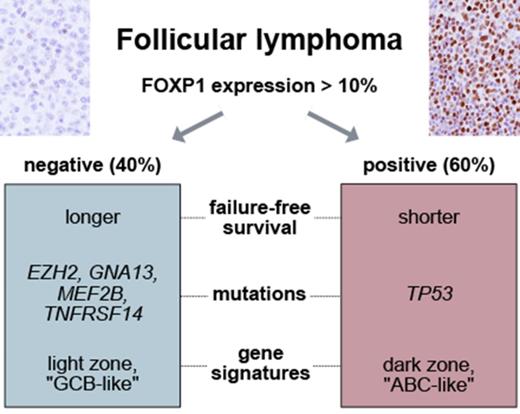
Key Points
High expression of FOXP1 predicts adverse FFS in patients with FL treated with immunochemotherapy.
FOXP1 high and low expressors differ in specific gene mutations and gene expression changes.
Abstract
Follicular lymphoma (FL) is a clinically and molecularly highly heterogeneous disease, yet prognostication relies predominantly on clinical tools. We recently demonstrated that integration of mutation status of 7 genes, including EZH2 and MEF2B, improves risk stratification. We mined gene expression data to uncover genes that are differentially expressed in EZH2- and MEF2B-mutated cases. We focused on FOXP1 and assessed its protein expression by immunohistochemistry (IHC) in 763 tissue biopsies. For outcome correlation, a population-based training cohort of 142 patients with FL treated with rituximab, cyclophosphamide, vincristine, and prednisone, and a clinical trial validation cohort comprising 395 patients treated with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) ± rituximab were used. We found FOXP1 to be significantly downregulated in both EZH2- and MEF2B-mutated cases. By IHC, 76 specimens in the training cohort (54%) had high FOXP1 expression (>10%), which was associated with reduced 5-year failure-free survival (FFS) rates (55% vs 70%). In the validation cohort, high FOXP1 expression status was observed in 248 patients (63%) and correlated with significantly shorter FFS in patients treated with R-CHOP (hazard ratio [HR], 1.95; P = .017) but not in patients treated with CHOP (HR, 1.15; P = .44). The impact of high FOXP1 expression on FFS in immunochemotherapy-treated patients was additional to the Follicular Lymphoma International Prognostic Index. High FOXP1 expression was associated with distinct molecular features such as TP53 mutations, expression of IRF4, and gene expression signatures reminiscent of dark zone germinal center or activated B cells. In summary, FOXP1 is a downstream phenotypic commonality of gene mutations and predicts outcome following rituximab-containing regimens.
Introduction
Follicular lymphoma (FL) is the second most common subtype of lymphoma,1 usually characterized by a slowly progressive disease course.2 However, most patients with FL present with advanced-stage disease and are considered to have an incurable illness with current standard immunochemotherapy regimens, because they will eventually experience progression and/or transformation to aggressive histologies. The translocation t(14;18)(q32;q21), resulting in the juxtaposition of the BCL2 gene under the control of the IGH promoter, is a genetic hallmark of FL, because it is present in 75% to 90% of cases.3,4 It has long been recognized that this translocation is insufficient for lymphomagenesis and that additional genetic alterations are required to develop overt FL.5-8 During the last few years, the advent of next-generation sequencing technology has led to dramatic improvements in our understanding of the genetics that underlie pathogenesis and disease evolution.9-14 Of particular relevance are genetic aberrations of histone modifiers and chromatin remodeling genes, which are among the most frequently mutated genes in FL and diffuse large B-cell lymphoma (DLBCL).14-17
Although FL is recognized as a clinically and molecularly highly heterogeneous disease, prognostication relies predominantly on clinical tools,18,19 and there is currently no consensus strategy that allows for risk-based treatment stratification. We have recently demonstrated that integration of the mutation status of 7 genes (including EZH2 and MEF2B) into a combined clinicogenetic risk model18 improves risk stratification and represents a promising approach to identify a subset of patients at highest risk of treatment failure.20 In particular, EZH2 mutations were associated with longer failure-free survival (FFS) in 2 independent cohorts of patients receiving immunochemotherapy, a finding that was recently validated in 2 additional series.21,22 EZH2 functions as a histone methyltransferase and is a component of the Polycomb repressive complex 2, mediating repression of gene expression by methylation of histone H3 on lysine 27 residues.23-26 EZH2 is frequently mutated in malignant lymphomas, and gain-of-function hotspot mutations (mostly affecting amino acid position Y646) have been identified in the germinal center B-cell–like (GCB) subtype of DLBCL and in FL.15,27-29 MEF2 proteins belong to a transcription factor family that has been shown to play a central role in the development of mesenchymal tissue, the central nervous system, and lymphoid cells.30 Virtually all MEF2B mutations detected in FL occur in the N-terminal domains, which are involved in DNA binding and homodimerization.16,31
The goals of the current study were to identify putative transcriptional targets of EZH2 and MEF2B and to assess their relevance for prognostication and molecular subtyping. We focused on FOXP1 because its expression was significantly downregulated in EZH2-mutated cases from 4 independent studies, as well as in MEF2B-mutated cases.20,21,28,29 We leveraged 2 large independent cohorts to assess the relationship between FOXP1 expression and outcome. In addition, we performed in-depth molecular characterization to answer the question whether select genotypic and phenotypic alterations are associated with FOXP1 expression. In aggregate, our findings illustrate the prognostic value of FOXP1 expression in FL, and open new avenues for pursuing distinct therapeutic targeting of vulnerabilities associated with FOXP1 expression status.
Methods
Patients and tissue biopsy materials
In total, we analyzed tissue biopsies from 763 patients with FL in this study. The training cohort consisted of 142 patients who had been treated with rituximab, cyclophosphamide, vincristine, and prednisone (R-CVP) at the BC Cancer Agency (BCCA) between 2003 and 2009. From 2006 onward, maintenance therapy with rituximab was given to those patients achieving a partial or a complete remission. Patients were retrospectively selected from the database of the Centre for Lymphoid Cancer at BCCA. The validation cohort consisted of 395 patients with advanced-stage FL who were prospectively included into trials from the German Low-Grade Lymphoma Study Group (GLSG). Patients were considered eligible for this study if they had been treated with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) as part of the GLSG1996 trial,32 or CHOP with rituximab (R-CHOP) or without rituximab as part of the GLSG2000 trial.33 In the latter trial, patients aged 60 years or younger achieving at least a partial remission could be randomly assigned to either total body irradiation, high-dose chemotherapy, and autologous stem cell transplantation (ASCT), or to maintenance therapy with interferon-α.33 In addition, for exploratory analyses, we used specimens from 350 BCCA patients, 124 of which overlapped with the aforementioned training cohort. All tissue biopsies were arranged in previously constructed tissue microarrays and were used for evaluation of FOXP1 protein expression by immunohistochemistry (IHC). All research described herein was approved by the respective Research Ethics Boards.
IHC and fluorescence in situ hybridization
IHC for FOXP1 was performed on tissue microarrays using a monoclonal mouse antibody against FOXP1 (clone JC12, ThermoFisher Scientific). IHC staining was performed on a Ventana BenchMark XT system (Ventana Medical Systems), and slides were independently evaluated by 2 hematopathologists (A.M. and P.F.). The percentage of tumor cells stained was recorded in 10% increments. All cases of the training cohort were scored again on a multiheaded microscope to derive 1 consensus score per case for cutoff determination. Discrepant cases (ie, positive vs negative) in the validation cohort were subjected to re-review and discussion of the cases on a multiheaded microscope until consensus was reached. In addition, the intensity of staining was recorded (weak, moderate, or strong). As a reference point for the assessment of staining intensity in the malignant cell population, we used small, reactive T cells in the tumor microenvironment, which typically display moderate to strong nuclear staining. We did not take staining intensity into consideration for outcome analyses.
Gene mutations and GSEA
Gene mutations and Illumina Whole-Genome DASL gene expression data were generated and analyzed as previously described.11,20 For gene expression analysis, we collapsed genes with multiple probes into 1 value per sample using the WGCNA R software package (version 1.51).34 Activated B-cell (ABC), GCB, and NF-κB signatures were obtained from the Web companion of Shaffer et al.35 We derived signatures from normal GC dark and light zone cells using the data set from Victora et al (National Center for Biotechnology Information's Gene Expression Omnibus data set GSE38697).36 Gene expression data were quantile normalized with use of the affy R package (version 1.52.0),37 and differential gene expression changes were determined with use of the limma R package (version 3.30.13).38 Genes were deemed to be differentially expressed if P value < .01 and absolute(log2(fold-change)) > 1. In order to test the association of gene mutations with FOXP1 expression, we performed pairwise Fisher's exact tests, with resulting P values adjusted for false discovery (fdr < 0.1). We performed gene set enrichment analysis (GSEA) using the Java-based Desktop application of GSEA (version 2.2.4).39 In order to ascertain deregulated pathways in low and high FOXP1 expressors, we performed differential gene expression analysis in extremes of FOXP1 expression (n = 58 with 0% FOXP1 expression and n = 30 with >50% FOXP1 expression). We used a P cutoff value of .05 for input into a custom R Reactome FI Cytoscape plugin wrapper.
Statistical analysis
To differentiate cases with high vs low FOXP1 expression, we chose the cutoff point that maximized the log-rank test statistic for FFS in the training cohort. The primary end point for this study was FFS, defined as the time between start of first induction treatment to either stable disease after first induction, progression, or death from any cause. For patients with stable disease after induction, progression was counted at the time of initiation of new treatment in the BCCA cohort, and at the time of documentation of stable disease that was considered an indication for second-line treatment in the GLSG cohort. In the validation cohort, patients receiving ASCT were censored for FFS at the stem cell reinfusion date. In a sensitivity analysis, we evaluated FFS without censoring for ASCT. Before testing the prognostic value of FOXP1 in the validation cohort, we performed a power calculation to determine whether enough events had been observed. Given 191 observed events for FFS, the power to detect a hazard ratio (HR) of at least 1.81 as observed in the training cohort was 98%. The effect of FOXP1 expression on FFS in the validation cohort was estimated with use of Cox regression analysis, also adjusting for binary Follicular Lymphoma International Prognostic Index (FLIPI; high vs low/intermediate) or for FLIPI factors. To account for different outcome according to use of rituximab in addition to CHOP during induction, we performed a formal interaction analysis of rituximab use and FOXP1 expression. We determined associations between categorical variables using Fisher’s exact test or the χ2 test for variables with more than 2 categories. We performed all analyses using R.40
Results
FOXP1 messenger RNA expression is downregulated in EZH2- and MEF2B-mutated cases
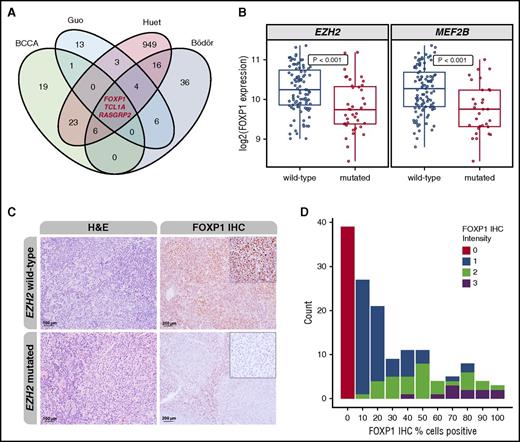
Because EZH2 mutations lead to increased repression of gene expression, we focused on genes that had lower expression in EZH2-mutated vs EZH2-wild-type cases. In our own gene expression data set20 and those from 3 additional studies,21,28,29 3 genes (FOXP1, TCL1A, and RASGRP2) were consistently downregulated in EZH2-mutated cases (Figure 1A-B). We concentrated on the forkhead transcription factor FOXP1 because assessment of its protein expression by IHC is widely available and has prognostic value in DLBCL.41 We had previously shown that MEF2B mutations are enriched in m7-FLIPI low cases, similar to EZH2 mutations.20 FOXP1 expression was also significantly downregulated in cases with mutated MEF2B (Figure 1B). In addition, we observed a highly significant overlap between genes downregulated in EZH2- and MEFB2-mutated cases, suggesting that the transcriptional footprints of these 2 genes mediate similar biological programs (P = 2.78 × 10−7; supplemental Table 1, available on the Blood Web site). Given the aforementioned associations, we hypothesized that high expression of FOXP1 correlates with adverse FFS.
EZH2 mutations are associated with transcriptional regulation of FOXP1. (A) Venn diagram displaying the respective overlaps of downregulated genes according to EZH2 mutation status. Three genes (FOXP1, TCL1A, and RASGRP2) were found to be consistently downregulated in EZH2-mutant cases. (B) Assessment of FOXP1 messenger RNA transcript levels showed significant downregulation of FOXP1 expression in EZH2- and MEF2B-mutated cases compared with wild-type. (C) Depicted are hematoxylin and eosin (H&E; left) and FOXP1 IHC (right) of an EZH2-unmutated and EZH2-mutated case, respectively. Original magnification ×200 (H&E) and ×100 (FOXP1 IHC; insets ×400). (D) Stacked bar plot showing the distribution of percentages of positively stained tumor cells and the staining intensity. Numbers on the y-axis represent the number of cases with the respective pattern.
EZH2 mutations are associated with transcriptional regulation of FOXP1. (A) Venn diagram displaying the respective overlaps of downregulated genes according to EZH2 mutation status. Three genes (FOXP1, TCL1A, and RASGRP2) were found to be consistently downregulated in EZH2-mutant cases. (B) Assessment of FOXP1 messenger RNA transcript levels showed significant downregulation of FOXP1 expression in EZH2- and MEF2B-mutated cases compared with wild-type. (C) Depicted are hematoxylin and eosin (H&E; left) and FOXP1 IHC (right) of an EZH2-unmutated and EZH2-mutated case, respectively. Original magnification ×200 (H&E) and ×100 (FOXP1 IHC; insets ×400). (D) Stacked bar plot showing the distribution of percentages of positively stained tumor cells and the staining intensity. Numbers on the y-axis represent the number of cases with the respective pattern.
FOXP1 staining in the training cohort
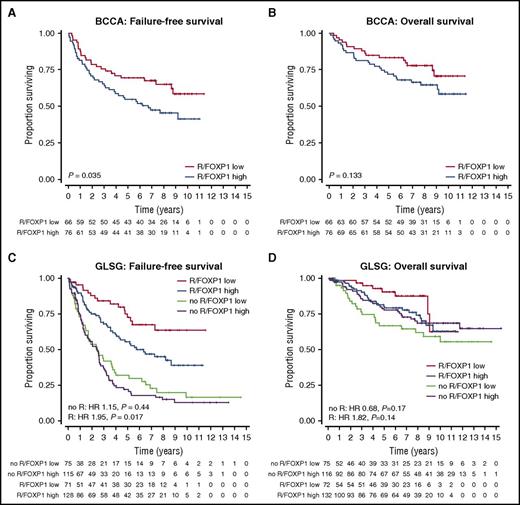
The training cohort consisted of 142 FL specimens from patients treated with R-CVP, with or without rituximab maintenance, and in which FOXP1 IHC (Figure 1C) was evaluable. Clinical characteristics are shown in supplemental Table 2. The distribution of percentage and intensity of staining is shown in Figure 1D. The cutoff value that best separated favorable from unfavorable FFS was determined to be >10% tumor cells expressing FOXP1 using the log-rank test statistic (P = .035). Based on this threshold, specimens from 76 patients in the training cohort (54%) had high FOXP1 expression. Clinical risk factors were similarly distributed among patients with FOXP1 high vs low expression, with the exception that elevated levels of lactate dehydrogenase (LDH) were more commonly observed in patients with high FOXP1 expression (27% vs 11%; P = .030) (Table 1). Median follow-up of living patients was 8.5 years, and 5-year FFS rates were 55% vs 70% for patients with high vs low FOXP1 expression, respectively (Figure 2A). The overall survival (OS) rate was lower in patients with high FOXP1 expression (72% vs 83%), but this difference was not statistically significant (P = .13) (Figure 2B).
Patient characteristics by FOXP1 expression status
| . | BCCA . | GLSG . | ||||||
|---|---|---|---|---|---|---|---|---|
| Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | |
| Age ≤60 y | 142 | 29 (44) | 41 (54) | 395 | 45 (31) | 67 (27) | ||
| Age >60 y | 37 (56) | 35 (46) | .245 | 102 (69) | 181 (73) | .42 | ||
| ECOG ≤1 | 140 | 54 (83) | 68 (91) | 389 | 138 (95) | 225 (93) | ||
| ECOG >1 | 11 (17) | 7 (9) | .100 | 8 (5) | 18 (7) | .60 | ||
| Stage I | 142 | 1 (2) | 1 (1) | .837 | 393 | — | — | <.001 |
| Stage II | 6 (9) | 6 (8) | — | — | ||||
| Stage III | 25 (38) | 24 (32) | 66 (45) | 68 (28) | ||||
| Stage IV | 34 (52) | 45 (59) | 81 (65) | 178 (72) | ||||
| No. of nodal sites ≤4 | 140 | 18 (28) | 16 (21) | 380 | 57 (40) | 54 (23) | ||
| No. of nodal sites >4 | 47 (72) | 59 (79) | .432 | 87 (60) | 182 (77) | <.001 | ||
| LDH normal | 135 | 55 (89) | 53 (73) | 387 | 112 (78) | 161 (66) | ||
| LDH elevated | 7 (11) | 20 (27) | .030 | 32 (22) | 82 (34) | .022 | ||
| Hemoglobin ≥120 g/L | 138 | 6 (9) | 10 (14) | 387 | 125 (86) | 181 (72) | ||
| Hemoglobin <120 g/L | 58 (81) | 64 (86) | .596 | 21 (14) | 70 (28) | .020 | ||
| FLIPI low/intermediate | 138 | 32 (49) | 36 (49) | 383 | 96 (67) | 110 (46) | ||
| FLIPI high | 33 (51) | 37 (51) | 1.000 | 47 (33) | 130 (54) | <.001 | ||
| m7-FLIPI low | 107 | 44 (85) | 39 (71) | 87 | 22 (81) | 41 (68) | ||
| m7-FLIPI high | 8 (15) | 16 (29) | .108 | 5 (19) | 19 (32) | .312 | ||
| R-chemotherapy* | 142 | 66 (100) | 76 (100) | 395 | 72 (49) | 132 (53) | ||
| Chemotherapy* | — | — | N/A | 75 (51) | 116 (47) | .48 | ||
| R maintenance | 142 | 57 (86) | 60 (79) | 395 | — | — | ||
| No R maintenance | 9 (14) | 16 (21) | .276 | 147 (100) | 248 (100) | N/A | ||
| . | BCCA . | GLSG . | ||||||
|---|---|---|---|---|---|---|---|---|
| Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | |
| Age ≤60 y | 142 | 29 (44) | 41 (54) | 395 | 45 (31) | 67 (27) | ||
| Age >60 y | 37 (56) | 35 (46) | .245 | 102 (69) | 181 (73) | .42 | ||
| ECOG ≤1 | 140 | 54 (83) | 68 (91) | 389 | 138 (95) | 225 (93) | ||
| ECOG >1 | 11 (17) | 7 (9) | .100 | 8 (5) | 18 (7) | .60 | ||
| Stage I | 142 | 1 (2) | 1 (1) | .837 | 393 | — | — | <.001 |
| Stage II | 6 (9) | 6 (8) | — | — | ||||
| Stage III | 25 (38) | 24 (32) | 66 (45) | 68 (28) | ||||
| Stage IV | 34 (52) | 45 (59) | 81 (65) | 178 (72) | ||||
| No. of nodal sites ≤4 | 140 | 18 (28) | 16 (21) | 380 | 57 (40) | 54 (23) | ||
| No. of nodal sites >4 | 47 (72) | 59 (79) | .432 | 87 (60) | 182 (77) | <.001 | ||
| LDH normal | 135 | 55 (89) | 53 (73) | 387 | 112 (78) | 161 (66) | ||
| LDH elevated | 7 (11) | 20 (27) | .030 | 32 (22) | 82 (34) | .022 | ||
| Hemoglobin ≥120 g/L | 138 | 6 (9) | 10 (14) | 387 | 125 (86) | 181 (72) | ||
| Hemoglobin <120 g/L | 58 (81) | 64 (86) | .596 | 21 (14) | 70 (28) | .020 | ||
| FLIPI low/intermediate | 138 | 32 (49) | 36 (49) | 383 | 96 (67) | 110 (46) | ||
| FLIPI high | 33 (51) | 37 (51) | 1.000 | 47 (33) | 130 (54) | <.001 | ||
| m7-FLIPI low | 107 | 44 (85) | 39 (71) | 87 | 22 (81) | 41 (68) | ||
| m7-FLIPI high | 8 (15) | 16 (29) | .108 | 5 (19) | 19 (32) | .312 | ||
| R-chemotherapy* | 142 | 66 (100) | 76 (100) | 395 | 72 (49) | 132 (53) | ||
| Chemotherapy* | — | — | N/A | 75 (51) | 116 (47) | .48 | ||
| R maintenance | 142 | 57 (86) | 60 (79) | 395 | — | — | ||
| No R maintenance | 9 (14) | 16 (21) | .276 | 147 (100) | 248 (100) | N/A | ||
Bold values indicate statistical significance.
ECOG, Eastern Cooperative Oncology Group performance status; N/A, not applicable; R, rituximab.
Chemotherapy refers to CVP for BCCA patients and CHOP for GLSG patients.
FOXP1 protein expression levels identify a group of patients with inferior failure-free survival. (A) In the training cohort, we applied log-rank statistics to determine the cutoff value of percent FOXP1-positive tumor cells at which the best separation between rates of FFS in 2 groups could be achieved. The cutoff point was subsequently locked at >10% and transferred to the validation cohort. Five-year FFS was 55% vs 70% in patients with high and low FOXP1 expression, respectively. (B) OS by FOXP1 expression status in the training cohort. Five-year OS was 72% vs 83% in patients with high and low FOXP1 expression, respectively. (C) FOXP1 protein expression was significantly associated with FFS in the validation cohort in R-CHOP–treated patients. Five-year FFS for high vs low FOXP1 expression was 22% vs 32% in CHOP-treated patients and 55% vs 72% in R-CHOP–treated patients. (D) No clear association between FOXP1 expression and OS was found in the validation cohort.
FOXP1 protein expression levels identify a group of patients with inferior failure-free survival. (A) In the training cohort, we applied log-rank statistics to determine the cutoff value of percent FOXP1-positive tumor cells at which the best separation between rates of FFS in 2 groups could be achieved. The cutoff point was subsequently locked at >10% and transferred to the validation cohort. Five-year FFS was 55% vs 70% in patients with high and low FOXP1 expression, respectively. (B) OS by FOXP1 expression status in the training cohort. Five-year OS was 72% vs 83% in patients with high and low FOXP1 expression, respectively. (C) FOXP1 protein expression was significantly associated with FFS in the validation cohort in R-CHOP–treated patients. Five-year FFS for high vs low FOXP1 expression was 22% vs 32% in CHOP-treated patients and 55% vs 72% in R-CHOP–treated patients. (D) No clear association between FOXP1 expression and OS was found in the validation cohort.
Correlation of FOXP1 staining with outcome in the validation cohort
We performed validation in an independent cohort of patients treated with R-CHOP or CHOP without rituximab within prospective phase 3 trials conducted by the GLSG. A total of 390 patients were evaluable for FFS (191 CHOP, 199 R-CHOP) and 395 patients for OS (191 CHOP, 204 R-CHOP). The validation cohort differed from the training cohort by several clinical risk factors, but neither by FLIPI nor by m7-FLIPI (supplemental Table 2). In the validation cohort, 248 patients (63%) had high expression of FOXP1, which was slightly higher than in the BCCA cohort (P = .058). Adverse clinical risk factors such as stage, number of nodal sites, LDH, and low hemoglobin levels were enriched in patients with high FOXP1 expression who, as a consequence, were more often categorized as having high-risk disease by FLIPI (54% vs 34%; P < .001) (Table 1).
Median follow-up was 7.8 years. The primary analysis with FFS and OS censored for ASCT and adjusted for rituximab revealed a significant association of high FOXP1 expression with unfavorable FFS (HR, 1.37; 95% confidence interval [CI], 1.02-1.86; P = .040), but not with OS (HR, 0.98; 95% CI, 0.63-1.53; P = .93). However, the interaction analysis revealed that high FOXP1 expression was associated with significantly shorter FFS in patients treated with R-CHOP (HR, 1.95; 95% CI, 1.13-3.38; P = .017), but not in patients treated with CHOP (HR, 1.15; 95% CI, 0.80-1.67; P = .44). Five-year FFS rates for high vs low FOXP1 expression were 22% vs 32% in CHOP-treated patients and 55% vs 72% in R-CHOP–treated patients, respectively (Figure 2C). No significant association was found for FOXP1 staining with OS, but we observed a potential effect of shortened OS with high FOXP1 expression among R-CHOP–treated patients (R-CHOP: HR, 1.82; 95% CI, 0.83-4.01; P = .14; CHOP: HR, 0.68; 95% CI, 0.39-1.18; P = .17), which was similar to the result observed in the training cohort (Figure 2D). We performed a sensitivity analysis in which those patients who underwent ASCT were not censored at the time of stem cell reinfusion. This analysis showed similar results for FFS (R-CHOP: HR, 1.81; 95% CI, 1.09-3.01; P = .022; CHOP: HR, 1.19; 95% CI, 0.84-1.68; P = .34) and OS (R-CHOP: HR, 1.43; 95% CI, 0.72-2.86; P = .31; CHOP: HR, 0.73; 95% CI, 0.43-1.23; P = .23).
Multivariate analyses
In order to take potential confounding by clinical covariates into account, we performed separate multivariate analyses in the 2 outcome cohorts (Table 2). In BCCA patients, high FOXP1 expression remained significantly associated with inferior FFS in a multivariate Cox regression model including rituximab maintenance (by intent to treat) and FLIPI high vs low/intermediate (HR, 1.84; 95% CI, 1.10-3.08; P = .021). In a similar manner, in the GLSG cohort, high FOXP1 expression was significantly associated with poor FFS in patients treated with R-CHOP, after adjustment for rituximab, binary FLIPI, and the interaction of FOXP1 with rituximab (HR, 1.84; 95% CI, 1.05-3.24; P = .035). No significant association was observed in patients treated with CHOP (HR, 1.06; 95% CI, 0.73-1.54; P = .76). Adjustment for individual FLIPI factors yielded qualitatively similar results in both the BCCA and GLSG cohorts (supplemental Table 3).
Multivariate analyses
| Cohort . | FFS . | OS . | |||||
|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||
| BCCA* | FOXP1 high | 1.84 | 1.10-3.08 | .021 | 2.04 | 1.08-3.86 | .029 |
| R maintenance | 0.89 | 0.49-1.61 | .700 | 1.53 | 0.69-3.38 | .292 | |
| FLIPI high risk | 2.19 | 1.31-3.66 | .003 | 4.23 | 2.07-8.61 | <.001 | |
| GLSG model without FLIPI† | FOXP1 high (no R) | 1.15 | 0.80-1.67 | .44 | 0.68 | 0.39-1.18 | .17 |
| FOXP1 high (R) | 1.95 | 1.13-3.38 | .017 | 1.82 | 0.83-4.01 | .14 | |
| First-line rituximab (FOXP1 low) | 0.25 | 0.14-0.44 | <.001 | 0.37 | 0.16-0.83 | .016 | |
| First-line rituximab (FOXP1 high) | 0.42 | 0.30-0.60 | <.001 | 0.99 | 0.58-1.67 | .96 | |
| FOXP1:rituximab (interaction term) | 1.69 | 0.98-3.27 | .12 | 2.68 | 1.02-7.02 | .045 | |
| GLSG model with FLIPI‡ | FOXP1 high (no R) | 1.06 | 0.73-1.54 | .76 | 0.62 | 0.36-1.08 | .092 |
| FOXP1 high (R) | 1.84 | 1.05-3.24 | .035 | 1.38 | 0.62-3.08 | .43 | |
| First-line rituximab (FOXP1 low) | 0.23 | 0.13-0.42 | <.001 | 0.38 | 0.17-0.86 | .019 | |
| First-line rituximab (FOXP1 high) | 0.41 | 0.28-0.58 | <.001 | 0.85 | 0.49-1.46 | .55 | |
| FOXP1:rituximab (interaction term) | 1.74 | 0.89-3.40 | .11 | 2.23 | 0.85-5.90 | .10 | |
| FLIPI high risk | 1.62 | 1.21-2.17 | .0014 | 2.78 | 1.74-4.43 | <.001 | |
| Cohort . | FFS . | OS . | |||||
|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||
| BCCA* | FOXP1 high | 1.84 | 1.10-3.08 | .021 | 2.04 | 1.08-3.86 | .029 |
| R maintenance | 0.89 | 0.49-1.61 | .700 | 1.53 | 0.69-3.38 | .292 | |
| FLIPI high risk | 2.19 | 1.31-3.66 | .003 | 4.23 | 2.07-8.61 | <.001 | |
| GLSG model without FLIPI† | FOXP1 high (no R) | 1.15 | 0.80-1.67 | .44 | 0.68 | 0.39-1.18 | .17 |
| FOXP1 high (R) | 1.95 | 1.13-3.38 | .017 | 1.82 | 0.83-4.01 | .14 | |
| First-line rituximab (FOXP1 low) | 0.25 | 0.14-0.44 | <.001 | 0.37 | 0.16-0.83 | .016 | |
| First-line rituximab (FOXP1 high) | 0.42 | 0.30-0.60 | <.001 | 0.99 | 0.58-1.67 | .96 | |
| FOXP1:rituximab (interaction term) | 1.69 | 0.98-3.27 | .12 | 2.68 | 1.02-7.02 | .045 | |
| GLSG model with FLIPI‡ | FOXP1 high (no R) | 1.06 | 0.73-1.54 | .76 | 0.62 | 0.36-1.08 | .092 |
| FOXP1 high (R) | 1.84 | 1.05-3.24 | .035 | 1.38 | 0.62-3.08 | .43 | |
| First-line rituximab (FOXP1 low) | 0.23 | 0.13-0.42 | <.001 | 0.38 | 0.17-0.86 | .019 | |
| First-line rituximab (FOXP1 high) | 0.41 | 0.28-0.58 | <.001 | 0.85 | 0.49-1.46 | .55 | |
| FOXP1:rituximab (interaction term) | 1.74 | 0.89-3.40 | .11 | 2.23 | 0.85-5.90 | .10 | |
| FLIPI high risk | 1.62 | 1.21-2.17 | .0014 | 2.78 | 1.74-4.43 | <.001 | |
Bold values indicate statistical significance.
R, rituximab.
n = 138 patients/64 events for FFS, 43 events for OS.
n = 390 patients/191 events for FFS, n = 395 patients/86 events for OS.
n = 381 patients/189 events for FFS, n = 383 patients/84 events for OS.
In an exploratory analysis, we assessed the relationship of FOXP1 IHC with the m7-FLIPI clinicogenetic risk model. High FOXP1 expression was more common in m7-FLIPI high-risk patients, but this association did not reach statistical significance (Table 1). To determine whether FOXP1 expression adds prognostic information independently of the m7-FLIPI, we performed multivariate analyses. Both variables were available for 107 patients from the BCCA cohort and 87 patients from the GLSG cohort. In these subgroups, the HR for FFS with high FOXP1 expression, when adjusted for binary m7-FLIPI status, was 1.74 in both cohorts (BCCA: 95% CI, 0.98-3.08; P = .059; GLSG: 95% CI, 0.79-3.11; P = .203).
Molecular characteristics by FOXP1 expression
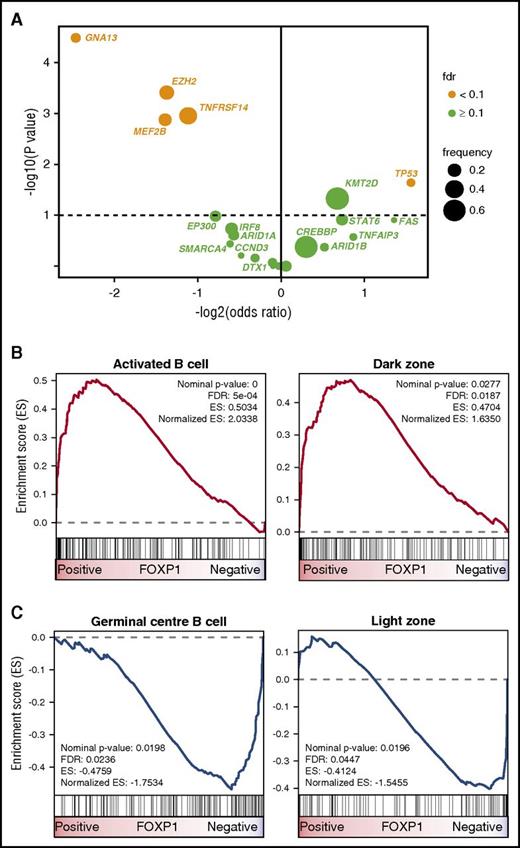
The correlation of high FOXP1 expression with unfavorable FFS in R-chemotherapy–treated patients prompted us to assess whether FOXP1 expression is a phenotypic marker underlying distinct biological FL subsets. First, we determined in a pairwise fashion whether recurrent gene mutations, in addition to EZH2 and MEF2B, are enriched in FOXP1 high or low expressors. Considering only genes that were mutated in >5% of patients in the data sets of either Pastore et al.20 or Kridel et al.11 (n = 22 genes; n = 324 FL samples) and adjusting for false discovery rate, we found 3 additional gene mutations to be significantly associated with FOXP1 expression: GNA13 and TNFRSF14 mutations were enriched in cases with low FOXP1 expression (odds ratio [OR], 0.18 and 0.46, respectively), whereas TP53 mutations were more commonly observed in cases with high FOXP1 expression (OR, 2.94) (Figure 3A). Several pathological characteristics were unevenly distributed across the FOXP1 expression spectrum, with high FOXP1 expression being significantly associated with the absence of BCL2 translocations and the presence of IRF4 protein expression. The association between FOXP1 and IRF4 expression was validated, whereas the association between FOXP1 expression and BCL2 translocation status did not reach statistical significance in GLSG patients (Table 3). A previous study reported that distinct subtypes exist in FL, one resembling normal centrocytes, and another one more closely related to in vitro–activated B cells.42 This finding led us to explore whether signatures indicative of discrete GC and post-GCB stages were associated with FOXP1 expression. GSEA showed that an ABC signature and a signature derived from normal GC dark zone cells were significantly enriched in cases with high FOXP1 expression (Figure 3B). Conversely, a GCB signature and genes related to normal GC light zone cells were significantly enriched in patients with low FOXP1 expression (Figure 3C). A broader analysis of gene expression changes in FOXP1 high vs low expressors showed enrichment of multiple gene sets related to cell cycle in FOXP1 high expressors (supplemental Table 2). Taken together, these observations suggest that FOXP1 expression delineates FL cases with distinct genomic and phenotypic characteristics.
FOXP1 expression identifies FL with distinct molecular features. (A) Volcano plot displaying gene mutations associated with FOXP1 expression. Only genes that were mutated in >5% of patients in the data sets of either Pastore et al20 or Kridel et al11 (n = 22 genes; n = 324 FL samples) were considered. After adjustment for false discovery, 5 gene mutations were found to be significantly associated with FOXP1 expression. GNA13, EZH2, TNFRSF14, and MEF2B mutations were enriched in cases with low FOXP1 expression (ORs: 0.18, 0.39, 0.46, and 0.38, respectively), whereas TP53 mutations were more commonly observed in cases with high FOXP1 expression (OR, 2.94). (B) GSEA shows that an activated B-cell signature and a signature derived from normal GC dark zone cells were significantly enriched in cases with high FOXP1 expression. (C) Conversely, a GCB signature and genes related to normal GC light zone cells were significantly enriched in cases with low FOXP1 expression. ES, enrichment score.
FOXP1 expression identifies FL with distinct molecular features. (A) Volcano plot displaying gene mutations associated with FOXP1 expression. Only genes that were mutated in >5% of patients in the data sets of either Pastore et al20 or Kridel et al11 (n = 22 genes; n = 324 FL samples) were considered. After adjustment for false discovery, 5 gene mutations were found to be significantly associated with FOXP1 expression. GNA13, EZH2, TNFRSF14, and MEF2B mutations were enriched in cases with low FOXP1 expression (ORs: 0.18, 0.39, 0.46, and 0.38, respectively), whereas TP53 mutations were more commonly observed in cases with high FOXP1 expression (OR, 2.94). (B) GSEA shows that an activated B-cell signature and a signature derived from normal GC dark zone cells were significantly enriched in cases with high FOXP1 expression. (C) Conversely, a GCB signature and genes related to normal GC light zone cells were significantly enriched in cases with low FOXP1 expression. ES, enrichment score.
Pathological characteristics in FOXP1 negative and positive FL cases
| . | BCCA . | GLSG . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . |
| Grade 1-2 | 252 | 91 (85) | 126 (87) | — | — | — | — | |
| Grade 3A | 16 (15) | 19 (12) | .715 | |||||
| BCL2 translocation negative | 221 | 7 (8) | 26 (20) | 322 | 14 (11) | 37 (18) | ||
| BCL2 translocation positive | 86 (92) | 102 (80) | .012 | 106 (89) | 165 (82) | .155 | ||
| BCL6 translocation negative | 232 | 84 (88) | 110 (81) | — | — | — | — | |
| BCL6 translocation positive | 12 (13) | 26 (19) | .210 | |||||
| MYC translocation negative | 197 | 77 (96) | 108 (92) | — | — | — | — | |
| MYC translocation positive | 3 (4) | 9 (8) | .366 | |||||
| CD10 IHC negative | 223 | 7 (7) | 11 (8) | — | — | — | — | |
| CD10 IHC positive | 99 (93) | 134 (92) | .810 | |||||
| BCL6 IHC negative | 252 | 4 (4) | 5 (3) | — | — | — | — | |
| BCL6 IHC positive | 103 (96) | 140 (97) | 1.000 | |||||
| IRF4 IHC negative | 252 | 106 (98) | 127 (88) | 359 | 123 (91) | 174 (78) | ||
| IRF4 IHC positive | 1 (2) | 18 (12) | .002 | 12 (9) | 50 (22) | .001 | ||
| TP53 IHC negative | 252 | 106 (99) | 139 (96) | — | — | — | — | |
| TP53 IHC positive | 1 (1) | 6 (4) | .244 | |||||
| . | BCCA . | GLSG . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . | Evaluable . | FOXP1 low, n (%) . | FOXP1 high, n (%) . | P . |
| Grade 1-2 | 252 | 91 (85) | 126 (87) | — | — | — | — | |
| Grade 3A | 16 (15) | 19 (12) | .715 | |||||
| BCL2 translocation negative | 221 | 7 (8) | 26 (20) | 322 | 14 (11) | 37 (18) | ||
| BCL2 translocation positive | 86 (92) | 102 (80) | .012 | 106 (89) | 165 (82) | .155 | ||
| BCL6 translocation negative | 232 | 84 (88) | 110 (81) | — | — | — | — | |
| BCL6 translocation positive | 12 (13) | 26 (19) | .210 | |||||
| MYC translocation negative | 197 | 77 (96) | 108 (92) | — | — | — | — | |
| MYC translocation positive | 3 (4) | 9 (8) | .366 | |||||
| CD10 IHC negative | 223 | 7 (7) | 11 (8) | — | — | — | — | |
| CD10 IHC positive | 99 (93) | 134 (92) | .810 | |||||
| BCL6 IHC negative | 252 | 4 (4) | 5 (3) | — | — | — | — | |
| BCL6 IHC positive | 103 (96) | 140 (97) | 1.000 | |||||
| IRF4 IHC negative | 252 | 106 (98) | 127 (88) | 359 | 123 (91) | 174 (78) | ||
| IRF4 IHC positive | 1 (2) | 18 (12) | .002 | 12 (9) | 50 (22) | .001 | ||
| TP53 IHC negative | 252 | 106 (99) | 139 (96) | — | — | — | — | |
| TP53 IHC positive | 1 (1) | 6 (4) | .244 | |||||
Bold values indicate statistical significance.
Discussion
Although the clinicogenetic risk stratifier m7-FLIPI was shown to be a promising approach to delineate patient populations at differential risk of treatment failure, the mechanism underlying the association between mutation of the 7 genes included in the m7-FLIPI and outcome has, thus far, not been elucidated.20 Moreover, determination of m7-FLIPI status relies on mutational analysis, which may not be routinely available to most clinicians. Herein, we extended the prognostic value of individual gene mutations to FOXP1 protein expression, a downstream phenotypic surrogate marker of EZH2 and/or MEF2B wild-type status. FOXP1 is a forkhead transcription factor that regulates normal B-cell development and is expressed in mantle zone rather than GC B cells.43,44 FOXP1 is also overexpressed in a subset of B-cell lymphomas, including the ABC subtype of DLBCL, and in lymphomas originating from mucosa-associated lymphoid tissue.45,46 Whereas FOXP1 correlates with inferior survival in DLBCL in most studies, possibly due to its association with the ABC subtype,45,47,48 its prognostic and correlative implications had not been comprehensively explored in FL.
In the present study, we showed that FOXP1 expression is surprisingly common in FL, in keeping with a small series of 13 FL cases reported by Brown et al.49 We report that high FOXP1 expression was associated with adverse FFS in 2 independent FL cohorts treated with upfront immunochemotherapy: a population-based series of patients treated with R-CVP (and rituximab maintenance in 82% of patients) from the BCCA and a GLSG clinical trial cohort treated with R-CHOP, but without rituximab maintenance. Outcomes of patients with high and low expression of FOXP1 were similar between the BCCA cohort and the GLSG R-CHOP–treated patients, despite differences in clinical characteristics and treatment between these 2 cohorts. On the other hand, FOXP1 did not separate distinct outcome groups in patients treated without rituximab, which suggests that FOXP1 is a predictive biomarker for outcome with immunochemotherapy.50 We documented that assessment of FOXP1 expression adds prognostic information in immunochemotherapy-treated patients, beyond well-established clinical risk factors. Indeed, in multivariate analyses, high FOXP1 expression remained significantly associated with FFS in such patients after adjustment for the FLIPI. OS was not significantly associated with FOXP1 expression, which may be explained by the effect of subsequent therapies after initial immunochemotherapy, or by insufficient statistical power to detect an OS effect. Although challenges with reproducibility of IHC are certainly recognized,51,52 it has advantages compared with genetic testing, being an indispensable part of the standard diagnostic lymphoma workup that can be performed at low cost and with rapid turnaround time. In addition, we show that IHC for FOXP1 serves as a phenotypic readout that integrates the effects from multiple upstream aberrations. However, our study was not sufficiently powered to determine whether FOXP1 expression adds prognostic information that is independent of the m7-FLIPI. This will have to be evaluated in larger data sets.
Reported mechanisms of FOXP1 overexpression include IGH and non-IGH rearrangements,53,54 trisomy of chromosome 3, and focal high-level amplifications.41 Conversely, microRNAs miR-34a and miR-150 have been reported to downregulate FOXP1 expression.55-57 Our data demonstrate a novel, epigenetic mechanism of regulation of FOXP1 function, as EZH2 mutations are robustly associated with decreased FOXP1 expression. In the B-cell lymphoma context, FOXP1 expression was shown to repress proapoptotic genes58 ; promote Wnt signaling59 ; suppress MHC class II expression60,61 ; and modulate B-cell surface markers such as CD19,60,61 which potentially underlie treatment resistance and contribute to early progression. Dekker et al61 showed that FOXP1 represses the expression of genes defining the GCB identity, while favoring the expression of genes contributing to plasma cell differentiation. It is tempting to speculate that low FOXP1 expression is characteristic of a phenotype that is more sensitive to rituximab, potentially explaining the lack of prognostic effect in patients treated without rituximab. Future studies are warranted to explore whether FOXP1 expression might rationally guide the use of anti-CD20–directed therapy, for example, with regard to the indication for maintenance therapy.
Lastly, we leveraged available patient data sets to correlate FOXP1 protein expression with well-characterized genetic and phenotypic alterations. We found that cases with low FOXP1 expression were enriched for GNA13 and TNFRSF14 mutations, whereas high FOXP1 expressers were more frequently positive for IRF4 and more frequently had TP53 mutation. Koues et al42 had previously identified distinct FL subtypes, with pathogenic regulatory circuitries resembling either centrocytes (subtype 1) or in vitro–activated B cells (subtype 2). Using GSEA, we showed that GCB and light zone signatures were enriched in patients with low FOXP1 expression, whereas ABC and dark zone signatures were enriched in those with high FOXP1 expression. Phenotypes of FOXP1 high and low expression are thus reflective of distinct sets of molecular alterations. Moreover, our findings are hypothesis-generating because they suggest that alternative treatment strategies should be explored in patients with high FOXP1 expression.
Our results illustrate the value of understanding the intricate relationships between lymphoma biology and response to treatment, at once providing a predictive biomarker to guide patient management and highlighting discrete biological contrasts within the heterogeneous molecular landscape that is inherent to FL. Our study shows that FOXP1 is a marker of higher-risk FL in patients receiving R-CVP and R-CHOP, but additional independent studies in other cohorts are needed to document whether such an outcome correlation holds true with regimens that contain other anti-CD20 antibodies (eg, obinutuzumab) and/or chemotherapy backbone (ie, bendamustine).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Program Project Grant from the Terry Fox Research Institute (http://www.tfri.ca, Grant No. 1023) (C.S., J.M.C., and R.D.G.); the Mildred-Scheel-Foundation for Cancer Research (Deutsche Krebshilfe) and the Michael Smith Foundation for Health Research and Lymphoma Canada (A.M.); the Max-Eder Program of the German Cancer Aid (110659) (O.W.); the German Research Foundation (DFG-SFB/CRC-1243, TP-A11); the Princess Margaret Cancer Centre; and the Leukemia & Lymphoma Society of Canada (R.K.).
Authorship
Contribution: A.M., V.J., E.H., O.W., and R.K. designed and performed the research, analyzed the data, and wrote the manuscript; P.F., E.L., and M.B. performed the research; and A.R., G.O., H.H., W.K., W.H., C.S., J.M.C., L.H.S., and R.D.G. collected and contributed samples and data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Kridel, Division of Medical Oncology and Hematology, Princess Margaret Cancer Centre, 610 University Ave, OPG 6th Floor, Suite 6-714, Toronto, ON M5G 2M9, Canada; e-mail: robert.kridel@uhn.ca.
References
Author notes
A.M., V.J., E.H., O.W., and R.K. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal