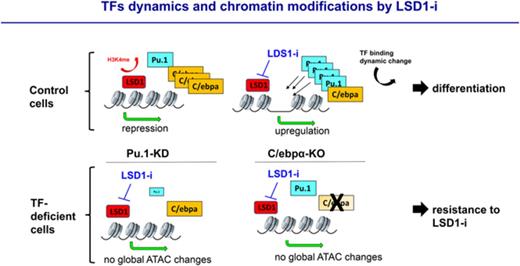
Key Points
LSD1 inhibition induces a global increase in chromatin accessibility, whereas DOT1L inhibition induces global decreases in accessibility.
Perturbation of PU.1 and C/EBPα expression renders AML cells more resistant to LSD1 inhibition.
Abstract
Epigenetic regulators are recurrently mutated and aberrantly expressed in acute myeloid leukemia (AML). Targeted therapies designed to inhibit these chromatin-modifying enzymes, such as the histone demethylase lysine-specific demethylase 1 (LSD1) and the histone methyltransferase DOT1L, have been developed as novel treatment modalities for these often refractory diseases. A common feature of many of these targeted agents is their ability to induce myeloid differentiation, suggesting that multiple paths toward a myeloid gene expression program can be engaged to relieve the differentiation blockade that is uniformly seen in AML. We performed a comparative assessment of chromatin dynamics during the treatment of mixed lineage leukemia (MLL)-AF9-driven murine leukemias and MLL-rearranged patient-derived xenografts using 2 distinct but effective differentiation-inducing targeted epigenetic therapies, the LSD1 inhibitor GSK-LSD1 and the DOT1L inhibitor EPZ4777. Intriguingly, GSK-LSD1 treatment caused global gains in chromatin accessibility, whereas treatment with EPZ4777 caused global losses in accessibility. We captured PU.1 and C/EBPα motif signatures at LSD1 inhibitor-induced dynamic sites and chromatin immunoprecipitation coupled with high-throughput sequencing revealed co-occupancy of these myeloid transcription factors at these sites. Functionally, we confirmed that diminished expression of PU.1 or genetic deletion of C/EBPα in MLL-AF9 cells generates resistance of these leukemias to LSD1 inhibition. These findings reveal that pharmacologic inhibition of LSD1 represents a unique path to overcome the differentiation block in AML for therapeutic benefit.
Introduction
Epigenetic dysregulation has been identified as a common feature of myeloid malignancies. Sequencing efforts aimed at characterizing the genomic landscape of acute myeloid leukemia (AML) have led to the discovery of recurrent mutations in epigenetic regulators.1,2 Genome-wide screens have also revealed epigenetic vulnerabilities in AML that can be exploited with therapies aimed at disarming leukemogenic gene expression programs by modulating the function of these chromatin modifiers.3 A number of compounds targeting epigenetic regulators are being developed and have entered early-phase clinical trials. These include inhibitors of histone methyltransferases such as DOT1L, which has been shown to be a dependency in AML driven by mixed lineage leukemia (MLL) rearrangements.4
Lysine-specific demethylase 1 (LSD1) similarly plays an important role in oncogenesis.5 LSD1 can demethylate mono- and dimethylated lysine residues 4 and 9 on histone H3 (H3K4me1/2 and H3K9me1/2, respectively).6 LSD1 is aberrantly expressed in cancer and is linked to poor clinical outcomes in solid malignancies.7-10 In hematologic malignancies, LSD1 is overexpressed in AML as well as lymphoid malignancies and myeloproliferative neoplasms.11 Multiple groups have demonstrated antitumor activity mediated either by inhibitors targeting LSD1 or by genetic depletion of LSD1 in solid tumors.12-15 Likewise, in hematologic malignancies, knockdown or inhibition of LSD1 has been shown to be a potential therapeutic strategy in mouse models and human cases of MLL-rearranged AML.5,16,17 Somervaille and colleagues have previously shown that pharmacologic LSD1 inhibition induced myeloid differentiation of AML cells, impairing their ability to cause leukemia in mouse models.5
One striking commonality among many of these epigenetic therapies for myeloid malignancies, such as those inhibitors targeting LSD1, DOT1L, or IDH1/2, is their shared ability to induce myeloid differentiation in addition to blocking the proliferative and self-renewal capacity of these malignant cells. This raises the possibility that there are multiple paths to relieve the differentiation blockade that is a hallmark feature of AML. In this study, we explore this hypothesis within the context of our characterization of an irreversible LSD1 inhibitor, GSK-LSD1, a compound that has potent efficacy in a highly penetrant and lethal mouse model of AML driven by MLL fusion proteins. We employed assays for transposase-accessible chromatin coupled with high-throughput sequencing (ATAC-seq) to detect changes in chromatin accessibility in AML cells caused by distinct targeted epigenetic therapies, LSD1 inhibitors (GSK-LSD1 and IMG-7289) and a DOT1L inhibitor (EPZ4777). While all compounds caused myeloid differentiation in MLL-AF9-driven AML cells, they induced starkly contrasting changes in chromatin accessibility. DOT1L inhibition caused a predominant loss in chromatin accessibility across the genome, whereas LSD1 inhibition induced gains in accessibility, with a strong enrichment of PU.1 and C/EBPα at these dynamic sites. Genetic loss of C/EBPα or depletion of PU.1 resulted in resistance of AML cells to LSD1 inhibition both in vitro and in vivo, thereby revealing the importance of recruiting a myeloid transcription factor (TF) network mediated by PU.1 and C/EBPα in modulating the antileukemic activity of GSK-LSD1. Our comparative investigations of this LSD1 inhibitor describe a means by which alterations in chromatin accessibility coincide with the engagement of a myeloid differentiation program that can be exploited as a therapeutic modality for an aggressive subtype of AML.
Methods
Small-molecule inhibitors
The irreversible LSD1 inhibitor GSK-LSD1 was kindly provided by GlaxoSmithKline. GSK-LSD1 was solubilized in sterile phosphate-buffered saline (PBS) for in vitro assays and in sterile NaCl 0.9% for in vivo experiments. The irreversible LSD1 inhibitor IMG-7289, kindly provided by Imago Biosciences, was solubilized in sterile dimethyl sulfoxide for in vitro assays. The DOT1L inhibitor EPZ4777 was kindly provided by Epizyme and was solubilized in dimethyl sulfoxide as the vehicle for these experiments.
Results
LSD1 inhibition with GSK-LSD1 has potent activity against MLL-AF9 leukemia
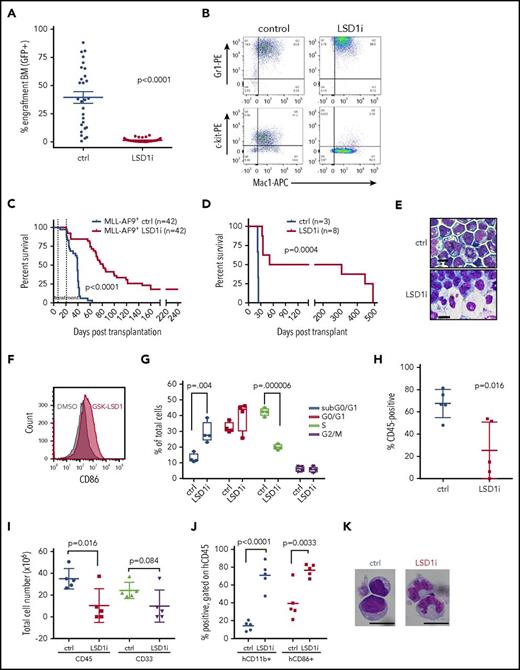
To assess the activity of LSD1 inhibition in vivo, secondary recipient mice engrafted with 1 × 105 MLL-AF9 primary AML cells were treated with GSK-LSD1. The drug was administered daily during a 14-day treatment window at a dose of 0.5 mg/kg. Treatment was initiated only after peripheral blood engraftment was confirmed (supplemental Figure 1A, available on the Blood Web site). After treatment, some mice were killed and analyzed using flow cytometric detection of GFP as a readout of MLL-AF9 allele burden. GSK-LSD1–treated mice exhibited a lower proportion of GFP+ cells in the bone marrow (Figure 1A), peripheral blood, and spleen (supplemental Figure 1B-C). Other measures of disease burden, including spleen weight, were markedly reduced in the setting of GSK-LSD1 treatment (supplemental Figure 1E). Mice treated with GSK-LSD1 exhibited a significant decline in platelet count (P = .003; supplemental Figure 1D), which is consistent with an on-target effect of LSD1 depletion.18 Immunophenotyping of bone marrow cells after 3 days of GSK-LSD1 treatment revealed a reduction of more primitive GFP+ leukemia cells coexpressing c-kit and Mac-1 (Figure 1B). GSK-LSD1–treated mice also had markedly improved survival (median survival, 78 days) compared with control mice (median survival, 39 days) (Figure 1C). Strikingly, a small proportion of treated mice had no detectable disease even 248 days after transplantation. In order to confirm this effect of LSD1 inhibition on survival, we performed serial transplantation of MLL-AF9 cells harvested from leukemic mice treated for 3 days with either vehicle alone or GSK-LSD1. Equivalent numbers of GFP+ cells purified from vehicle- or GSK-LSD1–treated mice were injected into sublethally irradiated mice. Tertiary recipient mice transplanted with cells harvested from GSK-LSD1–treated mice had improved survival when compared with vehicle-treated mice. While recipient mice transplanted with vehicle-treated cells had a median survival of 23 days, mice challenged with GSK-LSD1–treated leukemia cells had a median survival of 51 days (Figure 1D). Only 50% of the mice engrafted with GSK-LSD1–treated leukemia cells succumbed to AML. The remaining 50% of the mice transplanted with GSK-LSD1–treated cells remained healthy up to 308 days after transplantation and showed no signs of leukemia. These data suggest that LSD1 inhibition has potent antileukemic activity, improves overall survival, and occasionally causes complete disease eradication in an aggressive model of MLL-AF9–driven AML.
LSD1 inhibition with GSK-LSD1 has potent antileukemic activity. (A) GFP chimerism reporting MLL-AF9 allele burden in the bone marrow of GSK-LSD1–treated (red, n = 32) or vehicle-treated (blue, n = 28) leukemic mice following 2 weeks of treatment. (LSD1i = GSK-LSD1). t = 7.833; degrees of freedom (df) = 58. (B) Immunophenotype of GFP+ cells from bone marrow of mice after 3 days of vehicle (left) or GSK-LSD1 (right) administration. Double staining with antibodies recognizing Gr1, Mac-1, and c-kit cell surface markers are indicated. n = 3. (C) Kaplan-Meier survival curve of secondary MLL-AF9 leukemic mice. The interval between the dotted lines represents the period of treatment with GSK-LSD1 or vehicle. (D) Kaplan-Meier survival curve of tertiary sublethally irradiated mice transplanted with cells obtained from secondary mice treated with either vehicle (blue) or GSK-LSD1 (red) for 3 days. (E) Wright-Giemsa–stained cytospins of AML cells cultured after a 48-hour exposure to 0.5 µM GSK-LSD1. Scale bars, 10 µm. (F) Histogram plots demonstrating increased cell surface expression of CD86 after treatment of murine MLL-AF9 leukemia cells with 0.5 μM GSK-LSD1 (red) relative to vehicle control treatment (black). (G) Graph representing cell cycle states of murine AML cells treated with 0.5 µM GSK-LSD1 for 48 hours. n = 3; subG0/G1: t=4.49, df=6; G0/G1: P = .186 n.s.; S: t=14.9, df=6; G2/M: P = .776. (H) Chimerism of human CD45+ AML derived cells from bone marrow of PDX mice treated for 6 weeks with vehicle (blue) or GSK-LSD1 (red) at a dose of 0.5 mg/kg. n = 5. (I) Total cellularity of hCD45+ and hCD45+/hCD33+ cells derived from bone marrow of PDX mice treated with vehicle or GSK-LSD1 (0.5 mg/kg). PDX recipient mice were transplanted with cells from bone marrow of an AML patient harboring the MLL-AF9 gene rearrangement. n = 5. (J) Bar graphs representing the proportion of hCD45+ engrafted PDX cells expressing hCD11b or hCD86 after treatment with vehicle or GSK-LSD1. n = 5. (K) Morphology of Wright-Giemsa–stained hCD45+ bone marrow cells derived from PDX NSG mice treated with vehicle or GSK-LSD1. Bar, 10 µm. n = 3. APC, allophycocyanin; BM, bone marrow; ctrl, control; DMSO, dimethyl sulfoxide; PE, phycoerythrin.
LSD1 inhibition with GSK-LSD1 has potent antileukemic activity. (A) GFP chimerism reporting MLL-AF9 allele burden in the bone marrow of GSK-LSD1–treated (red, n = 32) or vehicle-treated (blue, n = 28) leukemic mice following 2 weeks of treatment. (LSD1i = GSK-LSD1). t = 7.833; degrees of freedom (df) = 58. (B) Immunophenotype of GFP+ cells from bone marrow of mice after 3 days of vehicle (left) or GSK-LSD1 (right) administration. Double staining with antibodies recognizing Gr1, Mac-1, and c-kit cell surface markers are indicated. n = 3. (C) Kaplan-Meier survival curve of secondary MLL-AF9 leukemic mice. The interval between the dotted lines represents the period of treatment with GSK-LSD1 or vehicle. (D) Kaplan-Meier survival curve of tertiary sublethally irradiated mice transplanted with cells obtained from secondary mice treated with either vehicle (blue) or GSK-LSD1 (red) for 3 days. (E) Wright-Giemsa–stained cytospins of AML cells cultured after a 48-hour exposure to 0.5 µM GSK-LSD1. Scale bars, 10 µm. (F) Histogram plots demonstrating increased cell surface expression of CD86 after treatment of murine MLL-AF9 leukemia cells with 0.5 μM GSK-LSD1 (red) relative to vehicle control treatment (black). (G) Graph representing cell cycle states of murine AML cells treated with 0.5 µM GSK-LSD1 for 48 hours. n = 3; subG0/G1: t=4.49, df=6; G0/G1: P = .186 n.s.; S: t=14.9, df=6; G2/M: P = .776. (H) Chimerism of human CD45+ AML derived cells from bone marrow of PDX mice treated for 6 weeks with vehicle (blue) or GSK-LSD1 (red) at a dose of 0.5 mg/kg. n = 5. (I) Total cellularity of hCD45+ and hCD45+/hCD33+ cells derived from bone marrow of PDX mice treated with vehicle or GSK-LSD1 (0.5 mg/kg). PDX recipient mice were transplanted with cells from bone marrow of an AML patient harboring the MLL-AF9 gene rearrangement. n = 5. (J) Bar graphs representing the proportion of hCD45+ engrafted PDX cells expressing hCD11b or hCD86 after treatment with vehicle or GSK-LSD1. n = 5. (K) Morphology of Wright-Giemsa–stained hCD45+ bone marrow cells derived from PDX NSG mice treated with vehicle or GSK-LSD1. Bar, 10 µm. n = 3. APC, allophycocyanin; BM, bone marrow; ctrl, control; DMSO, dimethyl sulfoxide; PE, phycoerythrin.
To explore the biological effects of LSD1 inhibition, we treated MLL-AF9 cells in vitro with GSK-LSD1. We observed morphologic evidence of myeloid differentiation even after 48 hours of treatment, with increased granule formation, nuclear condensation, and a markedly diminished nuclear to cytoplasmic ratio consistent with maturation (Figure 1E). Furthermore, LSD1 inhibitor treatment induced upregulation of myeloid differentiation markers like CD86 (Figure 1F), consistent with prior reports.19-21 Additionally, cell cycle analysis revealed a lower proportion of proliferating cells in S phase and an accumulation of cells in G0-G1 phase (Figure 1G), suggesting that LSD1 inhibition with GSK-LSD1 causes myeloid differentiation and cell cycle arrest of AML cells.
We next tested the efficacy of GSK-LSD1 on human AML cells using patient-derived xenografts (PDX) in vivo. PDX models were also generated using 2 MLL-rearranged AML samples and 2 nonrearranged samples engrafted into NSG mice. The characteristics of the leukemias tested and the treatment schedules are reported (supplemental Tables 1 and 2, respectively). PDX mice were treated with GSK-LSD1 for 10 days for up to 6 weeks (supplemental Figure 1F). After treatment, engrafted mice treated with GSK-LSD1 for 6 weeks showed a significant decrease in the frequency of human CD45+ bone marrow cells (25.4% hCD45+ for GSK-LSD1 treated vs 67.7% hCD45+ for vehicle treated, P = .016; Figure 1H), although mice treated for <6 weeks did not exhibit a treatment response. There was also a decrease in the absolute numbers of hCD45+ and hCD33+ cells (Figure 1I). The majority of hCD45+ cells were hCD11b+ and hCD86+ (Figure 1J; supplemental Table 2), and AML blasts also showed morphologic features of myelomonocytic differentiation (Figure 1K). These findings demonstrate antileukemic activity of GSK-LSD1 in MLL-rearranged and PDX mouse models.
Contrasting effects of LSD1 and DOT1L inhibition on chromatin accessibility
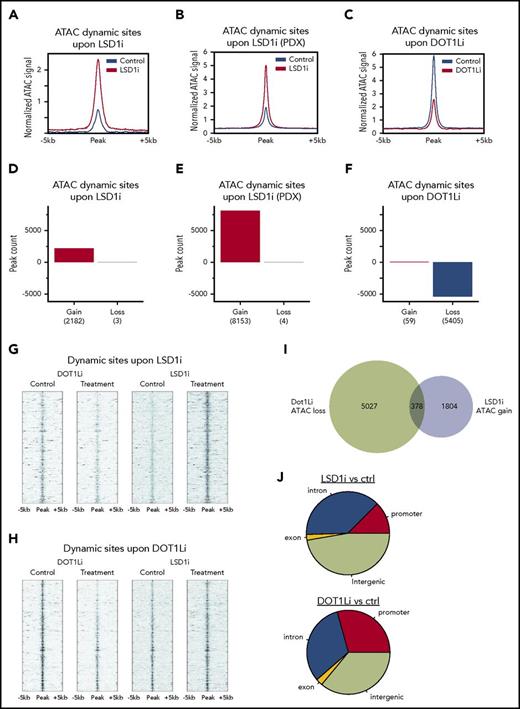
Previous work demonstrated the efficacy of inhibition of the H3K79 histone methyltransferase DOT1L against MLL-rearranged leukemias.22 Treatment of leukemia cells with the DOT1L inhibitor EPZ4777 induced myeloid differentiation, similar to what was observed with GSK-LSD1 treatment. To compare the impact of these distinct differentiation-inducing therapies targeting epigenetic regulators on chromatin accessibility, we performed ATAC-seq on murine MLL-AF9 cells treated with vehicle, EPZ4777, or GSK-LSD1. We also performed ATAC-seq on PDX AML cells derived from mice treated with GSK-LSD1. Analysis of all dynamic sites of chromatin accessibility induced by drug treatment revealed that GSK-LSD1 treatment generally resulted in gains in accessibility in both MLL-AF9 cells (Figure 2A) and PDX cells (Figure 2B). In contrast, treatment of MLL-AF9 cells with EPZ4777 caused a global decrease in chromatin accessibility (Figure 2C). Across all dynamic sites in GSK-LSD1–treated leukemia cells, sites with decreases in chromatin accessibility were vastly outnumbered by sites exhibiting increases in accessibility (Figure 2D-E). Likewise, for EPZ4777-treated cells, the number of sites with gains in accessibility was markedly less than those showing decreases in accessibility (Figure 2F).
Contrasting effects of LSD1 and DOT1L inhibition on chromatin accessibility. (A-C) Composite plots show normalized ATAC-seq signal at all dynamic sites in treatment and control for GSK-LSD1 in mouse (A) and PDX (B) and for EPZ4777 in mouse (C). (D-F) ATAC-seq peak dynamics in treatment and control for GSK-LSD1 in mouse (D) and PDX (E) and for EPZ4777 in mouse (F). (G) Heat map of normalized ATAC-seq read density at sites ranked by gain with GSK-LSD1 treatment, and the corresponding sites under control and EPZ4777 treatment. (H) Heat map of normalized ATAC-seq read density at sites ranked by loss with EPZ4777 treatment, and the corresponding sites under control and GSK-LSD1 treatment. (I) Venn diagram showing the overlap between dynamic ATAC-seq sites under treatment GSK-LSD1 or EPZ4777 relative to control. (J) Genome annotation of dynamic ATAC-seq sites under treatment GSK-LSD1 or EPZ4777 relative to control. ctrl, control.
Contrasting effects of LSD1 and DOT1L inhibition on chromatin accessibility. (A-C) Composite plots show normalized ATAC-seq signal at all dynamic sites in treatment and control for GSK-LSD1 in mouse (A) and PDX (B) and for EPZ4777 in mouse (C). (D-F) ATAC-seq peak dynamics in treatment and control for GSK-LSD1 in mouse (D) and PDX (E) and for EPZ4777 in mouse (F). (G) Heat map of normalized ATAC-seq read density at sites ranked by gain with GSK-LSD1 treatment, and the corresponding sites under control and EPZ4777 treatment. (H) Heat map of normalized ATAC-seq read density at sites ranked by loss with EPZ4777 treatment, and the corresponding sites under control and GSK-LSD1 treatment. (I) Venn diagram showing the overlap between dynamic ATAC-seq sites under treatment GSK-LSD1 or EPZ4777 relative to control. (J) Genome annotation of dynamic ATAC-seq sites under treatment GSK-LSD1 or EPZ4777 relative to control. ctrl, control.
By ranking those dynamic sites with the greatest increases in accessibility in response to LSD1 inhibition, we found that those corresponding sites were either unchanged or showed decreased accessibility after DOT1L inhibition (Figure 2G). When dynamic sites were ranked in order of greatest decreases in accessibility induced by EPZ4777, we similarly observed that those same sites under conditions of GSK-LSD1 treatment showed either no change or increases in accessibility (Figure 2H). The intersection of sites with dynamic accessibility upon DOT1L inhibition or LSD1 inhibition showed minimal overlap between the 2 treatments (Figure 2I). Characterization of the genomic regions occupied by these dynamic sites also revealed distinct distributions that are impacted by DOT1L or LSD1 inhibition, with DOT1L inhibition affecting a greater number of sites in promoter regions, whereas LSD1 inhibition affected a higher proportion of loci occupied within the intergenic space and intronic regions (Figure 2J). This is generally consistent with the functions of DOT1L in regulating transcriptional output and LSD1 histone demethylase activity in perturbing the cis-regulatory landscape of malignant cells. These findings demonstrate that 2 targeted epigenetic therapies sharing the ability to overcome the differentiation blockade that is uniformly seen in AML have dramatically contrasting effects on chromatin.
Enrichment of C/EBPα and PU.1 at dynamic sites of chromatin accessibility upon LSD1 inhibition
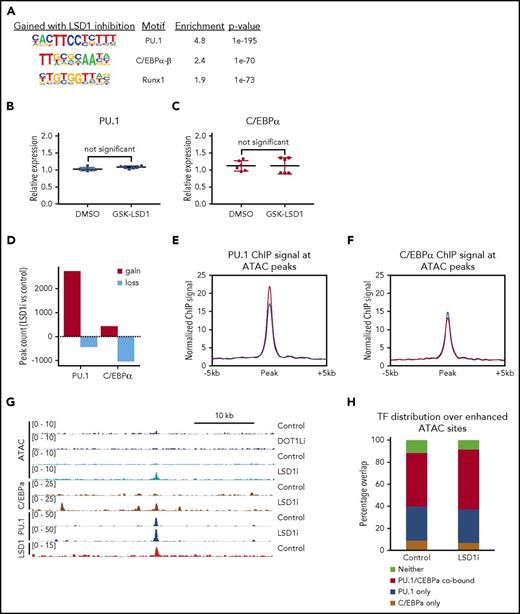
To further characterize the regions of altered chromatin accessibility affected by GSK-LSD1, we identified TF motif signatures at GSK-LSD1–induced dynamic sites. The top 3 most enriched motifs captured were those for myeloid TFs: PU.1, C/EBPα-β, and Runx1 (Figure 3A). We found no difference in the amount of PU.1 (Figure 3B) or C/EBPα (Figure 3C) mRNA expression between vehicle or GSK-LSD1–treated cells. We confirmed these findings by performing parallel ATAC-seq analyses on MLL-AF9 cells treated with a second LSD1 inhibitor, IMG-7289. Not only was the pattern of overall gain in chromatin accessibility observed when MLL-AF9 cells were treated with GSK-LSD1 also reproduced in the setting of IMG-7289 treatment (supplemental Figure 5A-B), but TF motif analysis of these datasets also revealed that C/EBPα, PU.1, and Runx1 were also the top 3 most enriched motifs in cells treated with IMG-7289 (supplemental Figure 5C). These findings using a separate LSD1 inhibitor strongly validate the core observations made with GSK-LSD1 and suggest that these are less likely to be related to off-target effects. Next, we performed chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) to identify sites of C/EBPα, PU.1, and LSD1 occupancy after GSK-LSD1 treatment and calculated genome-wide peak dynamics for sites changing by at least twofold in either direction. We observed that there was predominantly a global gain in PU.1 occupancy in response to GSK-LSD1 treatment relative to control. Conversely, there was a loss of C/EBPα signal after GSK-LSD1 treatment. (Figure 3D). When focusing specifically on dynamic ATAC-seq peaks in response to LSD1 inhibition, PU.1 ChIP-seq signal increased with GSK-LSD1 while C/EBPα occupancy was maintained at pretreatment levels (Figure 3E-F). Figure 3G shows an example of a region gaining chromatin accessibility after LSD1 inhibition, but not DOT1L inhibition. This dynamic intergenic site also demonstrates co-occupancy of PU.1 and C/EBPα, with LSD1 already bound at that same locus without GSK-LSD1 treatment (Figure 3G). Importantly, this pattern of dynamic chromatin changes is similarly observed at myeloid differentiation–related genes such as C/EBPβ, with increased ATAC-seq signal at these sites in response to LSD1 inhibition, but not DOT1L inhibition, and with LSD1 occupying the promoter region as well as nearby intergenic regions (supplemental Figure 4B). Figure 3H shows the distribution PU.1 and C/EBPα occupancy, as assessed by a binary “present/absent” call, at sites where chromatin accessibility is gained upon LSD1 inhibition. Even for the vehicle-only control group, the majority of these enhanced ATAC-seq sites are bound by PU.1, C/EBPα, or both. Moreover, GSK-LSD1 treatment did not alter the distribution of these sites. Taken together, this suggests that TF networks are already established with occupancy of PU.1, C/EBPα, or both prior to GSK-LSD1 treatment, and then after LSD1 inhibitor treatment, those sites with gains in accessibility exhibit more PU.1 binding, but not more C/EBPα binding, as shown in Figure 3E-F. From this, we infer that these sites of enhanced accessibility in the presence of LSD1 inhibition already have major drivers of myeloid differentiation present but are limited by the activity of LSD1.
Enrichment of C/EBPα and PU.1 at dynamic sites of chromatin accessibility upon LSD1 inhibition. (A) Top 3 motif signatures of GSK-LSD1–induced dynamic ATAC-seq peaks. (B) Plot showing the relative expression of PU.1 messenger RNA (mRNA) in MLL-AF9 leukemia cells treated with vehicle control (PBS) or 0.5 μM GSK-LSD1. (C) Plot showing the relative expression of C/EBPα mRNA in MLL-AF9 leukemia cells treated with vehicle control (PBS) or 0.5 μM GSK-LSD1. (D) Bar plot showing the number of peaks with either gains or losses of signal in ChIP-seq of PU.1 and C/EBPα in GSK-LSD1 treatment vs control. (E-F) Composite plots show normalized ChIP-seq signal at all dynamic ATAC sites in treatment and control for (E) PU.1 and (F) C/EBPα. (G) Genome browser track (mm9 coordinates chromosome 9: 14 425 541-14 466 000) showing normalized ATAC-seq and ChIP-seq signal under treatment and control conditions. (H) Percentage occupancy of PU.1 and C/EBPα in control and treatment conditions for ATAC-seq sites induced by GSK-LSD1. DMSO, dimethyl sulfoxide.
Enrichment of C/EBPα and PU.1 at dynamic sites of chromatin accessibility upon LSD1 inhibition. (A) Top 3 motif signatures of GSK-LSD1–induced dynamic ATAC-seq peaks. (B) Plot showing the relative expression of PU.1 messenger RNA (mRNA) in MLL-AF9 leukemia cells treated with vehicle control (PBS) or 0.5 μM GSK-LSD1. (C) Plot showing the relative expression of C/EBPα mRNA in MLL-AF9 leukemia cells treated with vehicle control (PBS) or 0.5 μM GSK-LSD1. (D) Bar plot showing the number of peaks with either gains or losses of signal in ChIP-seq of PU.1 and C/EBPα in GSK-LSD1 treatment vs control. (E-F) Composite plots show normalized ChIP-seq signal at all dynamic ATAC sites in treatment and control for (E) PU.1 and (F) C/EBPα. (G) Genome browser track (mm9 coordinates chromosome 9: 14 425 541-14 466 000) showing normalized ATAC-seq and ChIP-seq signal under treatment and control conditions. (H) Percentage occupancy of PU.1 and C/EBPα in control and treatment conditions for ATAC-seq sites induced by GSK-LSD1. DMSO, dimethyl sulfoxide.
LSD1 inhibitor–induced changes in chromatin accessibility are PU.1 and C/EBPα dependent
We next sought to delineate whether the TFs PU.1 and C/EBPα are critical for these dynamic changes in chromatin accessibility. We generated MLL-AF9 leukemias using an established mouse model, referred to in this study as PU.1 upstream regulatory element (URE) knockout (KO), in which heterozygous or homozygous genetic deletion of a PU.1 upstream regulatory element causes down-regulation (70% or 20% decrease, respectively) of PU.1.23 Additionally, we generated MLL-AF9 leukemias and then inactivated C/EBPα (referred to as C/EBPα KO) as previously described.24-26
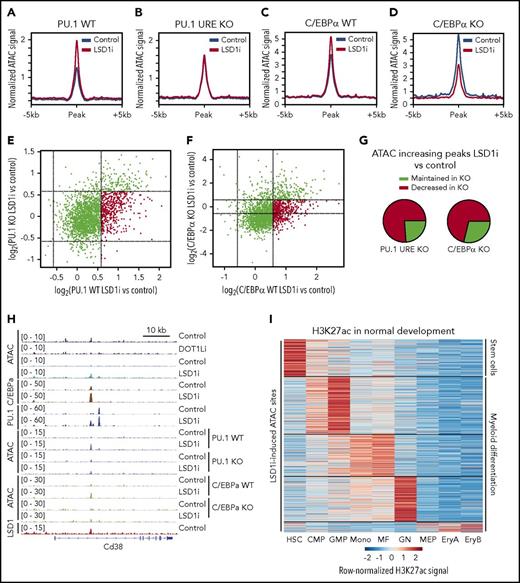
We performed ATAC-seq on PU.1 URE wild-type (WT) and PU.1 URE KO MLL-AF9 leukemias as well as C/EBPα WT and C/EBPα KO MLL-AF9 leukemias treated with vehicle or GSK-LSD1. WT MLL-AF9 leukemias treated with GSK-LSD1 exhibited a global increase in ATAC-seq signal at dynamic sites (Figure 4A,C). However, this increase in chromatin accessibility was abrogated in both PU.1 URE KO and C/EBPα KO MLL-AF9 leukemia cells (Figure 4B,D), suggesting that both of these TFs are necessary for the dynamic chromatin changes seen by ATAC-seq in the setting of LSD1 inhibition. Furthermore, not only was there no gain in chromatin accessibility seen after LSD1 inhibition in C/EBPα KO MLL-AF9 cells, there was additional loss of accessibility that was not seen in PU.1 URE KO cells, suggesting that C/EBPα loss may have additional effects on chromatin accessibility in the setting of LSD1 inhibition (Figure 4D). A log2 plot comparing dynamic changes in accessibility between PU.1 URE WT and PU.1 URE KO leukemia cells treated with vehicle or GSK-LSD1 revealed a preferential dropout of regions of increased accessibility with GSK-LSD1 treatment in the PU.1 URE KO leukemia cells (Figure 4E,G, top right quadrant). This was similarly observed in a log2 plot comparing dynamic sites between C/EBPα WT and C/EBPα KO MLL-AF9 leukemia cells (Figure 4F-G, top right quadrant). Figure 4H shows comprehensive TF ChIP-seq and ATAC-seq tracks at the CD38 locus as an illustrative example of the alterations in chromatin dynamics and TF networks that occur uniquely in the setting of LSD1 inhibition, but not with DOT1L inhibition. An intronic region within the CD38 locus harbored a site of increased accessibility and co-occupancy of PU.1 and C/EBPα in response to LSD1 inhibition, and, in the setting of decreased PU.1 expression or C/EBPα deficiency, this site exhibited decreased accessibility when compared with WT treated cells (Figure 4H). These findings suggest that the chromatin dynamics observed with LSD1 inhibition require the myeloid TFs PU.1 and C/EBPα.
Effect of TF loss on LSD1 inhibitor-induced changes in chromatin accessibility. (A-B) Composite plots show normalized ATAC-seq signal at GSK-LSD1-induced sites in PU.1 WT (A) and PU.1 depletion (B). (C-D) Composite plots show normalized ATAC-seq signal at GSK-LSD1–induced sites in C/EBPα WT (C) and C/EBPα KO (D). (E) Scatterplot of log2 fold changes in ATAC-seq signal of GSK-LSD1 treatment vs control for PU.1 WT (x axis) and PU.1 depletion (y axis). Red indicates sites that increase 1.5-fold in WT that fail to reach such levels in the PU.1-depleted setting. (F) Scatterplot of log2 fold changes in ATAC-seq signal of GSK-LSD1 treatment vs control for C/EBPα WT (x axis) and C/EBPα KO (y axis). Red indicates sites that increase 1.5-fold in WT that fail to reach such levels in the C/EBPα KO setting. (G) Pie charts representing the quantification of sites in panels E-F that either maintain or lose the increase in accessibility upon treatment with GSK-LSD1 in the setting of TF depletion. (H) Genome browser track showing normalized ATAC-seq and ChIP-seq signal under treatment and control conditions, centered on a site that fails to become induced in the setting of either PU.1 depletion or C/EBPα KO. (I) Heat map of normalized H3K27ac signal in normal hematopoietic development at all GSK-LSD1–induced ATAC-seq peaks, showing the majority of sites are active in cells associated with myeloid differentiation (clustered by k means with k = 5). CMP, common myeloid progenitor; EryA, erythroblasts A; EryB, erythroblasts B; GN, granulocyte; HSC, hematopoietic stem cell; MEP, megakaryocyte/erythrocyte progenitor; MF, macrophage; Mono, monocyte.
Effect of TF loss on LSD1 inhibitor-induced changes in chromatin accessibility. (A-B) Composite plots show normalized ATAC-seq signal at GSK-LSD1-induced sites in PU.1 WT (A) and PU.1 depletion (B). (C-D) Composite plots show normalized ATAC-seq signal at GSK-LSD1–induced sites in C/EBPα WT (C) and C/EBPα KO (D). (E) Scatterplot of log2 fold changes in ATAC-seq signal of GSK-LSD1 treatment vs control for PU.1 WT (x axis) and PU.1 depletion (y axis). Red indicates sites that increase 1.5-fold in WT that fail to reach such levels in the PU.1-depleted setting. (F) Scatterplot of log2 fold changes in ATAC-seq signal of GSK-LSD1 treatment vs control for C/EBPα WT (x axis) and C/EBPα KO (y axis). Red indicates sites that increase 1.5-fold in WT that fail to reach such levels in the C/EBPα KO setting. (G) Pie charts representing the quantification of sites in panels E-F that either maintain or lose the increase in accessibility upon treatment with GSK-LSD1 in the setting of TF depletion. (H) Genome browser track showing normalized ATAC-seq and ChIP-seq signal under treatment and control conditions, centered on a site that fails to become induced in the setting of either PU.1 depletion or C/EBPα KO. (I) Heat map of normalized H3K27ac signal in normal hematopoietic development at all GSK-LSD1–induced ATAC-seq peaks, showing the majority of sites are active in cells associated with myeloid differentiation (clustered by k means with k = 5). CMP, common myeloid progenitor; EryA, erythroblasts A; EryB, erythroblasts B; GN, granulocyte; HSC, hematopoietic stem cell; MEP, megakaryocyte/erythrocyte progenitor; MF, macrophage; Mono, monocyte.
LSD1 has previously been implicated in controlling tissue- and disease-specific gene expression programs through decommissioning of enhancers.27 We hypothesized that the myeloid differentiation program engaged by LSD1 inhibition may be in part mediated through the “recommissioning” of enhancers important for myeloid differentiation. To test this computationally, we leveraged previously generated datasets that defined H3K27ac enhancer peaks enriched in various stages of normal hematopoiesis.28 We then identified GSK-LSD1–induced sites of dynamic chromatin accessibility as assessed by ATAC-seq and intersected these sites with H3K27ac peaks present in normal hematopoiesis. We then used k means clustering of the H3K27ac signal in an attempt to infer a pattern of regulatory element activation in development. In Figure 4I, as identified by the labels to the right of the heat map, the bulk of the H3K27ac signal at GSK-LSD1–induced ATAC-seq sites are representative of regulatory elements engaged in myeloid differentiation. The preponderance of this intersection between GSK-LSD1–induced dynamic sites of chromatin accessibility and lineage-specific H3K27ac peaks occurred in more differentiated myeloid progenitors, in particular granulocyte-macrophage progenitors (GMP), monocytes, macrophages, and granulocytes (Figure 4I). Since these leukemia cells are known to be arrested at a stage of myeloid development close to GMPs, these data demonstrate that indeed LSD1 inhibitors are recommissioning enhancers that control a normal late-myeloid development program to induce AML cell differentiation.
Reduced expression of PU.1 confers resistance of MLL-AF9 leukemia cells to LSD1 inhibition
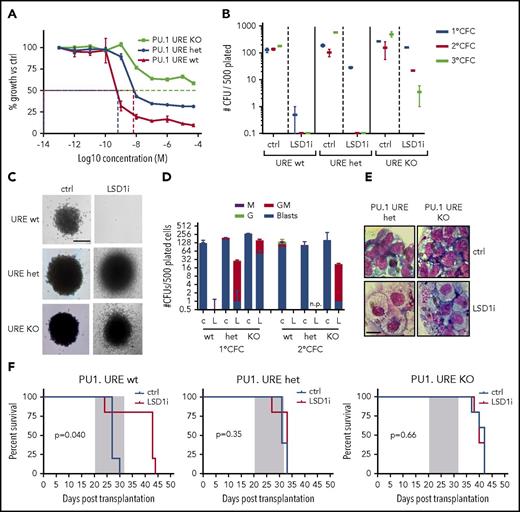
PU.1 regulates hematopoietic differentiation29 and also plays a role in leukemogenesis. Complete loss of PU.1 abrogates MLL-AF9 leukemogenicity, while decreased expression of PU.1 modulated by heterozygous or homozygous deletion of an upstream enhancer element (PU.1 URE KO) contributes to AML development.30,31 Given that the dynamic changes in chromatin accessibility in response to LSD1 inhibition are PU.1 dependent, we next employed this system to assess the impact of PU.1 depletion on sensitivity to LSD1 inhibition in vitro and in vivo. We treated PU.1 URE KO, PU.1 URE heterozygous, and WT MLL-AF9 primary leukemias cells with GSK-LSD1 for 48 hours in vitro. PU.1 URE KO AML cells were more resistant to LSD1 inhibition relative to PU.1 URE heterozygous and WT AML cells (Figure 5A). PU.1 URE heterozygous leukemia cells, which express PU.1 at ∼70% of WT levels (supplemental Figure 2A-C), were also more resistant to LSD1 inhibition than WT leukemia cells (Figure 5A). In a colony-formation assay, we similarly observed increased replating capacity in PU.1 URE KO cells treated with GSK-LSD1 up to the third replating (Figure 5B) with persistence of granulocyte-macrophage and blast-like colonies (Figure 5C-E). This resistance to LSD1 inhibition was observed in vivo in a secondary transplantation model. Primary PU.1 URE KO, PU.1 URE heterozygous, and WT MLL-AF9 leukemias were engrafted into secondary recipients subsequently treated with GSK-LSD1 during a 2-week window. Consistent with the in vitro findings, the survival advantage seen in mice engrafted with WT MLL-AF9 leukemias and treated with GSK-LSD1 was lost in recipients engrafted with PU.1 URE KO and PU.1 URE heterozygous MLL-AF9 leukemias cells (Figure 5F). These findings suggest that PU.1 levels influence the efficacy of LSD1 inhibition in vitro and in vivo.
LSD1 inhibition with GSK-LSD1 is PU.1 dependent. (A) In vitro IC50 growth curve of PU.1 URE WT, heterozygous (het), and KO MLL-AF9 leukemia cells treated with increasing concentrations of GSK-LSD1. n = 3. (B) Colony-formation assay performed by plating 500 GFP+ cells derived from primary MLL-AF9 PU.1 URE WT, heterozygous, or KO leukemic mice with or without GSK-LSD1. n = 3. (C) Representative 1° colony-forming unit (CFU) assay colonies showing persistence of more immature colonies in PU.1 URE het and KO cells. Bar, 0.1 mm. n = 3. (D) Representation of morphologies derived from 1° CFU and 2° CFU assays upon GSK-LSD1 treatment (c, control; G, granulocyte; GM, granulocyte/macrophage; L, LSD1i; M, macrophage). n = 3. (E) Cytospins of cells isolated from PU.1 URE heterozygous and KO 1° CFU assays. Scale bar, 10 µm. (F) Kaplan-Meier survival curves of PU.1 URE WT (left), PU.1 URE heterozygous (middle), and PU.1 URE KO (right) secondary transplanted mice treated with vehicle or GSK-LSD1 for 2 weeks. n = 5 mice per cohort. CFC, colony-forming cell.
LSD1 inhibition with GSK-LSD1 is PU.1 dependent. (A) In vitro IC50 growth curve of PU.1 URE WT, heterozygous (het), and KO MLL-AF9 leukemia cells treated with increasing concentrations of GSK-LSD1. n = 3. (B) Colony-formation assay performed by plating 500 GFP+ cells derived from primary MLL-AF9 PU.1 URE WT, heterozygous, or KO leukemic mice with or without GSK-LSD1. n = 3. (C) Representative 1° colony-forming unit (CFU) assay colonies showing persistence of more immature colonies in PU.1 URE het and KO cells. Bar, 0.1 mm. n = 3. (D) Representation of morphologies derived from 1° CFU and 2° CFU assays upon GSK-LSD1 treatment (c, control; G, granulocyte; GM, granulocyte/macrophage; L, LSD1i; M, macrophage). n = 3. (E) Cytospins of cells isolated from PU.1 URE heterozygous and KO 1° CFU assays. Scale bar, 10 µm. (F) Kaplan-Meier survival curves of PU.1 URE WT (left), PU.1 URE heterozygous (middle), and PU.1 URE KO (right) secondary transplanted mice treated with vehicle or GSK-LSD1 for 2 weeks. n = 5 mice per cohort. CFC, colony-forming cell.
C/EBPα deletion confers resistance of MLL-AF9 leukemia cells to LSD1 inhibition
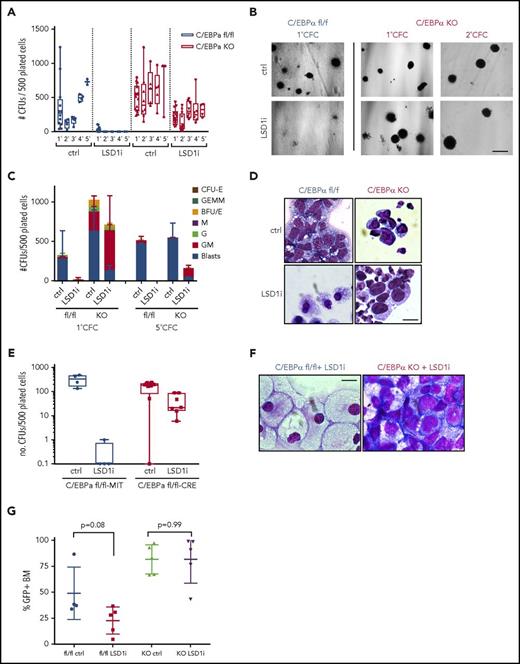
C/EBPα has been reported to be essential for initiation of Hoxa9/Meis1-mediated leukemogenesis32 and also for AML initiated by MLL-ENL.33 In MLL-AF9–driven AML, C/EBPα is essential for leukemia initiation, but not maintenance.25 Like the PU.1-dependent nature of the dynamic chromatin changes seen in response to LSD1 inhibition, these changes are also C/EBPα-dependent. We therefore generated C/EBPα-deficient MLL-AF9 cells to assess the effect of C/EBPα loss on sensitivity to LSD1 inhibition. C/EBPα KO MLL-AF9 leukemias were generated by retroviral delivery of the MLL-AF9 fusion into LSK cells from Mx-Cre × C/EBPαfl/fl mice previously treated with polyinosinic/polycytidylic acid to induce interferon-mediated induction of Cre and excision of the C/EBPα alleles. These preleukemic C/EBPα-deficient cells were then transplanted into recipient hosts and, in order to overcome loss of C/EBPα as previously reported, were supported with continuous administration of interleukin-3 and granulocyte colony-stimulating factor.25 C/EBPα KO and WT MLL-AF9 leukemia cells were serially replated up to 5 times in semisolid medium in the presence of 0.5 μM GSK-LSD1. Similar to what was observed for PU.1 URE KO cells, C/EBPα KO leukemia cells were more resistant to LSD1 inhibition than WT leukemia cells (Figure 6A-B). Only after the third replating of C/EBPα KO cells were more differentiated (GM-like) colonies observed with GSK-LSD1 treatment (Figure 6C). This was in contrast to WT MLL-AF9 cells, which were lost after the first replating. Cells with blast-like morphology persisted in the GSK-LSD1–treated C/EBPα KO cells even up to the fifth replating (Figure 6B-D).
LSD1 inhibition with GSK-LSD1 is C/EBPα-dependent. (A) Colony formation assay from WT or C/EBPα KO MLL-AF9 leukemia cells treated with vehicle or GSK-LSD1. n = 3. (B) Morphology of 1° CFU colonies derived from WT (left) or C/EBPα KO cells (right) treated with vehicle (top) or GSK-LSD1 (bottom). Bar, 0.5 mm. n = 3. (C) CFU assay with colony morphologies upon GSK-LSD1 treatment in WT and C/EBPα KO cells (BFU-E, burst-forming unit-erythroid; CFU-E, colony forming unit-erythroid; G, granulocyte; GEMM, granulocyte/erythrocyte/macrophage/megakaryocyte; GM, granulocyte/macrophage; M, macrophage). n = 3. (D) Representative morphology of cells from CFU assay with GSK-LSD1 treatment. Scale bar, 10 µm. (E) CFU assay with colony types after treatment with vehicle or GSK-LSD1 in C/EBPαfl/fl+MIT empty vector and C/EBPαfl/fl+MIT-Cre leukemia cells. n = 3. (F) Representative morphology of cells from 1° CFU assay from WT or C/EBPα-excised MLL-AF9 leukemia cells treated with 0.5 µM GSK-LSD1. Scale bar, 5 µm. (G) GFP engraftment in the BM of leukemic mice engrafted with WT or C/EBPα KO MLL-AF9 cells treated in vivo with vehicle or GSK-LSD1 for 2 weeks. n = 5. (H) Bar graphs representing spleen weights from leukemic mice engrafted with either WT or C/EBPα KO MLL-AF9 leukemia cells treated in vivo with vehicle or GSK-LSD1 for 2 weeks. n = 3.
LSD1 inhibition with GSK-LSD1 is C/EBPα-dependent. (A) Colony formation assay from WT or C/EBPα KO MLL-AF9 leukemia cells treated with vehicle or GSK-LSD1. n = 3. (B) Morphology of 1° CFU colonies derived from WT (left) or C/EBPα KO cells (right) treated with vehicle (top) or GSK-LSD1 (bottom). Bar, 0.5 mm. n = 3. (C) CFU assay with colony morphologies upon GSK-LSD1 treatment in WT and C/EBPα KO cells (BFU-E, burst-forming unit-erythroid; CFU-E, colony forming unit-erythroid; G, granulocyte; GEMM, granulocyte/erythrocyte/macrophage/megakaryocyte; GM, granulocyte/macrophage; M, macrophage). n = 3. (D) Representative morphology of cells from CFU assay with GSK-LSD1 treatment. Scale bar, 10 µm. (E) CFU assay with colony types after treatment with vehicle or GSK-LSD1 in C/EBPαfl/fl+MIT empty vector and C/EBPαfl/fl+MIT-Cre leukemia cells. n = 3. (F) Representative morphology of cells from 1° CFU assay from WT or C/EBPα-excised MLL-AF9 leukemia cells treated with 0.5 µM GSK-LSD1. Scale bar, 5 µm. (G) GFP engraftment in the BM of leukemic mice engrafted with WT or C/EBPα KO MLL-AF9 cells treated in vivo with vehicle or GSK-LSD1 for 2 weeks. n = 5. (H) Bar graphs representing spleen weights from leukemic mice engrafted with either WT or C/EBPα KO MLL-AF9 leukemia cells treated in vivo with vehicle or GSK-LSD1 for 2 weeks. n = 3.
Resistance to LSD1 inhibition in C/EBPα KO MLL-AF9 leukemia cells was also confirmed by deleting C/EBPα after primary MLL-AF9 had been generated. C/EBPαfl/fl primary MLL-AF9 leukemia cells were transduced with MIT-Cre vectors (supplemental Figure 3A). Similar to MLL-AF9 leukemias generated from LSK cells already harboring induced C/EBPα deletions, C/EBPα KO MLL-AF9 cells exhibited increased replating capacity and blast persistence upon LSD1 inhibition relative to WT MLL-AF9 leukemia cells (Figure 6E-F). These findings were also confirmed using short hairpin RNA constructs to knock down C/EBPα expression in primary MLL-AF9 leukemia cells (supplemental Figure 3B-E). Furthermore, we treated secondary recipients engrafted with primary C/EBPα KO MLL-AF9 leukemias with GSK-LSD1 (supplemental Figure 2D). After 2 weeks of treatment, C/EBPα KO leukemic mice showed no decrease in MLL-AF9 allele burden in the bone marrow as measured by the frequency of GFP+ leukemia cells (Figure 6G). Taken together, these findings suggest that, in addition to PU.1 levels, C/EBPα is important for the antileukemic activity of LSD1 inhibition in vitro and in vivo.
Discussion
We demonstrate the antileukemic activity of the LSD1 inhibitor GSK-LSD1 using a leukemia model driven by MLL-AF9. In a few cases, GSK-LSD1 was able to cause complete leukemia eradication in treated mice. We noted the ability of GSK-LSD1 treatment to induce immunophenotypic and morphologic evidence of myeloid differentiation in AML blasts, similar to the prodifferentiation effects of another targeted epigenetic therapy, the DOT1L inhibitor EPZ4777, which has been demonstrated by our group and others to also have activity against this group of MLL-rearranged leukemias. By performing ATAC-seq, we found that they had profoundly contrasting effects on chromatin accessibility. Whereas DOT1L inhibition predominantly caused losses in chromatin accessibility, LSD1 inhibition with GSK-LSD1 caused increased chromatin accessibility. Motif analysis of these GSK-LSD1–induced dynamic sites revealed a possible role for the myeloid TFs PU.1 and C/EBPα. This was then confirmed by TF ChIP-seq, which revealed common sites of GSK-LSD1–induced dynamic changes in chromatin accessibility and occupancy by PU.1 and C/EBPα. These sites also had a tendency to coincide with H3K27ac enhancer peaks particularly found in more differentiated myeloid progenitors rather than primitive hematopoietic stem cells.
The observation that DOT1L inhibition with EPZ4777 largely induced decreased chromatin accessibility across the genome is consistent with what is known about the pathogenesis of MLL-AF9–driven leukemogenesis. Our group previously demonstrated that H3K79 methylation states function to control gene expression during leukemogenesis, with higher level H3K79 methylation correlating with increased levels of gene expression, particularly in MLL-AF9 target genes, including the HOXA cluster genes and Meis1.34 With less H3K79 methylation, the oncogenic transcriptional program was shut down and reversed the differentiation blockade seen in AML. EPZ4777 treatment specifically ablates H3K79 methylation and mediates transcriptional silencing of these genes, which is consistent with the closing of chromatin accessibility that was observed with EPZ4777 treatment.
In contrast, the antileukemic activity mediated by LSD1 inhibition correlates instead with increased chromatin accessibility, and more importantly, requires the TFs PU.1 and C/EBPα. Our analyses examining the intersection between the TF ChIP-seq and ATAC-seq datasets revealed that PU.1 occupancy increased at sites of increased chromatin accessibility after treatment with GSK-LSD1, while C/EBPα binding was relatively unchanged at these sites. One interpretation of these observations is that C/EBPα may be important for defining the loci at which gains in accessibility occur, whereas increased PU.1 occupancy may be important for mediating this increased accessibility. Previous studies have established the necessity of engaging a myeloid differentiation program as a prerequisite for AML initiation by MLL-AF9.25 Genetic ablation of C/EBPα blocked GMP formation and was sufficient to prevent AML initiation.25 Here, we found that depletion of PU.1 or C/EBPα rendered MLL-AF9 cells resistant to LSD1 inhibition. We speculate that it may be just as important to engage this myeloid differentiation pathway to drive AML differentiation as it is to reverse stem cell–associated gene expression as happens with DOT1L inactivation. Furthermore, the increased chromatin accessibility seen after LSD1 inhibition is consistent with what has been described with other LSD1 inhibitors that have been studied in the context of AML-directed therapies. Two novel LSD1 inhibitors exhibited cytostatic activity against multiple leukemia subtypes, including MLL-AF9 leukemias and were found to activate superenhancers by increasing H3K27ac at these regions.21 It remains unclear precisely how pharmacologic inhibition of LSD1 catalytic activity or disruption of LSD1 complex formation via these compounds lead to increased chromatin accessibility, although other studies have revealed that LSD1 inhibitors can evict LSD1 from chromatin.19,35 By removing LSD1 repressive enzymatic activity from target genes, one could speculate that the net impact of this is a shift in the balance toward gene activation at these sites with occupancy of activating TFs, such as PU.1 and C/EBPα, that lead to increased accessibility in these regions. It will be informative to determine whether genetic depletion of LSD1 recapitulates the changes in chromatin dynamics seen with pharmacologic LSD1 inhibition. This will help clarify whether it is the loss of LSD1 more broadly or the inhibition of LSD1 enzymatic activity at chromatin-occupied repressive complexes that is important for mediating the epigenetic changes and anti-leukemic properties.
One limitation of this study was the use of only MLL-rearranged AML mouse models to assess the efficacy of LSD1 inhibition. It is clear that LSD1 inhibition may have antileukemic efficacy against multiple AML subtypes driven by other mutations.36-38 For this reason, it will be important to determine if similar effects on chromatin dynamics are seen and whether similar TF networks are engaged when non–MLL-rearranged AML cells are treated with GSK-LSD1. Consistent with this, a previous study demonstrated that knockdown of PU.1 modulates LSD1 inhibitor sensitivity in a non–MLL-rearranged human acute erythroid leukemia cell line.19 This further validates the important role of PU.1 in the antileukemic activity of LSD1 inhibitors in other AML subtypes. Additional investigations down this line will be informative for design of clinical trials using this class of epigenetic therapies.
Our investigations into the chromatin-modifying effects of 2 distinct epigenetic therapies targeting either the histone demethylase activity LSD1 or the histone methyltransferase DOT1L is illustrative of the concept that multiple paths toward myeloid differentiation can potentially be engaged to achieve therapeutic efficacy in AML. The first of the paths toward induction of myeloid differentiation and loss of leukemia self-renewal is the inhibition of stem cell–associated gene expression programs such inhibition of HOX gene expression via DOT1L inhibition. The second pathway, described in detail here, is the engagement of myeloid-associated enhancers through LSD1 inhibition to override the myeloid differentiation block and thus inhibit leukemia self-renewal. Combination therapies, such as LSD1 inhibition with all-trans-retinoic acid, have already identified synergies that could be achieved to augment these responses and are under investigation in clinical trials.38 Furthermore, based on the different mechanisms of action of LSD1 inhibition and DOT1L inhibition, this combination is potentially worth exploring.18 Additional mechanistic understanding of how these targeted agents are effective against AML is needed in order to identify optimal combination regimens that deepen remissions and improve outcomes for patients with AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their patients and their families for their participation in this study.
Technical assistance was provided by Katie Yang Li and Xujun Wang from the Innovation Center at the Center for Epigenetics Research, Memorial Sloan-Kettering Cancer Center. This work was supported by the German Research Council (DFG) within the Collaborative Research Centre (SFB) 1243 “Cancer Evolution” (startup funding 2017 to M.C.), a Young Investigator Award from the Conquer Cancer Foundation (S.F.C.), a Career Development Program Fellow Award from the Leukemia & Lymphoma Society (S.F.C.), a Memorial Sloan-Kettering Cancer Center Clinical Scholars Biomedical Research Fellowship (S.F.C.), National Institutes of Health (NIH), National Cancer Institute grants PO1 CA66996 and R01 CA140575 (S.A.A.), Gabrielle’s Angel Research Foundation (S.A.A.), and an NIH Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748) from the National Cancer Institute. This research was funded, in part, through the NIH/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Authorship
Contribution: M.C. and S.A.A. conceived of and designed the experiments; M.C., S.F.C., A.C., E.L., M.D.W., M.Y., and B.W. performed experiments; R.P.K. analyzed the data; S.A.A., S.F.C., A.K., K.N.S., R.G.K., H.P.M., D.G.T., U.S., R.L.L., and H.Y.R. provided oversight for critical aspects of the study; and M.C., R.P.K., S.F.C., and S.A.A. interpreted the data and results and wrote the manuscript.
Conflict-of-interest disclosure: S.A.A. is a consultant for Epizyme, Inc. S.F.C., R.L.L., and S.A.A. are consultants for Imago Biosciences, Inc. H.Y.R. is chief executive officer of Imago Biosciences, Inc. The remaining authors declare no competing financial interests.
Correspondence: Scott A. Armstrong, Department of Pediatric Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 1641, Boston, MA 02215; e-mail: scott_armstrong@dfci.harvard.edu; and Richard P. Koche, Center for Epigenetics Research, Memorial Sloan Kettering Cancer Center, 1275 York Ave, Box 20, New York, NY 10065; e-mail: kocher@mskcc.org.
References
Author notes
M.C. and S.F.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal