Others have identified yearly increase in the mean VAF of 3.95% for DNMT3A and of 9.98% for TET2 (P < .0001),38 which may indicate a greater proliferative rate for TET2 mutated cells over DNMT3A mutated cells.42 Furthermore, it was shown that the number of ARCH-related mutations per individual is increasing with age.36,41,42 DNMT3A hotspot mutations typically have significantly larger clone sizes than those of clones with non–hotspot mutations,36 suggesting that specific mutations within the same genes provide greater fitness than others. The fact that recurrent specific mutations in DNMT3A, TET2, and ASXL1 account for >90% of ARCH suggests that these mutations change a cellular phenotype that provides selective advantage to HSPCs. It is still unclear why these mutations are associated with better HSPCs fitness. It is important to consider that ARCH-related mutations may have distinct functional consequences during ARCH evolution, and later during leukemia progression and the evolution of leukemic clones. In addition, it is acceptable that single mutations are pleiotropic in nature, and in the context of ARCH, it is likely that the consequence of this pleiotropy may be distinct in different cell types. For example, TET2 mutations might provide a selective advantage to HSPCs while exerting a different effect on monocytes and T cells, calling for functional studies of hematopoietic cells of ARCH individuals. Adding more to the complex phenotype of ARCH evolution, it was recently suggested2 that early, developmental mosaic-CH and hemizygous selection can expand over time due to mechanisms that are not a selection of specific clones, but rather due to genetic drift. Finally, many ARCH-related mutations occur in proteins that play key roles in DNA methylation or its regulation (eg, DNMT3A, TET2, IDH1), providing a possible link between the genetics of ARCH and changes in the epigenome of blood cells, specifically in cytosine methylation (reviewed by Kunimoto and Nakajima43 ).
Clinical features
Although, by definition, ARCH occurs among the healthy elderly population, it is becoming clearer that ARCH is associated with a large number of pathological states, including an elevated risk for blood cancers (Figure 2). Whether ARCH is the cause of these pathologies, a contributor, a marker, or just correlated with them due to its high correlation with aging calls for further investigation.
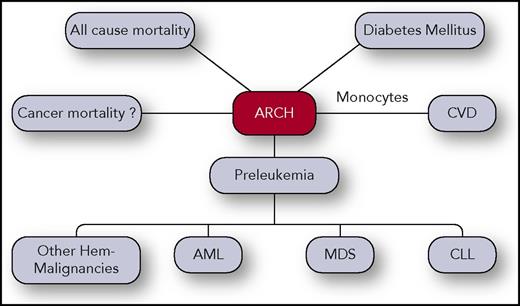
ARCH clinical correlations. ARCH was found to be associated with all-cause cancer mortality, CVD, hematological malignancies such as AML, CLL, MDS, and others. Association between ARCH and cancer mortality was also reported but might be confounded due to exposure to chemo/radiotherapy or due to other reasons. Professional illustration by Somersault18:24.
ARCH clinical correlations. ARCH was found to be associated with all-cause cancer mortality, CVD, hematological malignancies such as AML, CLL, MDS, and others. Association between ARCH and cancer mortality was also reported but might be confounded due to exposure to chemo/radiotherapy or due to other reasons. Professional illustration by Somersault18:24.
ARCH and mortality
A study of a cohort of 500 women aged 73 to 100 found that women with lower ratios of XIS had an increased mortality in comparison with women with higher XIS ratios,44 suggesting that ARCH due to specific mutations might be correlated with mortality whereas hemizygous selection might not.45 Studies on ARCH defined by LOY identified increased, all-cause mortality among individuals with LOY with a hazard ratio (HR) of 1.91 and increased nonhematological cancer mortality (HR 3.62; 132 cases). Importantly, LOY affected at least 8.2% of the entire population.46 The studies on ARCH based on specific mutations in exome sequencing data22,23 identified increased all-cause mortality among individuals with ARCH (HR 1.4). Increased mortality was mainly attributable to cardiovascular disease (CVD) in 1 of the studies23 and to cancer in the other study.22 A larger study on cancer-related mortality recently reported that individuals with ARCH exhibit higher mortality due to their original malignancy.47 Specifically, Coombs et al reported that increased mortality was more prominent among individuals with ARCH due to driver preleukemic mutations with larger clones.47 With regard to cancer mortality, it is hard to elucidate whether a confounder can be introduced in light of the increased frequency of specific somatic mutations such as PPM1D and TP53 following chemotherapy.48 It has been clearly established that TP53 mutations are present in HSPCs before chemotherapy, and that their VAF increases after therapy, which enables them to be detected by whole-exome sequencing.49 Accordingly, patients with cancer who were treated with chemotherapy will have more ARCH due to these mutations,47 and because getting chemotherapy in many cancers means worse prognosis, the correlation between ARCH and cancer mortality could be indirect. A good example for such a confounder arose around the discovery of somatic PPM1D mutations in the blood of patients with breast and ovary carcinoma.50 The authors identified PPM1D mutations in a proportion of blood cells and concluded that they are predisposed to developing cancer because controls had much lower prevalence of these mutations. However, many of the patients with cancer recruited for these studies received chemotherapy, which was subsequently enriched for specific ARCH mutations, and later studies proved that the increased prevalence of PPM1D mutations in the blood of patients with cancer is associated with exposure to chemotherapy.48 One has to be extremely careful when suggesting a cause-and-effect relationship between 2, well-correlated observations. To conclude this section, mortality is an outcome that is influenced by a complex set of parameters, such that the association between ARCH and mortality may be indirect and not always readily discernible.
ARCH, type 2 diabetes, and CVD
Inflammation plays a key role in the pathogenesis of atherosclerosis. It has been demonstrated that monocytes promote atherosclerotic plaque growth by production of inflammatory cytokines (reviewed by Pamukcu et al51 ). Recent studies from rodents suggest that TET2-mutated monocytes might play a role in the buildup of aortic atherosclerotic plaques.52 Jaiswal et al found that individuals with ARCH carry an increased risk for CVD (relative risk [RR] 1.9) due to mutations in DNMT3a, TET2, ASXL1, and JAK2.53 Taken together, it is not clear whether mechanisms related to TET2-deficient monocytes-altered function will be valid for other mutations. ARCH could also contribute to CVD in an indirect manner, through its correlations with type 2 diabetes mellitus (T2DM). ARCH defined by CNV was significantly associated with T2DM with an RR of 5.3 and even more so in nonobese individuals. Notably, individuals with ARCH had higher prevalence of vascular complications.54 Another possible confounder to the correlation between ARCH and CVD could be the correlation between ARCH and smoking.47 Altogether, a considerable amount of information has accumulated on the correlations between ARCH and CVD with some evidence of causation. However, the inconsistencies between the different studies suggest that either the definition of ARCH, selection biases, or other confounders might bias the results.
ARCH and preleukemia
Some ARCH cases are clearly preleukemic (will definitely evolve to leukemia) as was demonstrated by several studies19-21 and reviewed previously.55 Mutations described in ARCH were identified not only in AML but also in other types of hematological malignancies, including CLL,56 T-cell lymphomas,57 and MDS.58 However, the majority of individuals with ARCH will not develop leukemia during their lifetime. Recent studies identified increased prevalence of multiple types of leukemia among individuals with ARCH, with an RR of ∼10 to 35.16,22 These studies cannot be used for the prediction of different types of leukemia yet, because the number of leukemia cases in these studies was too low. Although the risk for de novo MDS/AML among ARCH individuals is low, it is better related to therapy-induced MDS/AML, and specifically, following autologous bone marrow transplantation (BMT). PPM1D and TP53 mutations were more common in patients with therapy-related MDS.59 Among elderly who received chemotherapy and developed therapy-induced MDS/AML, 62% had ARCH before chemotherapy, whereas only 15% of matched controls (who did not develop MDS/AML) had ARCH.60 In a recent study, 29.9% of the participants had ARCH at the time of autologous BMT. ARCH was associated with an increased rate of therapy-related leukemia; the 10-year risk among individuals with ARCH was 14.1% vs 4.3% among individuals without ARCH. Individuals with ARCH had also increased mortality rate at 10 years after autologous BMT,61 and among patients with allogeneic BMT, ARCH-related donor leukemia was described.62 Donor ARCH (DNMT3A mutations) was also identified in 5 out of 552 allogeneic BMT recipients with unexplained cytopenia.63 Specific ARCH mutations were identified among 64% of individuals with unexplained cytopenia and with no history of blood malignancies; however, many of these individuals eventually developed myeloid malignancies (∼50%). Carrying 1 somatic mutation with a VAF >10% or carrying 2 or more mutations had a positive predictive value for diagnosis of myeloid neoplasm equal to 0.86 and 0.88, resepectively.64 The majority of myeloid malignancies evolved in the first 5 years after the detection of ARCH. The authors concluded that mutational analysis could improve the prediction of malignancy among individuals with unexplained cytopenia.64 ARCH was also reported among survivors of aplastic anemia,65 which have increased risk for developing secondary AML and MDS. Specifically, the VAF of ARCH-related mutations increased over time, with high ASXL1 median VAF of ∼31%.65 These data suggest that the stressed BM positively selects ARCH clones, as the pattern of recurrent mutations cannot be explained by genetic drift. Altogether, ARCH plays an important role in the premalignant state and can be potentially used in the future as a predictive tool for hematological malignancies.
Laboratory features
Most ARCH studies could not detect clear laboratory changes in complete blood counts and chemistry. ARCH with TET2 mutations was associated with mild reductions in neutrophils, however, without neutropenia.38 ARCH was highly enriched among individuals with unexplained cytopenia, although not all individuals with mutations developed a myeloid leukemia during the study follow-up time.64 Accordingly, carrying a recurrent mutation and having unexplained cytopenia should prompt a closer follow-up, whereas currently no treatment is needed unless a diagnosis of MDS can be made based on the 2016 World Health Organization criteria,66 and prognostic criteria suggest treatment benefit. Future studies will better define the current ambiguity in the overlap between ARCH and MDS among individuals with unexplained cytopenia. ARCH combined with increased red cell distribution width (>14.5%) was associated with increased mortality.23 No clear correlations between high red cell distribution width and increased ARCH prevalence were documented so far. In a study on therapy-related ARCH, several correlations were identified with various blood count parameters47 ; however, it is still too early to determine whether these changes are specifically related to ARCH per se or to exposure to radio/chemotherapy.
Diagnosis and differential diagnosis
Suggested criteria for diagnosing ARCH were suggested by Steensma et al67 ; here these criteria are being updated based on recent advances in the field. ARCH should be diagnosed based on NGS of DNA extracted from bulk PB. A suggested list of regions that need to be sequenced by NGS is provided in Table 1 and is aimed at covering the majority of ARCH cases but not all. Any disruptive variant (nonsynonymous, nonsense, or frameshift) in the coding regions of the suggested target list should be considered as defining ARCH based on VAF cutoffs. It is still unclear whether some synonymous variants in these genes can be disruptive; however, for now synonymous variants do not define ARCH. This review aims at setting clear ARCH defining lesions so that results from different studies correlating ARCH with other phenotypes will be reproducible. ARCH should be diagnosed under the following settings: (1) At coverage > 100× and <1000 VAF should be >2% and <30% (even under these values, false positives can occur and different variant calling strategies are recommended). (2) At coverage (>1000×) VAFs >0.5% and <40% define ARCH.36 Variants with high VAF (>14%), which were reported as common single nucleotide polymorphism database (minor allele frequency of ≥0.01 in at least 1 major population), should be excluded. It is recommended that the list of genes/regions that define ARCH will be constantly updated as the original ARCH studies identified only variants with high VAFs.22-24 Accordingly, new recurrent ARCH-defining regions, which either occur at older age or do not give a strong selective advantage and therefore present at lower VAF (<2%), might have been missed. Furthermore, as sequencing technology and variant calling are constantly improving, it is crucial to constantly evaluate ARCH diagnosis. Until proven otherwise, any large somatic CNV or LOH (>2 Mb) should also be considered as diagnostic for ARCH. A repeated blood test is needed to provide evidence for stable or increasing ARCH after 6 months. Differential diagnosis of ARCH includes CH due to early developmental mosaic-CH and hemizygous selection.45 In this case, genetic variants can be present in a large proportion of blood cells because the variation occurred in 1 of the cells that contributed to the HSPC pool. Finding an ARCH mutation in a different tissue excludes ARCH. Another alternative diagnosis to ARCH is reduction in blood clonality due to aging-related processes in B and T cells. Reduced T-cell diversity due to various reasons might lead to the detection of CH. Nevertheless, it is unlikely that it will occur in ARCH-specific genes. If the expanding T-/B-cell clone does carry an ARCH somatic mutation, blood counts should rule out lymphocytosis and the restriction of the mutation to B- or T-cell lineages.
ARCH defining genes
| Gene . | Chromosome . | Region . | Gene . | Chromosome . | Region . |
|---|---|---|---|---|---|
| ASXL1 | 20 | Whole gene | NOTCH1 | 9 | Indel: exon 0 |
| BRAF | 5 | Missense: position 600 (aa) | NPM1* | 5 | Indel: position 288 (aa) |
| CALR | 12 | Indel: positions 360 to 380 (aa) | NRAS | 1 | Missense: positions 12, 13, 61 (aa) |
| CBL | 11 | Whole gene | PPM1D | 17 | Whole gene |
| DNMT3A | 2 | Whole gene | PTPN11 | 12 | Whole gene |
| EZH2 | 7 | Whole gene | RUNX1 | 21 | Whole gene |
| FLT3 | 12 | Missense: position 835 (aa) and exons 9-11 | SETBP1 | 18 | Missense: position 870 (aa) |
| GNAS | 20 | Whole gene | SF3B1 | 2 | Missense: positions 625, 666, 700 (aa) |
| GNB1 | 1 | Missense: positions 57, 76, 80 (aa) | SMC1A | X | Whole gene |
| IDH1 | 2 | Missense: positions 132 (aa) | SMC3 | 10 | Whole gene |
| IDH2 | 15 | Missense: positions 140, 172 (aa) | SRSF2 | 17 | Missense: position 95 (aa) |
| JAK2 | 9 | Missense: position 617 (aa) | STAG2 | X | Whole gene |
| KIT* | 4 | Missense: position 816 (aa) | TET2 | 4 | Whole gene |
| KRAS | 12 | Missense: positions 12, 13, 61 (aa) | TP53 | 17 | Whole gene |
| MPL | 1 | Indel: exon 10 | U2AF1 | 21 | Missense: positions 34, 154 (aa) |
| MYD88 | 3 | Missense: position 265 (aa) | WT1* | 11 | Whole gene |
| Gene . | Chromosome . | Region . | Gene . | Chromosome . | Region . |
|---|---|---|---|---|---|
| ASXL1 | 20 | Whole gene | NOTCH1 | 9 | Indel: exon 0 |
| BRAF | 5 | Missense: position 600 (aa) | NPM1* | 5 | Indel: position 288 (aa) |
| CALR | 12 | Indel: positions 360 to 380 (aa) | NRAS | 1 | Missense: positions 12, 13, 61 (aa) |
| CBL | 11 | Whole gene | PPM1D | 17 | Whole gene |
| DNMT3A | 2 | Whole gene | PTPN11 | 12 | Whole gene |
| EZH2 | 7 | Whole gene | RUNX1 | 21 | Whole gene |
| FLT3 | 12 | Missense: position 835 (aa) and exons 9-11 | SETBP1 | 18 | Missense: position 870 (aa) |
| GNAS | 20 | Whole gene | SF3B1 | 2 | Missense: positions 625, 666, 700 (aa) |
| GNB1 | 1 | Missense: positions 57, 76, 80 (aa) | SMC1A | X | Whole gene |
| IDH1 | 2 | Missense: positions 132 (aa) | SMC3 | 10 | Whole gene |
| IDH2 | 15 | Missense: positions 140, 172 (aa) | SRSF2 | 17 | Missense: position 95 (aa) |
| JAK2 | 9 | Missense: position 617 (aa) | STAG2 | X | Whole gene |
| KIT* | 4 | Missense: position 816 (aa) | TET2 | 4 | Whole gene |
| KRAS | 12 | Missense: positions 12, 13, 61 (aa) | TP53 | 17 | Whole gene |
| MPL | 1 | Indel: exon 10 | U2AF1 | 21 | Missense: positions 34, 154 (aa) |
| MYD88 | 3 | Missense: position 265 (aa) | WT1* | 11 | Whole gene |
Any nonsynonymous, nonsense, or frameshift mutations in the coding regions of these genes will define ARCH. For some genes, the entire sequence needs to be determined because mutations are found throughout the entire coding region. For other genes, determining the sequence of specific hotspots may suffice.
Genes are optional to rule out leukemia because they were not reported in ARCH.
Therapy and follow-up
Currently, no therapy for ARCH has been suggested, and the topic should be further studied. However, because ARCH was associated with hematological malignancies and all-cause mortality and because this state is prevalent in the general population, it is important that suggestions regarding therapy for this diagnosis will be made. To allow better understanding of ARCH and how it should be treated, it is suggested which individuals should be tested for ARCH in a research setting (Figure 3).
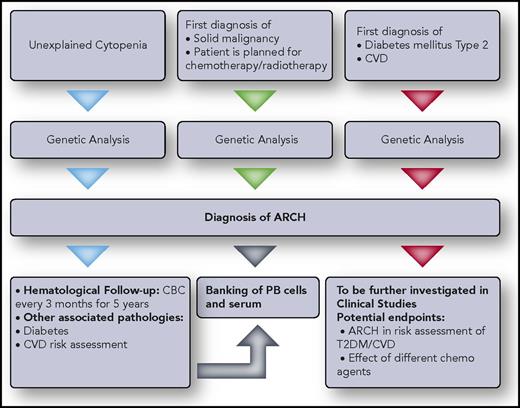
Clinical/research approach to ARCH. Recommendations for genetic testing for individuals with background disease are given. Individuals answering the inclusion criteria should undergo ARCH diagnostic testing and continue follow-up as recommended. Professional illustration by Somersault18:24.
Clinical/research approach to ARCH. Recommendations for genetic testing for individuals with background disease are given. Individuals answering the inclusion criteria should undergo ARCH diagnostic testing and continue follow-up as recommended. Professional illustration by Somersault18:24.
Summary
Humans carry the potential to live a longer life than they currently do. The major causes of death in the 21st century are vascular diseases and cancer, and remarkably, ARCH is associated with both, while no clear causation has been identified yet. ARCH is a heterogeneous entity with a common feature: the expansion of specific HSPCs and their progenies. Some ARCH-related mutations can increase the risk for leukemia, maybe others to heart disease and diabetes. Because ARCH diagnosis is based on calling small clones, rigorous sequencing methods should be used and standardized in the future. Not all CH is ARCH, and specific ARCH diagnosis is described here. Future studies should assume that although all ARCH-related mutations cause clonal expansion, each 1 of them could result in distinct phenotypes in diverse cell types, and in different individuals who carry diverse germline backgrounds, and have different background pathologies.
Acknowledgments
Thanks to Aviv de Morgan and all the Shlush lab members for thoughtful comments.
Authorship
Contribution: L.I.S. wrote the manuscript.
Conflict-of-interest disclosure: L.I.S. declares no competing financial interests.
Correspondence: Liran I. Shlush, Department of Immunology, Weizmann Institute of Science, Rehovot, Israel; e-mail: liran.shlush@weizmann.ac.il.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal