Key Points
tMNs after ASCT originate from HSCs bearing (pre-)tMN mutations that are present years before disease onset.
Post-ASCT treatment can influence selection and outgrowth of (pre)leukemic clones.
Abstract
Therapy-related myeloid neoplasms (tMNs) are severe adverse events that can occur after treatment with autologous hematopoietic stem cell transplantation (ASCT). This study aimed to investigate the development of tMNs following ASCT at the molecular level by whole-exome sequencing (WES) and targeted deep sequencing (TDS) in sequential (pre-) tMN samples. WES identified a significantly higher number of mutations in tMNs as compared with de novo myelodysplastic syndrome (MDS) (median 27 vs 12 mutations; P = .001). The mutations found in tMNs did not carry a clear aging-signature, unlike the mutations found in de novo MDS, indicating a different mutational mechanism. In some patients, tMN mutations were identified in both myeloid and T cells, suggesting that tMNs may originate from early hematopoietic stem cells (HSCs). However, the mutational spectra of tMNs and the preceding malignancies did not overlap, excluding common ancestry for these malignancies. By use of TDS, tMN mutations were identified at low variant allele frequencies (VAFs) in transplant material in 70% of the patients with tMNs. Reconstruction of clonal patterns based on VAFs revealed that premalignant clones can be present more than 7 years preceding a tMN diagnosis, a finding that was confirmed by immunohistochemistry on bone marrow biopsies. Our results indicate that tMN development after ASCT originates in HSCs bearing (pre-)tMN mutations that are present years before disease onset and that post-ASCT treatment can influence the selection of these clones. Early detection of premalignant clones and monitoring of their evolutionary trajectory may help to predict the development of tMNs and guide early intervention in the future.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 1877.
Disclosures
CME questions author Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Pfizer. Associate Editor Margaret A. Goodell and the authors declare no competing financial interests.
Learning objectives
Compare mutations in therapy-related myeloid neoplasms (tMNs) with those in de novo myelodysplastic syndrome, based on a case series studied using whole-exome sequencing and targeted deep sequencing in sequential (pre-)tMN samples.
Describe origination and development of tMN after autologous stem cell transplantation (ASCT).
Determine the clinical implications of molecular findings in development of tMNs after ASCT.
Release date: April 19, 2018; Expiration date: April 19, 2019
Introduction
Therapy-related acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS), together therapy-related myeloid neoplasms (tMNs), represent ∼10% to 20% of all AML and MDS cases.1 This serious condition is regarded as a late complication following cytotoxic exposure, including both chemotherapy and irradiation therapy.2 In particular, prior treatment with autologous hematopoietic stem cell transplantation (ASCT) increases the risk for tMN development significantly, with an estimated incidence of 5% to 10% at 10 years after transplantation.3-5 The World Health Organization recognizes tMN as a distinct disease entity,6 characterized by a high frequency of TP53 mutations; unfavorable karyotypes; and a poor response to conventional chemotherapeutic treatment, resulting in a very dismal prognosis.7,8
The use of highly sensitive next-generation sequencing techniques (NGS) has enabled identification of clonal hematopoiesis of indeterminate potential (CHIP) in healthy individuals; several genes, among which DNMT3A, SRSF2, SF3B1, TET2, and ASXL1 were found to be recurrently mutated at low variant allele frequencies (VAFs) in otherwise healthy adults.9-13 The presence of CHIP increases the risk of developing hematological neoplasms with a rate of 0.5% to 1% per year.14 It is interesting to note that recent studies reported that in 62% to 80% of the patients who developed a tMN after ASCT, CHIP was present at the time of transplantation.3,5,15 These findings raise the question of how CHIP can undergo malignant transformation to tMNs. Chemotherapy might lead to additional mutations that preleukemic cells need for transformation. Otherwise, chemotherapeutic treatment may create an evolutionary bottleneck, after which hematopoietic repopulation may occur by the fittest hematopoietic stem and progenitor cells (HSPCs) harboring preexisting mutations.
In this report, we studied a cohort of patients who developed tMN after ASCT by performing whole-exome sequencing (WES), targeted deep sequencing (TDS), and immunohistochemistry. We studied the mutational landscape of tMNs and the clonal dynamics preceding tMN diagnosis, thereby providing further insight into the mechanism of tMN development after ASCT.
Materials and methods
Patient samples
We studied 18 patients in whom tMN developed between 1999 and 2013; 17 patients were diagnosed with tMN after ASCT and 1 patient after peripheral blood stem cell (PBSC) apheresis. ASCT conditioning consisted of high-dose melphalan for patients with plasma cell dyscrasia (either multiple myeloma or amyloidosis, n = 5)16 or BEAM (carmustine, etoposide, cytarabine, and melphalan) combination therapy for patients with lymphoma (n = 13)17 (Table 1; supplemental Tables 1-3, available on the Blood Web site). A control group consisted of matched patients who did not develop tMN after ASCT (supplemental Methods). Bone marrow (BM), peripheral blood (PB), PBSCs, and lymphoma (L) samples from patients with tMNs were biobanked after informed consent for investigational use. The study was conducted in accordance with the Declaration of Helsinki and institutional guidelines and regulations. Morphologic and cytogenetic analyses were performed according to standard procedures.
Patient characteristics
| UPN . | Sex . | Age at ASCT . | Indication ASCT . | Therapy pre-ASCT . | ASCT conditioning . | Relapse post-ASCT . | Intensive therapy post-ASCT . | Interval to tMN (y) . | WHO . | Transformation to AML . | % blasts at tMN diagnosis . | Cytogenetics at tMN diagnosis . | Survival (y) after tMN diagnosis . | Cause of death . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPN01 | M | 62 | AL-amyloidosis | ch | HDM | 6.3 | MDS | Yes | 2 | Complex | 0.5 | tMN | ||
| UPN02 | F | 64 | NHL-R | ch | BEAM | 1.5 | AML | 23 | Complex | 0.2 | tMN | |||
| UPN03 | M | 49 | MM | ch | HDM | Yes | Yes | 7.3 | AML | 21 | Complex | 0.3 | tMN | |
| UPN04 | M | 40 | HL-R | ch + irr | BEAM | 0.8 | MDS | 3 | t(3;21) | 0.9 | Compl. allo-SCT | |||
| UPN06 | F | 40 | NHL-R | ch | BEAM | Yes | 2.0 | MDS | 1 | NK | 0.3 | Compl. allo-SCT | ||
| UPN07 | M | 50 | NHL-R | ch + irr | BEAM | Yes | Yes | 12.6 | MDS | Yes | 3 | NK | 0.3 | Compl. allo-SCT |
| UPN08 | F | 36 | HL | ch + irr | BEAM | 5.8 | MDS | 2 | −7 | n.a. | n.a. | |||
| UPN10 | M | 52 | NHL-R | ch | BEAM | Yes | 1.4 | MDS | Yes | 1 | Complex | 0.8 | tMN | |
| UPN11 | M | 47 | MM | ch | HDM | Yes | 2.9 | MDS | 4 | n.a. | 0.1 | tMN | ||
| UPN12 | M | 56 | NHL | ch | BEAM | 3.0 | MDS | 1 | -Y and −7 | 2.4 | Compl. allo-SCT | |||
| UPN13 | M | 62 | NHL-R | ch | BEAM | 0.8 | MDS | 3 | −7 | 1.3 | Compl. allo-SCT | |||
| UPN14 | F | 64 | NHL-R | ch +irr | BEAM | 1.3 | MDS | 1 | n.a. | 0.5 | tMN | |||
| UPN15 | F | 53 | NHL-R | ch | BEAM | Yes | Yes | 7.3 | MDS | 1 | −7 | n.a. | n.a. | |
| UPN17 | M | 70 | NHL-R | ch +irr | BEAM | 1.1 | MDS | 2 | Complex | 0.1 | tMN | |||
| UPN18 | F | 64 | AL-amyloidosis | ch | 1.2* | MDS | Yes | 2 | −7 | 2.3 | tMN | |||
| UPN19 | M | 54 | NHL-R | ch | BEAM | Yes† | Yes | 16.2 | AML | 20 | Complex | 0.5 | tMN | |
| UPN20 | M | 52 | NHL-R | ch | BEAM | 7.3 | MDS | 3 | Complex | 1.4 | Compl. allo-SCT | |||
| UPN21 | M | 62 | MM | ch +irr | HDM | Yes | Yes | 6.8 | MDS | 3 | Complex | 0.5 | tMN |
| UPN . | Sex . | Age at ASCT . | Indication ASCT . | Therapy pre-ASCT . | ASCT conditioning . | Relapse post-ASCT . | Intensive therapy post-ASCT . | Interval to tMN (y) . | WHO . | Transformation to AML . | % blasts at tMN diagnosis . | Cytogenetics at tMN diagnosis . | Survival (y) after tMN diagnosis . | Cause of death . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPN01 | M | 62 | AL-amyloidosis | ch | HDM | 6.3 | MDS | Yes | 2 | Complex | 0.5 | tMN | ||
| UPN02 | F | 64 | NHL-R | ch | BEAM | 1.5 | AML | 23 | Complex | 0.2 | tMN | |||
| UPN03 | M | 49 | MM | ch | HDM | Yes | Yes | 7.3 | AML | 21 | Complex | 0.3 | tMN | |
| UPN04 | M | 40 | HL-R | ch + irr | BEAM | 0.8 | MDS | 3 | t(3;21) | 0.9 | Compl. allo-SCT | |||
| UPN06 | F | 40 | NHL-R | ch | BEAM | Yes | 2.0 | MDS | 1 | NK | 0.3 | Compl. allo-SCT | ||
| UPN07 | M | 50 | NHL-R | ch + irr | BEAM | Yes | Yes | 12.6 | MDS | Yes | 3 | NK | 0.3 | Compl. allo-SCT |
| UPN08 | F | 36 | HL | ch + irr | BEAM | 5.8 | MDS | 2 | −7 | n.a. | n.a. | |||
| UPN10 | M | 52 | NHL-R | ch | BEAM | Yes | 1.4 | MDS | Yes | 1 | Complex | 0.8 | tMN | |
| UPN11 | M | 47 | MM | ch | HDM | Yes | 2.9 | MDS | 4 | n.a. | 0.1 | tMN | ||
| UPN12 | M | 56 | NHL | ch | BEAM | 3.0 | MDS | 1 | -Y and −7 | 2.4 | Compl. allo-SCT | |||
| UPN13 | M | 62 | NHL-R | ch | BEAM | 0.8 | MDS | 3 | −7 | 1.3 | Compl. allo-SCT | |||
| UPN14 | F | 64 | NHL-R | ch +irr | BEAM | 1.3 | MDS | 1 | n.a. | 0.5 | tMN | |||
| UPN15 | F | 53 | NHL-R | ch | BEAM | Yes | Yes | 7.3 | MDS | 1 | −7 | n.a. | n.a. | |
| UPN17 | M | 70 | NHL-R | ch +irr | BEAM | 1.1 | MDS | 2 | Complex | 0.1 | tMN | |||
| UPN18 | F | 64 | AL-amyloidosis | ch | 1.2* | MDS | Yes | 2 | −7 | 2.3 | tMN | |||
| UPN19 | M | 54 | NHL-R | ch | BEAM | Yes† | Yes | 16.2 | AML | 20 | Complex | 0.5 | tMN | |
| UPN20 | M | 52 | NHL-R | ch | BEAM | 7.3 | MDS | 3 | Complex | 1.4 | Compl. allo-SCT | |||
| UPN21 | M | 62 | MM | ch +irr | HDM | Yes | Yes | 6.8 | MDS | 3 | Complex | 0.5 | tMN |
Interval to tMN represents interval between ASCT and diagnosis of tMN.
AL, amyloid light-chain; ch, chemotherapy; compl. allo, complications allogeneic; HDM, high-dose melphalan; HL, Hodgkin lymphoma; irr, radiotherapy (treatment details are listed in supplemental Table 1); MM, multiple myeloma; n.a.: not applicable; NHL, non-Hodgkin lymphoma; R, relapsed disease; WHO, World Health Organization.
For patient UPN18, interval between apheresis and tMN diagnosis, apheresis was followed by consolidation with melphalan and lenalidomide, no ASCT possible because of medical conditions.
Patient UPN19 was diagnosed with HL after ASCT as treatment of R-NHL.
DNA isolation and amplification
We extracted genomic DNA from PB mononuclear cells (MNCs), BM-MNCs, or PBSCs using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. DNA isolation from BM morphological slides and BM sediments is described in the supplemental Methods. DNA from lymphoma material was isolated using the QIAamp DNA FFPE (formalin-fixed paraffin-embedded) Tissue kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocol. When the extraction yield was insufficient, 80 ng of DNA was amplified using the Qiagen REPLI-g kit (Qiagen) in 4 parallel reactions (20 ng per reaction), according to the manufacturer’s protocol.
Cell culturing
We cultured T cells and mesenchymal stem cells (MSCs) in order to obtain germline reference material. Culturing protocols are described in supplemental Methods. DNA was isolated using the NucleoSpin Tissue kit (Machery-Nagel).
WES
WES to an average depth of 110× was performed on a BM-MNC (n = 6) or PB-MNC sample (n = 1) at the time of tMN. In addition, WES was performed on a sample from the primary malignancy (lymphoma, n = 3). We performed exome capture using SureSelect Human All Exon V5 (Agilent Technologies, Santa Clara, CA). Enriched exome fragments were then subjected to massively parallel sequencing using the HiSeq 2500 platform (illumina, San Diego, CA). We then performed sequence alignment and mutation calling using our in-house pipelines, as previously described,18,19 with minor modifications. The filtering strategy and variant calling are described in supplemental Methods.
Targeted amplicon-based deep sequencing
All of the acquired candidate somatic variants detected by WES were validated and quantified by amplicon-based deep sequencing at high depth (aim: 10.000× coverage). The median validation rate per patient was 92.6%. Using this approach, we measured mutational burdens in all available samples for each patient, including the transplant material (PBSCs). Amplicon-based deep sequencing was performed as previously described19 (supplemental Methods). For clonal reconstruction, all variants clearly distinctive from the background were considered, typically with a VAF >0.2% (detailed information is described in supplemental Methods). Reconstruction of clonal evolution was performed as previously described19 (supplemental Methods).
Mutational signature analysis
We performed a mutational signature analysis on patients with tMN (n = 7) and patients with de novo MDS (n = 11) using pmsignature.20 Confirmed passenger mutations (n = 261) from all 18 samples were used for analysis, in which substitution type, 2 bases 3′ and 5′ of the substitution and transcription strand biases were used as mutation features.
TDS using a myeloid gene panel
For cases from which only DNA extracted from fixated BM sediments was available at the time of tMNs (n = 4), we used a myeloid gene panel (TruSight, illumina) (supplemental Table 4) to screen for driver mutations. A VAF threshold of 10% was used to exclude sequencing artifacts. We validated the identified mutations using targeted amplicon-based deep sequencing. Subsequently, PBSCs were analyzed for the presence of the identified driver mutations.
Immunohistochemistry analysis
We evaluated p53 protein expression by performing immunohistochemistry on paraffin-embedded BM tissue sections, using an antibody that recognized both wild-type and mutant protein (clone Bp53-11, Ventana Medical Systems, Tucson, AZ). Staining was performed on a Ventana Medical Systems automated slide stainer. Detailed information about scoring is described in supplemental Methods.
Statistical analysis
Clinical characteristics and group comparisons were studied by use of a Mann-Whitney U test for continuous variables and a Fisher’s exact test for categorical variables. Bivariate correlations were made with a Pearson correlation (continuous variables) or Spearman correlation (categorical variables). A P value < .05 was used to define statistical significance. We performed statistical calculations using SPSS version 23 (IBM, Armonk, NY).
Results
Patient characteristics
All studied patients (n = 18) were diagnosed with tMNs after an ASCT procedure for another hematological malignancy (Table 1). The median interval between ASCT and tMN diagnosis was 3.0 years (range, 0.8-16.2 years), and the median age at tMN diagnosis was 61 years (age range, 41-71 years). The most frequent cytogenetic aberrancies were complex karyotypes (44%), followed by isolated abnormalities of chromosome 7 (28%) (Table 1; supplemental Table 2).
Mutational analysis of patients with tMNs
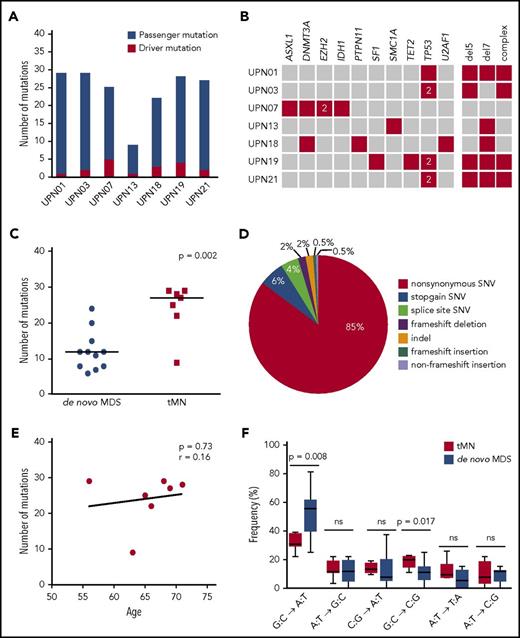
To identify mutations that contributed to malignant transformation post-ASCT, we performed WES on tMN diagnostic material from 7 patients. T cells and, where possible, cultured MSCs were used as germline controls. WES revealed a median of 27 somatically acquired mutations (range, 9-29 mutations) (Figure 1A; supplemental Table 10), with a median number of 3 mutations (range, 1-5 mutations) in known AML/MDS driver genes per patient (Figure 1B). TP53 was most frequently mutated in this cohort (in 4 of 7 patients). In all patients, both alleles of TP53 appeared to be affected, as 2 different mutations in TP53 were identified in 3 of these patients and a homozygous TP53 mutation was found in 1 patient. In addition, these samples were analyzed for the occurrence of germline mutations in 29 known cancer susceptibility genes (supplemental Table 5). However, in these genes, no germline mutations were found.
Genetic defects in patients with tMNs and comparison with patients with de novo MDS. (A) Number of acquired mutations in 7 patients in whom tMNs developed after ASCT, as determined by WES and confirmed by amplicon-based deep sequencing. In red, the number of mutations in genes previously implicated in the pathogenesis of myeloid malignancies is indicated (driver mutations); in blue, the number of mutations not previously implicated in myeloid malignancies (putative passenger mutations). (B) For each patient, all mutations in genes known to be recurrently mutated in myeloid malignancies are depicted as well as all cytogenetic defects detected by karyotype analysis (red). The “2” indicates 2 different mutations affecting the same gene. (C) Significantly more mutations could be detected in patients with tMNs compared with patients with de novo MDS. (D) Distribution of the different types of alterations detected in the total set of patients with tMNs. (E) No correlation could be observed between age and the number of genetic defects (genetic and cytogenetic defects) in patients with tMNs. The Pearson correlation coefficient was determined. (F) Different types of single nucleotide changes detected in all patients. In patients with tMNs, less G:C→A:T transitions are present than in patients with de novo MDS.
Genetic defects in patients with tMNs and comparison with patients with de novo MDS. (A) Number of acquired mutations in 7 patients in whom tMNs developed after ASCT, as determined by WES and confirmed by amplicon-based deep sequencing. In red, the number of mutations in genes previously implicated in the pathogenesis of myeloid malignancies is indicated (driver mutations); in blue, the number of mutations not previously implicated in myeloid malignancies (putative passenger mutations). (B) For each patient, all mutations in genes known to be recurrently mutated in myeloid malignancies are depicted as well as all cytogenetic defects detected by karyotype analysis (red). The “2” indicates 2 different mutations affecting the same gene. (C) Significantly more mutations could be detected in patients with tMNs compared with patients with de novo MDS. (D) Distribution of the different types of alterations detected in the total set of patients with tMNs. (E) No correlation could be observed between age and the number of genetic defects (genetic and cytogenetic defects) in patients with tMNs. The Pearson correlation coefficient was determined. (F) Different types of single nucleotide changes detected in all patients. In patients with tMNs, less G:C→A:T transitions are present than in patients with de novo MDS.
A comparison was made between cases of tMN (n = 7 patients; median age, 66 years) and de novo MDS (n = 11 patients; median age, 64 years; Revised International Prognostic Scoring System [IPSS-R] very low 18%, low 27%, intermediate 45% and high 9%) that were sequenced and analyzed using the same workflow.19 For de novo MDS cases, no correlation was observed between the IPSS-R and the total number of mutations (median, 12 [IPSS-R very low/low] vs 12 [IPSS-R intermediate/high]; P = .9266). A significantly higher number of mutations was identified in tMN cases (median, 27 vs 12 mutations in de novo MDS; P = .001) (Figure 1C). Similar to de novo MDS, the most frequent type of mutations in tMNs were missense mutations (Figure 1D). However, in contrast to de novo MDS cases, the number of mutations did not correlate with the age of the patient at tMN diagnosis (Pearson r = 0.16; P = .73; Figure 1E), consistent with the possibility that treatment of the primary malignancy contributed to increased mutagenesis in tMNs. In line with this, comparison of the different types of mutations showed a lower frequency of G:C→A:T transitions (mean, 33% vs 53%; P = .008) in tMNs, whereas the frequency of G:C→C:G transversions was higher (mean, 20% vs 11%; P = .017; Figure 1F).
Mutational signature analysis
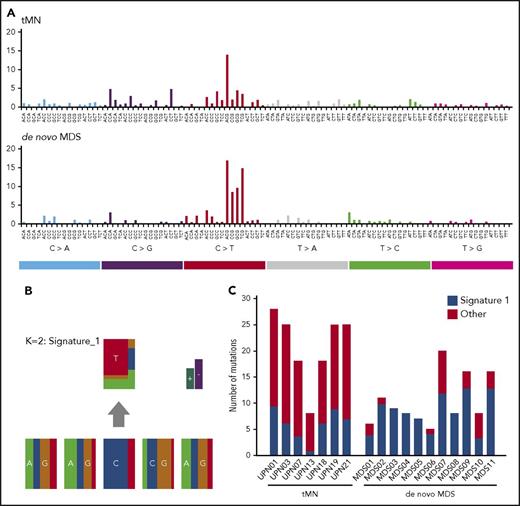
To find a further explanation for the observed difference in the number and type of mutations between tMN and de novo MDS cases, we performed a mutational signature analysis.20 Only mutations that had been successfully validated by TDS were included in the analysis. A mutational signature was created for both de novo MDS and tMN cases (Figure 2A). A distinctive mutational pattern, related to aging (signature 1) could be identified in both groups (Figure 2B). However, this aging-related signature represented a median of 27% (range, 8%-35%) of the mutations in tMN cases, compared with a median of 82% (range, 40%-100%) in de novo MDS cases (P = .0019; Figure 2C). For a high percentage of SNVs observed in tMN cases (median, 72%; range, 65%-92%), no signature could be identified, a finding that might be explained by the heterogeneous background of tMN cases, such as differences in initial diagnosis or prior treatment.
Mutational signature analysis. (A) A mutational signature was created for both de novo MDS and tMN patients, based on nondriver mutations that had been validated by TDS (n = 152 for tMN patients vs n = 145 for de novo MDS patients). (B) The distinctive mutational pattern, related to aging (signature 1: C > T@CpG) that was identified in both groups. (C) Representation of the number of mutations explained by signature 1 per individual patient. All other mutations are marked as “other” mutations.
Mutational signature analysis. (A) A mutational signature was created for both de novo MDS and tMN patients, based on nondriver mutations that had been validated by TDS (n = 152 for tMN patients vs n = 145 for de novo MDS patients). (B) The distinctive mutational pattern, related to aging (signature 1: C > T@CpG) that was identified in both groups. (C) Representation of the number of mutations explained by signature 1 per individual patient. All other mutations are marked as “other” mutations.
Clonal hematopoiesis in PBSCs
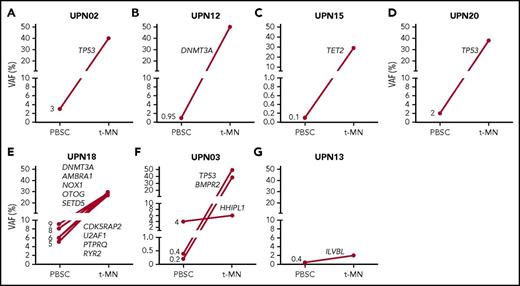
For obtaining insight regarding the existence of CHIP at the time of ASCT, the presence of mutations identified in tMN diagnostic material was assessed in the corresponding PBSCs. For 6 cases analyzed by WES, and 4 cases sequenced with use of a myeloid gene panel consisting of 54 known driver genes, PBSCs were available for analysis. CHIP in the autologous transplant could be demonstrated in 7 of 10 cases (70%), with VAFs between 0.1% and 8% (Figure 3). In 6 cases, the identified mutations present at low levels at the time of ASCT were also present in tMN cells that developed afterward (Figure 3A-F). In 2 cases, (Figure 3F-G), a “bystander clone” that was not part of the later-observed major leukemic clone could be identified.
Detection of preleukemic clones in mobilized PBSCs. The presence of preleukemic mutations was determined in PBSC material of 10 patients for whom mutational data were available at the time point of tMN diagnosis. Preleukemic clones could be determined in 7 of 10 analyzed cases at indicated VAFs. Panels A-D were analyzed by a myeloid gene panel, and panels E-G were analyzed by WES. In patients UPN02, UPN12, UPN15, UPN20, and UPN18 (A-E), a mutation present in the leukemic clone could be detected in PBSCs, whereas the detected variant in the PBSCs of patient UPN13 represents a bystander clone (G). For patient UPN03 (F), mutations that represent both a leukemic and bystander clone were detected in PBSCs.
Detection of preleukemic clones in mobilized PBSCs. The presence of preleukemic mutations was determined in PBSC material of 10 patients for whom mutational data were available at the time point of tMN diagnosis. Preleukemic clones could be determined in 7 of 10 analyzed cases at indicated VAFs. Panels A-D were analyzed by a myeloid gene panel, and panels E-G were analyzed by WES. In patients UPN02, UPN12, UPN15, UPN20, and UPN18 (A-E), a mutation present in the leukemic clone could be detected in PBSCs, whereas the detected variant in the PBSCs of patient UPN13 represents a bystander clone (G). For patient UPN03 (F), mutations that represent both a leukemic and bystander clone were detected in PBSCs.
tMN cases display impaired PBSC collection and regeneration after ASCT
Because preleukemic mutations were a frequent finding in transplant material of tMN cases, we wondered whether this trend correlated with impaired graft function. Therefore, we compared graft characteristics in tMN cases with matched control cases (supplemental Table 7). CD34+ PBSC collection was impaired in tMN cases (median, 4.98 × 106/kg vs 15.85 × 106/kg of PBSCs in control cases; P = .005), whereas the number of reinfused CD34+ cells did not differ (median, 4.55 × 106/kg vs 4.71 × 106/kg of PBSCs; P = .40) (supplemental Figure 1). At 3 months after ASCT, tMN cases displayed delayed platelet regeneration compared with controls (median, 44 × 109/L vs 95 × 109/L; P = .033) and lower hemoglobin levels (median, 8.9 g/dL vs 10.6 g/dL; P = .028) with an increased mean corpuscular volume (MCV) at 1 year after transplantation (median, 101.4 fL vs 96.0 fL; P = .001) (supplemental Figure 2). These observations are suggestive for an impaired stem cell function after transplantation in tMN cases, because a comparable number of CD34+ cells were reinfused while regeneration was delayed. Furthermore, tMN cases in which CHIP was identified in PBSCs showed a trend towards impaired regeneration after ASCT (supplemental Figure 3).
Preleukemic clonal dynamics
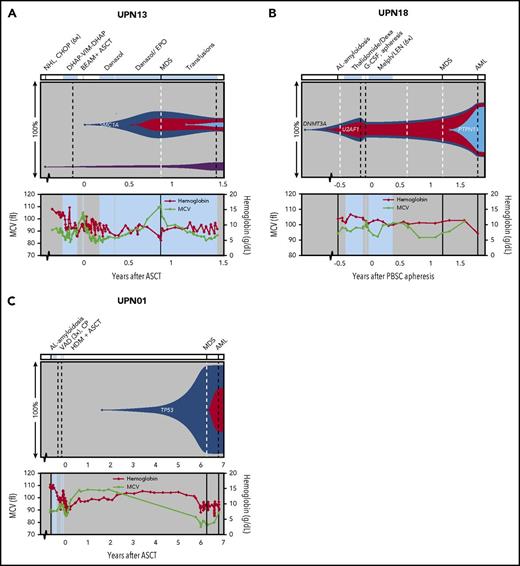
To gain insight into the dynamics of preleukemic clones, we tracked back all mutations detected by WES at tMN diagnosis (in 7 cases) in sequential preleukemic samples using TDS. Based on the VAFs detected at all different time points, clonal evolutionary patterns were reconstructed (Figure 4 and 5; supplemental Figure 4).
Preleukemic clonal dynamics. Representation of (preleukemic) clonal evolution occurring in patients UPN13 (A), UPN18 (B), and UPN01 (C). The upper panel shows percentages of clones detected at different sampling moments. The x-axis represents the time before and after ASCT (years). Dashed lines represent sampling moments; black lines for DNA derived from PBSC, BM-MNC, or PB-MNC samples; and white for DNA derived from BM slide or BM sediment material. Known leukemic driver genes are indicated in corresponding clones. Diagnosis details and treatment are indicated above the figure panels. Values for hemoglobin and MCV are depicted in the bottom section of each panel. For other PB values, see supplemental Figure 5. Periods of treatment are indicated with light blue planes. For UPN18 (B), the time after PBSC apheresis is plotted because this patient did not undergo ASCT. CAD, cyclophosphamide, Adriamycin, and dexamethasone; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisone; CP, cyclophosphamide; CVD, cyclophosphamide, bortezomib, and dexamethasone; Dexa, dexamethasone; DHAP-VIM-DHAP, dexamethasone, cytarabine, cisplatin, etoposide, ifosfamide, and methotrexate; EPO, erythropoietin; G-CSF, granulocyte colony-stimulating factor; HDM, high-dose melphalan; IDM, intermediate-dose melphalan; LEN, lenalidomide; Melph, melphalan; VAD, vincristine, Adriamycin, and dexamethasone.
Preleukemic clonal dynamics. Representation of (preleukemic) clonal evolution occurring in patients UPN13 (A), UPN18 (B), and UPN01 (C). The upper panel shows percentages of clones detected at different sampling moments. The x-axis represents the time before and after ASCT (years). Dashed lines represent sampling moments; black lines for DNA derived from PBSC, BM-MNC, or PB-MNC samples; and white for DNA derived from BM slide or BM sediment material. Known leukemic driver genes are indicated in corresponding clones. Diagnosis details and treatment are indicated above the figure panels. Values for hemoglobin and MCV are depicted in the bottom section of each panel. For other PB values, see supplemental Figure 5. Periods of treatment are indicated with light blue planes. For UPN18 (B), the time after PBSC apheresis is plotted because this patient did not undergo ASCT. CAD, cyclophosphamide, Adriamycin, and dexamethasone; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisone; CP, cyclophosphamide; CVD, cyclophosphamide, bortezomib, and dexamethasone; Dexa, dexamethasone; DHAP-VIM-DHAP, dexamethasone, cytarabine, cisplatin, etoposide, ifosfamide, and methotrexate; EPO, erythropoietin; G-CSF, granulocyte colony-stimulating factor; HDM, high-dose melphalan; IDM, intermediate-dose melphalan; LEN, lenalidomide; Melph, melphalan; VAD, vincristine, Adriamycin, and dexamethasone.
Preleukemic dynamics in patients who received additional chemotherapy after ASCT. Representation of (preleukemic) clonal evolution of patients UPN07 (A), UPN03 (B), UPN21 (C), and UPN19 (D). For figure details and abbreviations, see Figure 4 legends. For UPN03 (B), post-ASCT treatment with thalidomide* 200 mg/week, later reduced to 150 mg/week, thalidomide** 50 mg/week, later reduced to 100 mg/3 weeks. Aza, azacitidine; LOPP, chlorambucil, vincristine, procarbazine, and prednisone; LPP, chlorambucil, procarbazine, and prednisone; MM, multiple myeloma; r, relapsing disease; R, rituximab; RT, radiotherapy.
Preleukemic dynamics in patients who received additional chemotherapy after ASCT. Representation of (preleukemic) clonal evolution of patients UPN07 (A), UPN03 (B), UPN21 (C), and UPN19 (D). For figure details and abbreviations, see Figure 4 legends. For UPN03 (B), post-ASCT treatment with thalidomide* 200 mg/week, later reduced to 150 mg/week, thalidomide** 50 mg/week, later reduced to 100 mg/3 weeks. Aza, azacitidine; LOPP, chlorambucil, vincristine, procarbazine, and prednisone; LPP, chlorambucil, procarbazine, and prednisone; MM, multiple myeloma; r, relapsing disease; R, rituximab; RT, radiotherapy.
In 3 cases that were analyzed with use of WES, CHIP could be detected in the PBSC material (Figure 4A-B and 5B). Patient UPN13 showed signs of graft failure after ASCT and was therefore treated with androgens and erythropoietin (Figure 4A), which resulted in a marked increase in MCV. At MDS diagnosis, only a small clone, which contained a mutation in the leukemic driver gene SMC1a, was present. The ILVBL mutation that was detected in PBSCs did not expand in time. In contrast, the detected mutations in PBSCs of patients UPN03 and UPN18 did expand during tMN development. In patient UPN18 (Figure 4B), a clone (containing a DNMT3A mutation) and subclone (containing a U2AF1 mutation) were present at diagnosis of the primary malignancy, before any treatment was given. This patient did not undergo ASCT but received intensive chemotherapy before and after PBSC collection; MDS was diagnosed 1.3 years later. Transformation to AML was accompanied by the acquisition of additional mutations, including a mutation in PTPN11.
Transformation from MDS to AML was also accompanied by the acquisition of additional mutations in 2 other cases. Patient UPN01 developed an MDS at 6 years after ASCT (Figure 4C). The major leukemic clone contained a homozygous TP53 mutation and could be detected 2.5 years before the onset of MDS (Figure 6B). Transformation to AML was accompanied by the acquisition of mutations in ACAD8 and SLC23A2. Patient UPN07 (Figure 5A) suffered from a relapse of non-Hodgkin lymphoma (NHL) at 7 years after ASCT. Already before the start of NHL-directed chemotherapy following ASCT, a clone containing several mutations, including mutations in DNMT3A, ASXL1, and EZH2 (VAFs <1%), was present. Seven years later, at MDS diagnosis, this same clone had grown out (VAFs ∼35%). Progression to AML coincided with the acquisition of an IDH1 mutation.
Visualization of clonal expansion of TP53-mutated cells. Immunohistochemical staining for p53 protein expression on BM biopsies at tMN diagnosis and preleukemic time points for patients UPN17 (A), UPN01 (B), and UPN03 (C). Time points of biopsy and magnification are indicated in the figure panels. (D) Percentage of cells showing high p53 protein expression (p53++ cells) were scored at different time points (pre- and post-ASCT) for the 3 patients depicted in panels A-C; last time point is moment of tMN diagnosis.
Visualization of clonal expansion of TP53-mutated cells. Immunohistochemical staining for p53 protein expression on BM biopsies at tMN diagnosis and preleukemic time points for patients UPN17 (A), UPN01 (B), and UPN03 (C). Time points of biopsy and magnification are indicated in the figure panels. (D) Percentage of cells showing high p53 protein expression (p53++ cells) were scored at different time points (pre- and post-ASCT) for the 3 patients depicted in panels A-C; last time point is moment of tMN diagnosis.
For patient UPN07, the additional chemotherapy after ASCT might have provided an extra selective pressure causing selection of the preleukemic clone. Similar patterns were identified in patients UPN03, UPN21, and UPN19. In Patient UPN03 (Figure 5B), 2 mutations, including one in TP53, were detected in ∼2% of the transplant cells harvested 7.3 years before AML diagnosis. This finding suggests that a small ancestral clone later acquired additional mutations, including a second TP53 mutation, resulting in AML development. In addition, a small HHIPL1-mutated clone was present in PBSCs but appeared not to be involved in the AML clone. As in UPN07, the pre-tMN clone expanded after administration of chemotherapy. Four years after ASCT, all of the mutations that were present at AML diagnosis could be detected at VAFs of 12% to 25%. The clone size remained stable with thalidomide treatment and expanded after dose reduction and addition of cyclophosphamide. It is striking that, despite the large clone size, PB values were not significantly affected until AML diagnosis (Figure 5B; supplemental Figure 5). Also, patient UPN21 experienced a relapse of the primary malignancy 2 years after ASCT, for which intensive chemotherapy and, later, a combination of lenalidomide and dexamethasone were started (Figure 5C). Although intermediate moments of sampling between ASCT and tMN diagnosis are lacking, PB count dynamics indicate that this additional treatment may have affected the preleukemic BM; with intensive chemotherapy, the MCV increased, whereas this value decreased with lenalidomide treatment. MDS with a complex karyotype, including −5q, was diagnosed 1.5 years after the start of lenalidomide treatment. Finally, in patient UPN19 (Figure 5D), the interval between ASCT and tMN diagnosis was 16 years. This patient was initially diagnosed with NHL, for which several rounds of treatment including ASCT were applied. Fifteen years later, patient UPN19 was diagnosed with Hodgkin lymphoma (HL), for which chemotherapy was started. AML was diagnosed one year later. The major leukemic clone (with mutations in TP53, TET2 and SF1) that was detectable at HL diagnosis had expanded at that time, while a subclone arose after HL-directed chemotherapeutic treatment.
Together, the patients represented in Figure 4 and 5 suggest that preleukemic clones can be present for more than 7 years before onset of tMNs without majorly affecting PB cell counts and that treatment may be of influence on clonal dynamics, both by causing stabilization or outgrowth of preleukemic clones.
Presence of preleukemic mutations in T cells
Our analysis revealed that CHIP is not always restricted to the myeloid compartment. In patient UPN19 (Figure 5D), the mutations that were detected in the ancestral blue clone resulting in AML were also present in the T cells obtained at tMN diagnosis (VAF ∼18% in T cells). By contrast, the mutations solely present in the subclone were not detectable in the T cells (supplemental Figure 6A). Leukemic mutations were absent in autologous MSCs. For patient UPN18 (Figure 4B), the 5 mutations that represented the ancestral blue clone were also found in T cells obtained from PBSC material (VAF ∼2%). Mutations that were present in the two later subclones, were absent in the T cells (supplemental Figure 6B). These results suggest that the ancestral leukemic clone originated from an early hematopoietic stem cell, and that later mutations led to a block in lymphoid differentiation or, alternatively, were acquired in more committed myeloid progenitor cells.
No overlap between mutations detected in primary malignancy and tMNs
The presence of 2 different hematological malignancies in the same patient, and the observation that T cells may sometimes be part of the clonally expanded population, made us wonder whether the primary malignancy and the tMNs could be derived from a common ancestor. To answer this question, we performed WES on primary lymphoma samples. In patients UPN07 and UPN13, we studied NHL samples at relapse; in patient UPN19, we sequenced both the relapsed NHL and the HL that developed after ASCT. A total of 351 mutations were detected in the 4 lymphoma samples (data not shown), of which 41 mutations occurred in known cancer driver genes (supplemental Table 11). Using both autologous T cells and MSCs as reference, we identified a completely different set of acquired mutations in the NHL samples compared with the tMN samples, indicating that the primary disease and tMNs were not derived from a common ancestor. Exemplary for this finding are the different EZH2 mutations that were identified in patient UPN07: the NHL sample contained a lymphoma-specific activating mutation, while the tMN sample contained 2 inactivating mutations (supplemental Table 10 and 11).
Interestingly, mutations that were present in the ancestral clone that had evolved into AML of UPN19 were identified in the HL-lymph node biopsy (∼8% of the cells) 1 year before tMN diagnosis. These mutations were also observed in T cells at tMN diagnosis (supplemental Figure 6A). As classic-type HL is characteristically composed of a very low percentage of malignant cells surrounded by lymphocytic infiltration, the cells in the HL biopsy carrying the mutations most likely represent nonmalignant T cells.
Immunohistochemical staining of TP53-mutated cells in BM biopsies
As the majority of TP53 mutations result in overexpression of the p53 protein, their presence often can be detected by immunohistochemistry.21 Therefore, we used this technique to confirm the sequencing results. The presence of high p53 protein–expressing cells (p53++) was specific for patients with tMN (50% scored “p53-positive”) since these cells could not be found in BM biopsies of matched post-ASCT control patients who had received comparable treatment (n = 18; supplemental Figure 7A-C). For cases displaying ≥5% p53++ cells, the presence of a TP53 mutation could be confirmed (supplemental Figure 7D). In patient UPN19, the TP53 p.Y73X and p.W52X nonsense mutations could not be detected by immunohistochemistry (supplemental Figure 7E).
Available preleukemic BM biopsies were evaluated for p53 expression. For 3 of 9 “p53-positive” cases, a BM biopsy was taken between ASCT and tMN diagnosis. Of note is that in all 3 cases, clear clusters of p53++ cells could be detected up to almost 3 years before tMN diagnosis (Figure 6). This finding was in accordance with the sequencing results for patient UPN03 (Figure 5B). In patient UPN17, clear clusters were already visible 3 months before ASCT, as well as in the regenerating BM at 4 months after transplantation (Figure 6A). These results confirm the finding that small preleukemic clones are present in the BM of patients years before tMN diagnosis.
Discussion
In this report, we describe the comprehensive mutational analysis of patients who developed a therapy-related myeloid neoplasm after undergoing ASCT. We observed that tMNs are characterized by a significantly increased mutation load compared with de novo MDS. In addition, in tMNs, a low proportion of aging-related mutations were found, whereas mutations in the tumor suppressor gene TP53 were frequent. The lack of overlap between the mutational spectra observed in the tMN and lymphoma samples of the same case confirms that both hematological malignancies were not derived from a common ancestor.
An impaired CD34+ PBSC collection and engraftment after ASCT was observed in tMN cases. Gibson et al5 have recently also associated diminished PBSC collection with the presence of CHIP in PBSC material. However, regarding the low frequencies at which mutations are present, it is unlikely that CHIP is responsible for the impaired engraftment and poor PBSC mobilization. Instead, the presence of CHIP may correlate with a more general dysfunction of the HSC compartment and the surrounding microenvironment that may favor the outgrowth of preleukemic clones. This hypothesis is supported by findings that telomere shortening,22 mitochondrial dysfunction,23 increased HSPC proliferation,24 and microenvironmental defects25 have been observed with respect to delayed regeneration of PB cells in patients who developed tMNs after ASCT.
TDS of sequential (pre-)leukemic samples and immunohistochemistry revealed that the preleukemic clones had been present at low frequencies for up to 7 years before onset of tMNs. Expansion of preleukemic clones was particularly noticed in patients who needed additional (chemotherapeutic) treatment after ASCT. This observation is in line with the finding that heterozygous TP53-mutated cells have a selective advantage vs wild-type cells under both irradiation and chemotherapeutic treatment in mouse models.26,27 Besides the direct selective pressure induced by cytotoxic therapy, prolonged stress after ASCT, such as proliferative and replicative stress24 and higher levels of reactive oxygen species,23,28 might contribute to accelerated mutagenesis in HSPCs harboring preleukemic mutations. Indeed, we observed a significantly higher mutation load in tMNs compared with de novo MDS. This finding is in contrast to a report by Wong et al,26 who did not observe a difference in mutational burden between therapy-related and de novo leukemias. A possible explanation for this might be differences in the analytical approach, as we restricted our analysis to WES-identified mutations that could be confirmed by TDS, whereas Wong et al considered mutations found by whole-genome sequencing.
The identification of clonally related mutations in both T cells and myeloid cells, but not in MSCs, in patients UPN18 and UPN19 demonstrates that in these patients, the preleukemic clonal disorder arises from an early stem cell still capable of both myeloid and lymphoid differentiations. This observation is in agreement with recent reports in which authors have described the presence of lymphomyeloid clonal hematopoiesis in association with AMLs with complex karyotypes and TP53 mutations,29 as well as secondary AMLs and unfavorable treatment outcomes.30
In our cohort, CHIP could be detected in the transplant in 70% of the patients with tMNs, a finding that is in line with other reports.3,5,15 However, other authors have also recently reported that a substantial proportion of patients in whom a tMN did not develop after ASCT show the presence of CHIP at transplantation.3,5,15 In addition, our results show that mutations detected in PBSCs can also represent clones that are not involved in leukemic transformation. Future research, preferentially based on prospective studies using large patient numbers, is necessary to identify mutations and clinical parameters that are predictive for the risk for tMN development after ASCT. Our results would argue for regular monitoring of patients’ PB counts after ASCT. Patients who display aberrant PB cell regeneration (eg, unexplained persistent high MCV or prolonged low platelet counts) would require additional examinations including molecular analyses and careful monitoring. Our data suggest that determination of CHIP after ASCT becomes even more important when there is the intention to treat the patient with agents that could impose an extra-selective pressure on the hematopoietic system. For the future, NGS techniques may be routinely implemented for ASCT follow-up for the creation of individual risk profiles, based on types and combinations of mutations, VAFs, and clinical parameters. These data may not only be informative for risk assessment of tMN development, but also for events not related to tMNs. Of interest, other authors have reported that the presence of CHIP in PBSCs is associated with a significantly increased incidence of nonrelapse mortality, in particular of cardiovascular events,3,5,15,31 which suggests that CHIP might be a useful prognostic biomarker for the long-term follow-up of patients who have undergone ASCT.
In conclusion, our study demonstrates that tMNs arise from mutated early HSPCs and that outgrowth of preleukemic clones and transformation toward tMNs is accompanied by an increased amount of mutations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank K. Chiba, H. Tanaka, and A. Okada for help with bioinformatics analysis and A. Diepstra for help with immunohistochemistry analysis.
This work was supported by grants from ERA-NET JCT 2012 (TRIAGE-MDS) and HORIZON 2020 MDS-RIGHT; Grants-in-Aid from the Ministry of Health, Labour and Welfare of Japan, Japan Agency for Medical Research and Development (Health and Labour Sciences Research Expenses for Commission, Applied Research for Innovative Treatment of Cancer, and Project for Cancer Research and Therapeutic Evolution [P-CREATE]); and Grants-in-Aid for Scientific Research (KAKENHI), Japan Society for the Promotion of Science (JSPS) (grants 26221308, 15H05909, and 26890016).
Authorship
Contribution: G.B., L.I.K., K.Y., H.S., B.A.v.d.R., S.O., E.V., and J.H.J. designed the study; E.v.d.B. provided patient material and performed cytogenetic analysis; K.Y., H.U., Y.S., and S.M. performed WES analysis and mutational signature analysis; L.I.K., T.N.K.-S., and A.O.d.G. performed deep sequencing; G.B., L.I.K., and T.N.K.-S. performed reconstruction of clonal evolution; and G.B., L.I.K., E.V., and J.H.J. wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edo Vellenga, Department of Hematology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: e.vellenga@umcg.nl; and Joop H. Jansen, Department of Laboratory Medicine, Laboratory of Hematology, Radboud University Medical Center, Geert Grooteplein zuid 8, Nijmegen, 6525 GA, The Netherlands; e-mail: joop.jansen@radboudumc.nl.
REFERENCES
Author notes
G.B. and L.I.K. are joint first authors.
E.V. and J.H.J. are joint senior authors.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal