Key Points
SF3B1 mutations in MDS-RS have a multipotent lymphomyeloid origin.
Transplantation of SF3B1 mutated MDS-RS HSCs into immune-deficient mice results in generation of characteristic ring sideroblasts.
Abstract
Mutations in the RNA splicing gene SF3B1 are found in >80% of patients with myelodysplastic syndrome with ring sideroblasts (MDS-RS). We investigated the origin of SF3B1 mutations within the bone marrow hematopoietic stem and progenitor cell compartments in patients with MDS-RS. Screening for recurrently mutated genes in the mononuclear cell fraction revealed mutations in SF3B1 in 39 of 40 cases (97.5%), combined with TET2 and DNMT3A in 11 (28%) and 6 (15%) patients, respectively. All recurrent mutations identified in mononuclear cells could be tracked back to the phenotypically defined hematopoietic stem cell (HSC) compartment in all investigated patients and were also present in downstream myeloid and erythroid progenitor cells. While in agreement with previous studies, little or no evidence for clonal (SF3B1 mutation) involvement could be found in mature B cells, consistent involvement at the pro–B-cell progenitor stage was established, providing definitive evidence for SF3B1 mutations targeting lymphomyeloid HSCs and compatible with mutated SF3B1 negatively affecting lymphoid development. Assessment of stem cell function in vitro as well as in vivo established that only HSCs and not investigated progenitor populations could propagate the SF3B1 mutated clone. Upon transplantation into immune-deficient mice, SF3B1 mutated MDS-RS HSCs differentiated into characteristic ring sideroblasts, the hallmark of MDS-RS. Our findings provide evidence of a multipotent lymphomyeloid HSC origin of SF3B1 mutations in MDS-RS patients and provide a novel in vivo platform for mechanistically and therapeutically exploring SF3B1 mutated MDS-RS.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 955.
Disclosures
Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Alnylam, Biogen, and Pfizer. Associate Editor David M. Bodine and the authors declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Identify recurrently mutated genes in the mononuclear cell fraction of patients with myelodysplastic syndrome with ring sideroblasts (MDS-RSs), based on a study of bone marrow hematopoietic stem cell (HSC) and progenitor cell compartments.
Evaluate the origin of the SF3B1-mutated clone and stem cell function of patients with MDS-RSs in vitro, as well as in vivo, based on a study of bone marrow HSC and progenitor cell compartments.
Compare differentiation of SF3B1-mutated MDS-RS bone marrow HSCs and progenitor cells when transplanted into immune-deficient mice.
Release date: August 17, 2017; Expiration date: August 17, 2018
Introduction
The pathogenesis of myelodysplastic syndromes (MDS) involves a spectrum of genetic alterations and varies considerably between disease subgroups.1 Recurrent mutations in lower-risk MDS are largely confined to core components of the spliceosome (SF3B1, SRSF2, U2AF35, and ZRSR2), epigenetic modifiers (TET2, DNMT3A, IDH1/2, and ASXL1), and genes involved in pathways such as signal transduction and transcriptional regulation.2-4 While many mutations coexist with each other and may be found in various myeloid malignancies, splice factor mutations occur in a mutually exclusive manner and are much more frequent than in other hematological tumors.5,6 Remarkably, highly recurrent heterozygous mutations in the RNA splicing gene SF3B1 are found in as many as 70% to 85% of patients with lower-risk MDS and ring sideroblasts (MDS-RS)2,3,7 but are relatively rare in other MDS subtypes. Recurrent SF3B1 mutations are therefore likely to play a distinct biological role in MDS-RS pathogenesis.8 Other studies have identified recurrent SF3B1 mutations in otherwise healthy elderly individuals as evidence that SF3B1 mutations are also involved in premalignant clonal hematopoiesis.9
In MDS-RS, mitochondrial ferritin accumulates in the mitochondria of the erythroblasts, resulting in accumulation of characteristic ring sideroblasts (RSs), ineffective erythropoiesis, and anemia. We previously reported that MDS-RS erythroblasts display reduced expression of ABCB7, a gene involved in the regulation of mitochondrial iron homeostasis, and that the expression level of ABCB7 is inversely associated with the percentage of marrow RS. We also demonstrated that SF3B1-mutated MDS-RS progenitors display impaired splicing with potential downstream consequences for genes of key importance for hemoglobin synthesis and terminal erythroid differentiation.10 Although these and other findings implicate an impact of recurrent SF3B1 mutations primarily in the erythroid lineage, the primary cellular target(s) of these recurrent SF3B1 mutations remain to be elucidated. While having a low propensity for leukemic transformation, these patients typically suffer from severe anemia requiring regular transfusion therapy, and there is currently no curative treatment available for them.11 Importantly, in addition to negatively affecting erythroid lineage development, recurrent SF3B1 mutations provide a competitive clonal advantage in MDS-RS, but it remains unclear at which level in the hematopoietic hierarchy this advantage is achieved. Establishing this, as well as the identity of the hematopoietic cells capable of propagating and sustaining the SF3B1-mutated clone, will be key toward developing more targeted and potentially curative therapeutic strategies for SF3B1-mutated MDS-RS. Recent studies have suggested that SF3B1 mutations might primarily target the phenotypic hematopoietic stem cell (HSC) compartment.12 However, it is widely recognized that any phenotypically defined HSC population also contains a substantial fraction of non-HSCs, and since the lymphoid lineages were not found to be involved in the SF3B1-mutated clones,3,12,13 definitive evidence for recurrent SF3B1 mutations targeting true lymphomyeloid HSCs in MDS-RS is lacking.
Here, we provide evidence that recurrent SF3B1 mutations target the multipotent lymphomyeloid HSC compartment and that only these targeted HSCs are able to propagate long-term the SF3B1-mutated clone in vitro and in vivo. Importantly, we also demonstrate in immune-deficient mice the in vivo differentiation of purified HSCs from SF3B1-mutated MDS-RS patients to characteristic RSs, providing a valuable in vivo platform for unraveling and targeting the dysregulated erythroid development in SF3B1-mutated MDS-RS.
Materials and methods
Patient samples
Bone marrow (BM) and peripheral blood (PB) samples were obtained before treatment of MDS-RS patients, as defined in the World Health Organization 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia, diagnosed using a multiprofessional conference approach.14 BM and PB were collected from 15 healthy subjects in the Hematology Department at Karolinska University Hospital, Karolinska, Sweden. Informed consent was obtained from patients and healthy controls, and the study was approved by the Ethics Research Committee at Karolinska Institutet (2010/427-31/1 and 2011/1257-31). BM mononuclear cells (MNCs) were isolated by Lymphoprep (Fresenius, Oslo, Norway) gradient centrifugation.
Flow cytometry and fluorescent-activated cell sorting (FACS)
Stem and progenitor cell, mature lymphoid, myeloid, and erythroid cell, as well as nonobese diabetic/LtSz-scid IL2Rγc−/− (NSG) human engraftment analysis and purification were performed as described in supplemental Methods (available on the Blood Web site).
Functional stem and progenitor cell assays
Detailed methods for colony-forming cell (CFC) and long-term culture colony-forming cell (LTC-CFC) assays are described in supplemental Methods.
DNA mutational analysis
Targeted DNA sequencing of DNA isolated from the bulk BM from all 40 subjects was performed using Haloplex selector probes15 and prepared according to manufacturer’s instructions (Agilent Technologies, Santa Clara, CA). The sequencing platform and mutational call analysis have been previously described15 and are explained briefly in supplemental Methods. For a number of patients, computational prediction analysis was performed as previously described for the targeted sequenced samples.16 Briefly, after identifying and selecting the reliable driver mutations, we corrected the variant allele frequency (VAF) for gene mutations mapping on the X chromosome for the patients. We also assessed loss of heterozygosity for genes such that the 95% confidence interval of the estimated VAF is higher than 65%. The distribution of the VAF is drawn from a β-binomial distribution with the parameters number of mutated reads (Nmut) and number of unmutated reads (Nref = Ntot − Nmut), and we corrected the VAF accordingly, as previously described.16 We then assessed precedence between gene mutations for each patient by comparing the 95% confidence interval of the VAF between genes.
To identify mutations in LTC-CFC and CFC colonies, genomic DNA from individual colonies was isolated using isopropanol/ethanol precipitation. Pyrosequencing was applied to detect the heterozygous SF3B1 single-nucleotide mutations previously found by targeted sequencing using Pyromark Q24 system (QIAGEN, Hilden, Germany). DNA from healthy normal bone marrow (NBM) LTC-CFCs and CFCs was used as wild-type control. The colonies were only scored as positive or negative due to their assumed clonal origin. However, as there is a potential for contamination of cells from other colonies when picking colonies, we considered the colonies with a VAF >25% as positive, those with a VAF <5% as negative, and colonies with a VAF between 6% to 24% as inconclusive. See supplemental Methods for a more detailed description.
To quantify mutations in purified stem and progenitor cells, we subjected isolated DNA to whole-genome amplification using a Genomi-PHI V2 kit according to the manufacturer’s instructions (GE Healthcare, Chicago, IL). For pro–B cells, we performed pyrosequencing to identify the mutations previously found by targeted sequencing, and to analyze the rest of the populations, we used the Sequenom platform according to manufacturer’s instructions at the mutational analysis facility at Karolinska Institute. Briefly, the assays were set up in multiplexed reactions with primer pairs designed to amplify the desired point mutations. The polymerase chain reaction (PCR) was then cleaned up by enzymatic reaction, and extension primers were designed to anneal directly adjacent to the point mutation of interest. The extended primers were then subjected to matrix-assisted laser desorption/ionization time-of-flight spectrometry analysis on Sequenom MassARRAY Analyzer (Agena Bioscience, San Diego, CA). For the allelic discrimination and quantitative estimation of the mutated allelic frequency, SpectroTyper 4.0 software was used. The sensitivity of the analysis was validated by serial dilution spiked-in samples of patient BM MNCs. Our validation experiment revealed that for <5% of cells containing a mutation, the Sequenom assay could not confidentially predict the presence or absence of a mutation (data not shown).
For the analysis of purified peripheral mature and xenotransplanted cells, we directly lysed the cells in 4 μl D2 lysis buffer from a single-cell REPLI-g kit (QIAGEN), and the DNA was amplified according to manufacturer’s instructions17,18 prior to droplet digital PCR analysis. PCR mixture preparation, droplet generation procedures, droplet analysis for emission in the HEX or FAM fluorescent signal channels using the QX200 Droplet Digital PCR System (Bio-Rad, Berkeley, CA) and QuantaSoft version 1.15 (Bio-Rad), and validation experiments are described in detail in supplemental Methods.
Xenograft transplantation
NSG mice 8 to 10 weeks of age were given sublethal irradiation by exposure of 2 doses of 1.25 Gy (Cs source) 4 hours apart. Cells were injected intrafemorally within 24 hours of the last irradiation dose. 475 to 20 000 purified stem and progenitor cells were injected per mouse. Human engraftment and lineage distribution were analyzed by flow cytometry 20 to 22 weeks after injection. Reconstitution <0.1% was considered to be below the specificity of the method as previously described.19,20 See supplemental Methods for flow cytometry analysis from NSG BM. All mice were bred and maintained at the Oxford Biomedical Services, and all experiments were done with the approval of the UK Home Office.
CellProfiler image analysis software to measure RSs
RS counting from engrafted paraffin-embedded NSG femur legs was performed using the automated open-source program CellProfiler (http://www.cellprofiler.org).21 The original color images were converted into grayscale images from scanned paraffin-embedded sections using the Panoramic MIDI II scanner and Panoramic Viewer software (3D Histech, Budapest, Hungary). The images were taken using an AxioCam MRm (Carl Zeiss, Oberkochen, Germany) at 40× magnification. Segmentation was performed by the program’s automatic thresholding algorithm to identify objects that were darker than the surrounding environment; these objects were potential RS. Filters were used to exclude objects that were either too big or too small, which usually represented various staining artifacts or debris. Accepted RS were highlighted by blue borders. These output images were manually inspected as a measure of quality assurance, and they also provided guidance for optimizing the program settings. For each erythrocyte with RS morphology, 5 basic parameters were measured: RS area, perimeter, major axis, minor axis, and form factor. Form factor was calculated by the equation (4 × π × area / perimeter2 ) as a measurement of “roundness.” It equals unity (1) for perfectly round objects and decreases to 0 for more irregular or elongated objects. Image segmentation was done by manually picking a few representative RSs, which served as color references and were processed by the program’s proprietary algorithm.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software. For individual comparisons, a nonparametric Mann-Whitney test was used, and P values <.05 were considered significant.
Results
Patient characteristics
In total, we used BM samples from 40 patients with MDS-RS selected from the Karolinska Institutet MDS biobank for the various analyses. All had a normal karyotype; 39 patients were SF3B1 mutated and 1 was SF3B1 wild-type. Additional mutations found in these patients, as well as their clinical characteristics, are shown in Table 1.
Clinical, hematologic, and cytogenetic characteristics of MDS-RS patients
| Patient number . | Sex . | Age (y) . | Hb (g/L) . | Transfusion . | SF3B1 mutation . | Other mutations (targeted sequence) . | BM cellularity (%) . | RS (%) . | Blasts (%) . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 72 | 102 | No | p.K700E | STAG2 (340del) | 70 | 29 | 1 |
| 2 | F | 69 | 89 | No | p.K700E | TET2 (p.F125fs) | 60 | 24 | 4.5 |
| 3 | F | 83 | 88 | Yes | p.K700E | TET2 (Q916*) | 90 | 30 | 4 |
| 4 | M | 78 | 97 | No | p.K666R | c-KIT, TET2 (R1404*, 690*) | 50 | 39 | 3 |
| 5 | F | 69 | 93 | No | p.H662Q | TET2 (H1881Y) | 80 | 33 | 5.5 |
| 6 | M | 76 | 97 | No | p.H662D | DNMT3A (W305*) | 60 | 23 | 4.5 |
| 7 | F | 78 | 103 | No | p.E622D | B-COR (R63K) | 60 | 21 | 2 |
| 8 | M | 87 | 80 | Yes | p.R625L | IDH2 (R172K) | 70 | 22 | 2 |
| 9 | M | 81 | 106 | No | p.E622D | None | 60 | 33 | 2.5 |
| 10 | F | 65 | 77 | No | p.K700E | PRPF40B (669D) | 90 | 39 | 2 |
| 11 | F | 81 | 107 | No | p.K700E | EZH2 (T306fs), DNMT3A (E285*) | 70 | 85 | 0.5 |
| 12 | M | 64 | 132 | No | p.N626D | None | 50 | 42 | 5.5 |
| 13 | M | 70 | 117 | No | No mutation | SRSF2 (95_103del) | 90 | 73 | 1.5 |
| 14 | F | 71 | 98 | Yes | p.K700E | TET2 (Q746X) | 80 | 55 | 2 |
| 15 | F | 77 | 97 | Yes | p.E622D | JAK2 (V617F) | 100 | 77 | 1 |
| 16 | F | 80 | 115 | No | p.K700E | DNMT3A (V296M) | 50 | 36 | 2.5 |
| 17 | F | 60 | 113 | No | p.K700E | None | 100 | 25 | 1.5 |
| 18 | M | 79 | 108 | No | p.E622D | DNMT3A (I643M) | 80 | 16 | 2 |
| 19 | F | 54 | 125 | Yes | p.K700E | DNMT3A (F868S) | 90 | 10 | 0 |
| 20 | M | 67 | 97 | Yes | p.K700E | None | 65 | 35 | 0 |
| 21 | M | 67 | 100 | Yes | p.K700E | None | 60 | 40 | 1 |
| 22 | M | 68 | 101 | No | p.K700E | TET2 (E1555fs*22) | 70 | 40 | 0 |
| 23 | F | 74 | 106 | No | p.K666T | None | 70 | 20 | 2.5 |
| 24 | M | 76 | 107 | No | p.N626D | TET2 (R1452*) | — | 16 | 2 |
| 25 | F | 82 | 99 | No | p.E622D | None | 50 | 24 | 1.5 |
| 26 | M | 77 | 115 | Yes | p.E666R | None | 60 | 45 | 3 |
| 27 | F | 84 | 93 | Yes | p.K700E | EZH2 (K661E) | 40 | 33 | 2 |
| 28 | F | 75 | 107 | No | p.K700E | None | 40 | 28 | 1.5 |
| 29 | F | 76 | 105 | No | p.Y623C | None | 40 | 49 | 1.5 |
| 30 | F | 80 | 97 | No | p.K700E | None | 60 | 16 | 3.5 |
| 31 | F | 73 | 102 | No | p.E666R | None | 50 | 53 | 1 |
| 32 | F | 79 | 135 | No | p.K700E | TET2 (Q958fs*14) | — | 35 | 0 |
| 33 | M | 77 | 87 | Yes | p.R625L | None | — | 26 | 0 |
| 34 | M | 73 | 87 | Yes | p.R625L | TET2 (F1300I), SMC3 (L836X) | — | 16 | 2 |
| 35 | M | 64 | 101 | No | p.K666R | DNMT3A (Y735C) | — | 23 | 2 |
| 36 | F | 79 | 101 | No | p.H662Q | IDH2 (C402Y), TET2 (T1122fs*8) | — | 30 | 1 |
| 37 | F | 46 | 109 | No | p.K700E | None | 50 | 44 | 1 |
| 38 | M | 68 | 101 | Yes | p.K700E | IDH2 (R140Q), EZH2 (R732I), ZRSR2 (E120fs*24) | — | 16 | 1 |
| 39 | F | 77 | 111 | Yes | p.K700E | RUNX1 (R107S) | — | 31 | 2 |
| 40 | M | 49 | 121 | No | p.T663I | TET2 (P319fs*28) | 90 | 70 | 3 |
| Patient number . | Sex . | Age (y) . | Hb (g/L) . | Transfusion . | SF3B1 mutation . | Other mutations (targeted sequence) . | BM cellularity (%) . | RS (%) . | Blasts (%) . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 72 | 102 | No | p.K700E | STAG2 (340del) | 70 | 29 | 1 |
| 2 | F | 69 | 89 | No | p.K700E | TET2 (p.F125fs) | 60 | 24 | 4.5 |
| 3 | F | 83 | 88 | Yes | p.K700E | TET2 (Q916*) | 90 | 30 | 4 |
| 4 | M | 78 | 97 | No | p.K666R | c-KIT, TET2 (R1404*, 690*) | 50 | 39 | 3 |
| 5 | F | 69 | 93 | No | p.H662Q | TET2 (H1881Y) | 80 | 33 | 5.5 |
| 6 | M | 76 | 97 | No | p.H662D | DNMT3A (W305*) | 60 | 23 | 4.5 |
| 7 | F | 78 | 103 | No | p.E622D | B-COR (R63K) | 60 | 21 | 2 |
| 8 | M | 87 | 80 | Yes | p.R625L | IDH2 (R172K) | 70 | 22 | 2 |
| 9 | M | 81 | 106 | No | p.E622D | None | 60 | 33 | 2.5 |
| 10 | F | 65 | 77 | No | p.K700E | PRPF40B (669D) | 90 | 39 | 2 |
| 11 | F | 81 | 107 | No | p.K700E | EZH2 (T306fs), DNMT3A (E285*) | 70 | 85 | 0.5 |
| 12 | M | 64 | 132 | No | p.N626D | None | 50 | 42 | 5.5 |
| 13 | M | 70 | 117 | No | No mutation | SRSF2 (95_103del) | 90 | 73 | 1.5 |
| 14 | F | 71 | 98 | Yes | p.K700E | TET2 (Q746X) | 80 | 55 | 2 |
| 15 | F | 77 | 97 | Yes | p.E622D | JAK2 (V617F) | 100 | 77 | 1 |
| 16 | F | 80 | 115 | No | p.K700E | DNMT3A (V296M) | 50 | 36 | 2.5 |
| 17 | F | 60 | 113 | No | p.K700E | None | 100 | 25 | 1.5 |
| 18 | M | 79 | 108 | No | p.E622D | DNMT3A (I643M) | 80 | 16 | 2 |
| 19 | F | 54 | 125 | Yes | p.K700E | DNMT3A (F868S) | 90 | 10 | 0 |
| 20 | M | 67 | 97 | Yes | p.K700E | None | 65 | 35 | 0 |
| 21 | M | 67 | 100 | Yes | p.K700E | None | 60 | 40 | 1 |
| 22 | M | 68 | 101 | No | p.K700E | TET2 (E1555fs*22) | 70 | 40 | 0 |
| 23 | F | 74 | 106 | No | p.K666T | None | 70 | 20 | 2.5 |
| 24 | M | 76 | 107 | No | p.N626D | TET2 (R1452*) | — | 16 | 2 |
| 25 | F | 82 | 99 | No | p.E622D | None | 50 | 24 | 1.5 |
| 26 | M | 77 | 115 | Yes | p.E666R | None | 60 | 45 | 3 |
| 27 | F | 84 | 93 | Yes | p.K700E | EZH2 (K661E) | 40 | 33 | 2 |
| 28 | F | 75 | 107 | No | p.K700E | None | 40 | 28 | 1.5 |
| 29 | F | 76 | 105 | No | p.Y623C | None | 40 | 49 | 1.5 |
| 30 | F | 80 | 97 | No | p.K700E | None | 60 | 16 | 3.5 |
| 31 | F | 73 | 102 | No | p.E666R | None | 50 | 53 | 1 |
| 32 | F | 79 | 135 | No | p.K700E | TET2 (Q958fs*14) | — | 35 | 0 |
| 33 | M | 77 | 87 | Yes | p.R625L | None | — | 26 | 0 |
| 34 | M | 73 | 87 | Yes | p.R625L | TET2 (F1300I), SMC3 (L836X) | — | 16 | 2 |
| 35 | M | 64 | 101 | No | p.K666R | DNMT3A (Y735C) | — | 23 | 2 |
| 36 | F | 79 | 101 | No | p.H662Q | IDH2 (C402Y), TET2 (T1122fs*8) | — | 30 | 1 |
| 37 | F | 46 | 109 | No | p.K700E | None | 50 | 44 | 1 |
| 38 | M | 68 | 101 | Yes | p.K700E | IDH2 (R140Q), EZH2 (R732I), ZRSR2 (E120fs*24) | — | 16 | 1 |
| 39 | F | 77 | 111 | Yes | p.K700E | RUNX1 (R107S) | — | 31 | 2 |
| 40 | M | 49 | 121 | No | p.T663I | TET2 (P319fs*28) | 90 | 70 | 3 |
F, female; Hb, hemoglobin; M, male.
Functional characteristics of MDS-RS stem and progenitor cells
To gain information regarding the frequencies of the most primitive stem cells in MDS-RS patients, we used flow cytometry based on previously described immunophenotypic marker protocols.20,22-26 We isolated phenotypically defined HSCs (7AAD−Lin−CD34+CD38−CD90+CD45RA−), common myeloid progenitors (CMPs; 7AAD−Lin−CD34+CD38+CD90-CD123+CD45RA−), granulocyte-macrophage progenitors (GMPs; 7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA+), and megakaryocyte-erythroid progenitors (MEPs; 7AAD−Lin−CD34+CD38+CD90−CD123−CD45RA−) from primary BM aspirates and compared stem and progenitor cell immunophenotypes from 9 primary MNC MDS-RS samples with those from 4 healthy subjects. The immunophenotypic gating strategy, illustrated in Figure 1A, was validated by post–FACS-sort purity analysis (supplemental Figure 1). The size of the Lin−CD34+CD38−CD90+CD45RA− HSC compartment in MDS-RS patients, representing on average 0.13% of total BM cells (Figure 1B), was comparable to that in healthy NBM (0.08%, P = .6). No significant differences were found between the MDS-RS and healthy erythroid and myeloid progenitor compartments (Figure 1B).
Analysis of hematopoietic stem and progenitor cells in MDS-RS. (A) Representative FACS analysis of SF3B1-mutated MDS-RS BM (patient 4). Shown are FACS analyses of viable, lineage-negative, CD34-enriched cells further gated as CMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA−), MEPs (7AAD−Lin−CD34+CD38+CD90−CD123−CD45RA−), GMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA+), and HSCs (7AAD−Lin−CD34+CD38−CD90+CD45RA−). (B) Mean (standard error of the mean [SEM]) frequencies of HSCs and progenitors in SF3B1-mutated MDS-RS patients (n = 9) and healthy NBM (n = 4). (C) Mean (SEM) erythroid and myeloid colony formation from purified CMPs, GMPs, and MEPs from 8 MDS-RS patients and 4 NBMs. (D) Representative images of an erythroid colony (BFU-E) from an MDS-RS patient and a healthy NBM control. Original magnification for light microscopy ×10. (E) LTC-CFC activity in HSCs, CMPs, GMPs, and MEPs from MDS-RS patients (n = 8) and healthy NBM controls (n = 5). Results are mean (SEM) values for all patients and healthy controls, in each case derived from the mean of 3 technical replicates for each cell population and patient. **P < .005.
Analysis of hematopoietic stem and progenitor cells in MDS-RS. (A) Representative FACS analysis of SF3B1-mutated MDS-RS BM (patient 4). Shown are FACS analyses of viable, lineage-negative, CD34-enriched cells further gated as CMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA−), MEPs (7AAD−Lin−CD34+CD38+CD90−CD123−CD45RA−), GMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA+), and HSCs (7AAD−Lin−CD34+CD38−CD90+CD45RA−). (B) Mean (standard error of the mean [SEM]) frequencies of HSCs and progenitors in SF3B1-mutated MDS-RS patients (n = 9) and healthy NBM (n = 4). (C) Mean (SEM) erythroid and myeloid colony formation from purified CMPs, GMPs, and MEPs from 8 MDS-RS patients and 4 NBMs. (D) Representative images of an erythroid colony (BFU-E) from an MDS-RS patient and a healthy NBM control. Original magnification for light microscopy ×10. (E) LTC-CFC activity in HSCs, CMPs, GMPs, and MEPs from MDS-RS patients (n = 8) and healthy NBM controls (n = 5). Results are mean (SEM) values for all patients and healthy controls, in each case derived from the mean of 3 technical replicates for each cell population and patient. **P < .005.
The MDS-RS CMP, GMP, and MEP subpopulations revealed normal capacity to generate colonies with mixed, myeloid-, and erythroid-restricted lineage potentials, respectively, but the efficacy of GMPs and MEPs to generate myeloid and erythroid colonies was significantly reduced, as previously reported22 (Figure 1C). MDS-RS erythroid burst forming unit-erythroid (BFU-E) colonies were characterized by poor hemoglobinization when compared with BFU-E colonies derived from normal healthy control individuals (Figure 1D). We then investigated the self-renewal potential in vitro of MDS-RS stem and progenitor cells using LTC-CFCs as an assay for stem cell activity,20,27 and we compared them with healthy NBM. LTC-CFC activity in MDS-RS was significantly reduced but restricted to the MDS-RS HSC population, with no LTC-CFC activity in any of the lineage-restricted cell populations investigated (CMPs, GMPs, and MEPs; Figure 1E), thus suggesting that the self-renewal potential in MDS-RS is confined to the primitive stem cell compartment.
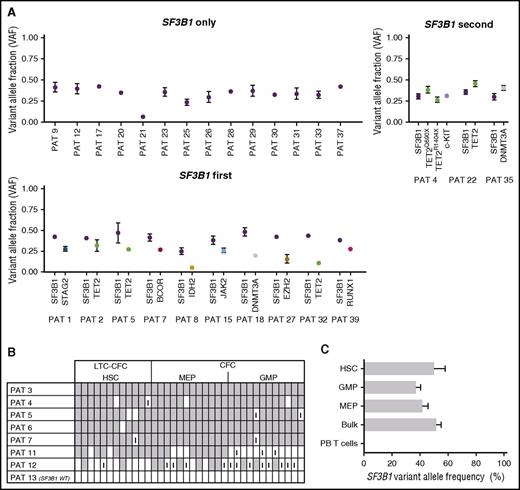
Initiating SF3B1 spliceosome lesions originate from HSCs and are retained during differentiation
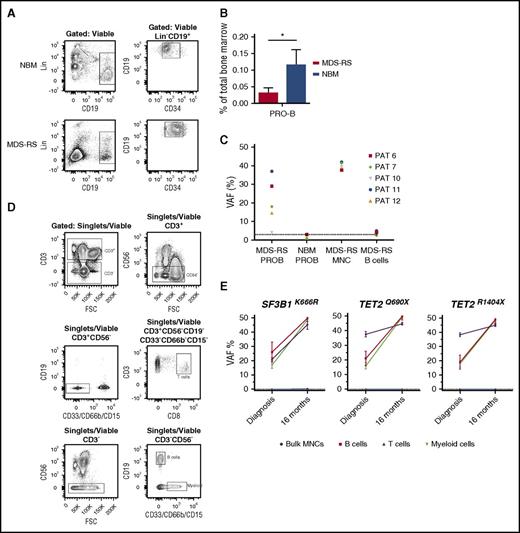
Computational prediction in the 39 cases studied of MDS-RS patients with ≥1 recurrent driver mutations based on targeted sequencing data (Figure 2A) demonstrated that the SF3B1 mutation occurred alone in 14 cases, was predicted to be the first event in 10 cases, and was predicted to be secondary to other oncogenic mutations in only 3 cases. The remaining 12 cases were inconclusive, as the computational analysis failed to reliably unravel the sequential acquisition pattern data (supplemental Figure 2). We also studied in vitro–cultured individually picked LTC-CFC–derived colonies from FACS-purified stem cells as well as colonies generated by purified progenitor cells. As expected, we were able to identify the SF3B1 mutation at the HSC level from LTC-CFCs in all 7 SF3B1-mutated patients (Figure 2B). These findings further support that recurrent SF3B1 mutations typically target the rare self-renewing HSC compartment in MDS-RS. Next, we extended this analysis to FACS-purified stem and progenitor cells, which were investigated for the presence of the SF3B1 mutation using Sequenom analysis.28 In agreement with the LTC-CFC results, in the 4 MDS-RS patients examined, the SF3B1 mutation could be backtracked to the HSC compartment and was also found in downstream GMPs and MEPs, but not in PB T cells (Figure 2C). In order to further explore the potential multipotent (lymphomyeloid) HSC origin of SF3B1 mutations in MDS-RS, we next performed pyrosequencing of FACS-purified pro–B cells (CD34+CD19+) (Figure 3A; supplemental Figure 3A), which were found to be reduced in MDS-RS BM compared with healthy controls (3.8-fold reduction, P < .05) (Figure 3B). In 4 of 5 MDS-RS patients analyzed, the SF3B1 mutation was distinctly present in highly purified CD34+CD19+ BM progenitors (Figure 3C). In contrast, in the same patients, the involvement of mature B cells was at or below detection level (Figure 3C-D; supplemental Figure 3B), in agreement with previous studies.12,13 However, in 1 MDS-RS case (patient 4) harboring both SF3B1 and TET2 mutations, mature B cells were also highly clonally involved at diagnosis, and the allelic burden of both mutations increased with time in myeloid as well as B cells (Figure 3E; supplemental Figure 4; supplemental Table 1). The SF3B1 mutation was not detected in T cells in any of the MDS-RS cases examined. Therefore, SF3B1 mutations target lymphomyeloid HSCs in MDS-RS and appear to negatively impact lymphoid development.
Evidence that SF3B1 mutations are initiating mutations targeting rare HSCs in MDS-RS. (A) Computational prediction of fraction of cells with identified genomic lesions within total BM MNCs from SF3B1-mutated MDS-RS patients based on VAF. Error bars indicate 95% confidence intervals. Shown are cases where SF3B1 is the only identified recurrent mutation, SF3B1 is predicted to be the first of multiple recurrent mutations, or SF3B1 is predicted to be secondary to other mutations. Inconclusive results (overlapping 95% confidence intervals) are included in supplemental Figure 2. (B) Tracking of SF3B1 mutations in individually picked HSC-derived LTC-CFCs or MEP- and GMP-derived CFCs. 7 SF3B1 mutated and 1 SF3B1 wild-type (WT) patients were analyzed using pyrosequencing to screen for identified SF3B1 mutations in each case and scored as positive (gray), negative (white), or inconclusive (I) (see supplemental Methods for definitions and cutoffs). (C) Sequenom analysis from 4 MDS-RS samples of FACS-purified stem and progenitor cell populations to assess SF3B1 VAF. PAT, patient.
Evidence that SF3B1 mutations are initiating mutations targeting rare HSCs in MDS-RS. (A) Computational prediction of fraction of cells with identified genomic lesions within total BM MNCs from SF3B1-mutated MDS-RS patients based on VAF. Error bars indicate 95% confidence intervals. Shown are cases where SF3B1 is the only identified recurrent mutation, SF3B1 is predicted to be the first of multiple recurrent mutations, or SF3B1 is predicted to be secondary to other mutations. Inconclusive results (overlapping 95% confidence intervals) are included in supplemental Figure 2. (B) Tracking of SF3B1 mutations in individually picked HSC-derived LTC-CFCs or MEP- and GMP-derived CFCs. 7 SF3B1 mutated and 1 SF3B1 wild-type (WT) patients were analyzed using pyrosequencing to screen for identified SF3B1 mutations in each case and scored as positive (gray), negative (white), or inconclusive (I) (see supplemental Methods for definitions and cutoffs). (C) Sequenom analysis from 4 MDS-RS samples of FACS-purified stem and progenitor cell populations to assess SF3B1 VAF. PAT, patient.
SF3B1 mutations in lymphoid and myeloid progenitors in MDS-RS. (A) Representative FACS analysis of BM MNCs from healthy NBM and SF3B1-mutated MDS-RS patient 6, showing gating strategy for viable Lin−CD34+CD19+ pro–B cells. (B) Mean (SEM) frequencies of pro–B cells in SF3B1-mutated MDS-RS patients (n = 9) and healthy NBM (n = 4). (C) VAF of SF3B1 mutations in MDS-RS pro–B cells (MDS-RS PROB, n = 5) and mature PB B cells from MDS-RS (MDS-RS B cells, n = 3) compared with MDS-RS bulk BM MNCs (n = 5). Healthy NBM pro–B cells (NBM PROB, n = 4) were included as a negative control for pyrosequencing. PAT, patient. (D) Representative gating strategy for FACS purification of PB CD3+CD8+ T cells, CD19+ B cells, and CD33/66B/15+ myeloid cells from MDS-RS patient 4. (E) VAF of identified mutations in FACS-purified PB myeloid, B, and T cells from MDS-RS patient 4 at diagnosis and 16 months later using droplet digital PCR. Healthy normal DNA was negative, as indicated by the dotted line (<0.1%).
SF3B1 mutations in lymphoid and myeloid progenitors in MDS-RS. (A) Representative FACS analysis of BM MNCs from healthy NBM and SF3B1-mutated MDS-RS patient 6, showing gating strategy for viable Lin−CD34+CD19+ pro–B cells. (B) Mean (SEM) frequencies of pro–B cells in SF3B1-mutated MDS-RS patients (n = 9) and healthy NBM (n = 4). (C) VAF of SF3B1 mutations in MDS-RS pro–B cells (MDS-RS PROB, n = 5) and mature PB B cells from MDS-RS (MDS-RS B cells, n = 3) compared with MDS-RS bulk BM MNCs (n = 5). Healthy NBM pro–B cells (NBM PROB, n = 4) were included as a negative control for pyrosequencing. PAT, patient. (D) Representative gating strategy for FACS purification of PB CD3+CD8+ T cells, CD19+ B cells, and CD33/66B/15+ myeloid cells from MDS-RS patient 4. (E) VAF of identified mutations in FACS-purified PB myeloid, B, and T cells from MDS-RS patient 4 at diagnosis and 16 months later using droplet digital PCR. Healthy normal DNA was negative, as indicated by the dotted line (<0.1%).
In vivo RS formation from SF3B1-mutated MDS-RS HSCs
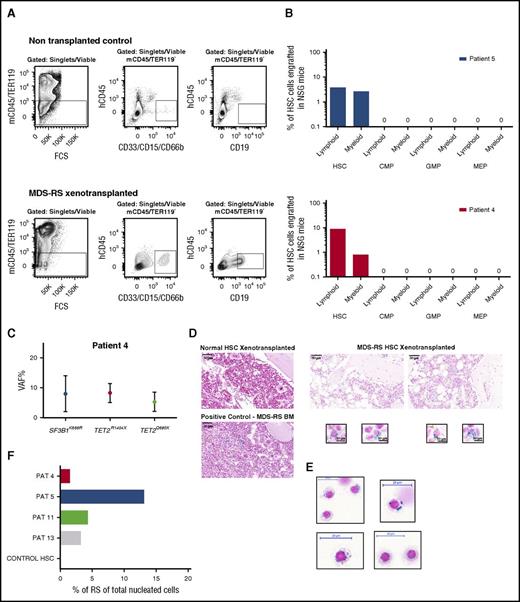
In order to assess the in vivo MDS-RS propagating ability of distinct MDS-RS stem and committed progenitor subsets, HSCs, CMPs, GMPs, and MEPs were purified from 2 MDS-RS patients (patients 4 and 5) and transplanted into NSG mice, and their ability to give sustained engraftment was analyzed after 20 to 22 weeks. In both cases, transplanted HSCs, but not CMPs, GMPs, or MEPs, resulted in sustained myeloid and B-cell engraftment (Figure 4A-B). Sensitive mutation detection analysis by droplet digital PCR established the SF3B1 and TET2 mutation clonal involvement of the myeloid cells reconstituted by the HSCs from patient 4 (Figure 4C; supplemental Table 2; supplemental Figure 5). In both of these cases, as well as in 2 other cases (patients 11 and 13), characteristic RSs developed in the BM of NSG mice transplanted with SF3B1-mutated HSCs, while no RSs were detected in mice transplanted with the progenitor cells from the same patients or in mice with engraftment following transplantation of healthy HSCs (Figure 4D-F). Importantly, since we, in agreement with the known mutually exclusive expression of CD45 and GPA in normal erythroid development,29 found RSs in SF3B1-mutated MDS-RS patients to be exclusively CD45−GPA+(supplemental Figures 6 and 7), we investigated the whole BM of transplanted NSG mice for the presence of RSs.
In vivo reconstitution of RSs from MDS-RS HSCs. (A) Representative FACS gating strategy used for analysis of human myeloid (CD33/CD66b/CD15+) and lymphoid (CD19+) engraftment in xenotransplanted nonobsese diabetic/LtSz-scid IL2Rγc−/− (NSG) mice transplanted with healthy or SF3B1-mutated MDS-RS human stem and progenitor cells. The number of purified HSCs, CMPs, GMPs, and MEPs transplanted into NSG mice was according to their relative ratios in the patient BM. Patient 4: 1400, 5000, 21 000, and 1400 cells; patient 5: 25 000, 55 000, 49 300, and 25 200 cells, respectively. Top panels show gating in a nontransplanted NSG mice (negative control); bottom panels show analysis in an NSG mouse transplanted with MDS-RS HSCs from patient 4. (B) In vivo human B-lymphoid and myeloid engraftment in BM of NSG mice 20 to 22 weeks posttransplantation of FACS-purified HSCs or indicated progenitors (mean values from 2 or 3 mice per cell population per patient) from 2 SF3B1-mutated MDS-RS. (C) Mean VAF in myeloid cells derived from patient 4 HSCs transplanted in NSG mice. (D) Prussian blue stains from sections of paraffin-embedded BM tissue from NSG mice with human reconstitution from healthy (top left) or SF3B1-mutated MDS-RS HSCs (top right; patient 5). The bottom left panel shows positive control from BM of MDS-RS patient; scale bar, 50 μm. The bottom right panel shows characteristic Prussian blue–positive cells in BM of NSG mice transplanted with MDS-RS HSCs from patient 5; scale bar, 10 μm. (E) Cytospins of erythroid cells purified from BM of patient 5 (supplemental Figure 6), stained with Prussian blue; scale bar, 20 μm. (F) Percentage of RSs of total nucleated BM cells in BM of NSG mice transplanted with purified HSCs. PAT, patient.
In vivo reconstitution of RSs from MDS-RS HSCs. (A) Representative FACS gating strategy used for analysis of human myeloid (CD33/CD66b/CD15+) and lymphoid (CD19+) engraftment in xenotransplanted nonobsese diabetic/LtSz-scid IL2Rγc−/− (NSG) mice transplanted with healthy or SF3B1-mutated MDS-RS human stem and progenitor cells. The number of purified HSCs, CMPs, GMPs, and MEPs transplanted into NSG mice was according to their relative ratios in the patient BM. Patient 4: 1400, 5000, 21 000, and 1400 cells; patient 5: 25 000, 55 000, 49 300, and 25 200 cells, respectively. Top panels show gating in a nontransplanted NSG mice (negative control); bottom panels show analysis in an NSG mouse transplanted with MDS-RS HSCs from patient 4. (B) In vivo human B-lymphoid and myeloid engraftment in BM of NSG mice 20 to 22 weeks posttransplantation of FACS-purified HSCs or indicated progenitors (mean values from 2 or 3 mice per cell population per patient) from 2 SF3B1-mutated MDS-RS. (C) Mean VAF in myeloid cells derived from patient 4 HSCs transplanted in NSG mice. (D) Prussian blue stains from sections of paraffin-embedded BM tissue from NSG mice with human reconstitution from healthy (top left) or SF3B1-mutated MDS-RS HSCs (top right; patient 5). The bottom left panel shows positive control from BM of MDS-RS patient; scale bar, 50 μm. The bottom right panel shows characteristic Prussian blue–positive cells in BM of NSG mice transplanted with MDS-RS HSCs from patient 5; scale bar, 10 μm. (E) Cytospins of erythroid cells purified from BM of patient 5 (supplemental Figure 6), stained with Prussian blue; scale bar, 20 μm. (F) Percentage of RSs of total nucleated BM cells in BM of NSG mice transplanted with purified HSCs. PAT, patient.
The percentage of RSs in total nucleated cells in the BM of the engrafted mice was calculated using CellProfiler image analysis software and ranged from 1.5% to 13.1% (Figure 4F), demonstrating robust in vivo generation of RSs from transplanted MDS-RS SF3B1-mutated HSCs in NSG mice.
Discussion
Herein, we provide phenotypic and functional evidence that the most primitive lymphomyeloid HSCs (Lin−CD34+CD38−CD90+CD45RA−) represent the origin of the mutated SF3B1 clone in MDS-RS and also represent the rare MDS-RS propagating cells. In all the samples studied, we identified the SF3B1 mutation in the majority of the in vitro–generated colonies from LTC-CFCs, compatible with the driver mutation SF3B1 providing HSCs in MDS-RS with a considerable clonal advantage at the HSC level. However, it is important to highlight the coexistence of wild-type LTC-CFCs, which could represent a potential therapeutic approach to change the balance of mutated vs wild-type HSCs. By contrast, self-renewal potential in vitro and in vivo was not observed by any of the committed progenitor populations, suggesting that only the rare HSC compartment can propagate the SF3B1-mutated RS clone over the long-term.
Previous studies of MDS-RS found that the B-cell lineage was not part of the SF3B1 clone,12,13 failing to support that SF3B1 mutations in MDS-RS target lymphomyeloid HSCs. Also in our studies, in all cases but one did we find mature B cells to be minimally or not involved in the SF3B1-mutated clone. In contrast, in the same patients, we found definitive evidence for involvement at the early CD34+CD19+ pro–B-cell stage. Together with high clonal involvement at the Lin−CD34+CD38−CD90+CD45RA− HSC level, these findings provide evidence for SF3B1 targeting lymphomyeloid HSCs in MDS-RS and at the same time suggest that the SF3B1 mutation and/or accompanying genomic lesions negatively impact B-lymphoid development. This seems to contrast with the observation in chronic lymphocytic leukemia (CLL), in which the same SF3B1 mutations do not appear to negatively impact on the B-cell lineage.30 This might reflect that SF3B1 mutations in CLL, unlike those in MDS-RS, typically are subclonal, suggesting that they are secondary to other driver lesions in CLL.31,32 Moreover, the impact of SF3B1 mutations on the B-cell lineage might be affected by the context of co-occurring driver mutations, which are distinct in MDS-RS and CLL. Finally, SF3B1 mutations might target an earlier and/or distinct progenitor in MDS-RS than in CLL.
Also, our consistent finding that the T-lymphoid lineage is not part of the SF3B1 clone is in line with recurrent SF3B1 mutations being incompatible with normal lymphoid development. However, similar findings have been made in del5q MDS33 and might also reflect that T cells are long lived and T lymphopoiesis is inactive in the elderly.34-36
Our computational prediction analyses are most compatible with a model in which mutations in SF3B1 in MDS-RS typically is the initiating recurrent driver mutation targeting HSCs in most MDS-RS cases, in accordance with previous studies.5,20 Importantly, our finding that SF3B1 recurrent mutations target the rare Lin−CD34+CD38−CD90+CD45RA− HSCs and provide these with a competitive advantage over healthy HSCs seems to be the rule rather than exception in low- to intermediate-risk MDS,20 most likely reflecting that MDS-initiating lesions do not typically confer self-renewal properties to downstream progenitors. Also, most MDS-initiating mutations targeting Lin−CD34+CD38−CD90+CD45RA− HSCs seem to negatively affect lymphoid development, as our studies are among the few that have been able to demonstrate significant clonal involvement also of lymphoid lineages.33
Notably, analysis of bone biopsy specimens of NSG mice established that transplanted SF3B1-mutated MDS-RS HSCs had differentiated into the erythroid lineage and generated morphological distinct RSs, the hallmark of MDS-RS. While RS engraftment was not reported in a previous study in which purified HSCs from a SF3B1-mutated MDS-RS case gave myeloid engraftment, this would have been missed, since apparently only engraftment of cells expressing the common leukocyte CD45 antigen was investigated,12 whereas the mature erythroid lineage including RSs does not express CD45.29
While initial studies suggested that SF3B1 haploinsufficient mice have increased frequencies of RS in the BM,37,38 subsequent studies of SF3B1K700E knockin mice as well as SF3B1+/− mice failed to confirm an increase in characteristic RSs or circulating siderocytes, despite other signs of progressive macrocytic anemia and myelodysplasia.39-42 This is consistent with the lack of RS formation in other mouse models resulting in sideroblastic anemia.43 The cellular and molecular basis for this possible species difference remains unclear. It cannot be excluded that this might be explained by the requirement for additional genetic or epigenetic mutations and dysregulation to develop the full phenotype and clinical picture of RS anemia. Regardless, the finding that characteristic RSs developed in NSG mice transplanted with SF3B1-mutated HSCs from MDS-RS patients, but not healthy human subjects, highlights the potential usefulness of exploring the cellular and molecular basis as well as therapeutic targeting of MDS-RS in this in vivo xenograft model. Moreover, it suggests that the difference observed in development of RS in SF3B1K700E knockin mice and SF3B1 mutated MDS-RS patients is likely to be explained by a hematopoietic intrinsic rather than extrinsic mechanism.
In conclusion, our findings provide evidence of a lymphomyeloid stem cell origin of the SF3B1 mutation in MDS-RS patients and a novel in vivo platform for mechanistically and therapeutically exploring MDS-RS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the WIRM flow cytometry facility (supported by the Knut and Alice Wallenberg Foundation) for technical assistance with the sorting experiments at Karolinska Institute (KI), Johanna Ungerstedt for BM sampling from healthy controls (KI), Asmaa Ben Azenkoud for processing the BM MNC and biobanking (KI), Gunilla Waldin for handling of clinical data for the patients (KI), Onima Chowdhury for technical support with the xenotransplantation experiments performed at the Weatherall Institute of Molecular Medicine, and Raoul Kuiper and Tarja Schröder from the core facility for morphological phenotype analysis (FENO) at the department of Laboratory Medicine (KI) for help with mice histopathology.
E.H.-L. is funded through the Swedish Cancer Society (Cancerfonden, 150269), the Cancer Research Foundations of Radiumhemmets (Radiumhemmets Forskningsfonder, 151103), and the Swedish Research Council (Vetenskapsrådet, 521-2013-3577). S.E.W.J. is supported by an international recruitment grant from the Swedish Medical Research Council, a grant from the Tobias Foundation, and a grant from the Center for Innovative Medicine at KI.
Authorship
Contribution: T.M.-B. performed and analyzed experiments and wrote the manuscript; E.H.-L. and S.E.W.J. designed the study and wrote the manuscript; M.D. and P.S.W. performed experiments, analyzed results, and contributed to writing of the manuscript; I.D. performed parts of the sorting experiments; M.D., M.K., M.M. and E.P. were involved in the generation, analysis or meta-analysis of sequencing data; E.E. and S.C. performed experiments; M.T. and M.J. coordinated the sampling of healthy donors and biobanking; L.S. performed the pathology analysis; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eva Hellström-Lindberg, Karolinska Institute, Department of Medicine, Center for Hematology and Regenerative Medicine, Karolinska University Hospital Huddinge, 141 86 Stockholm, Sweden; e-mail: eva.hellstrom-lindberg@ki.se; and Sten Eirik W. Jacobsen, Karolinska Institute, Department of Medicine, Center for Hematology and Regenerative Medicine, Karolinska University Hospital Huddinge, 141 86 Stockholm, Sweden; e-mail: sten.eirik.jacobsen@ki.se.
References
Author notes
S.E.W.J. and E.H.-L. contributed equally to this study.

![Figure 1. Analysis of hematopoietic stem and progenitor cells in MDS-RS. (A) Representative FACS analysis of SF3B1-mutated MDS-RS BM (patient 4). Shown are FACS analyses of viable, lineage-negative, CD34-enriched cells further gated as CMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA−), MEPs (7AAD−Lin−CD34+CD38+CD90−CD123−CD45RA−), GMPs (7AAD−Lin−CD34+CD38+CD90−CD123+CD45RA+), and HSCs (7AAD−Lin−CD34+CD38−CD90+CD45RA−). (B) Mean (standard error of the mean [SEM]) frequencies of HSCs and progenitors in SF3B1-mutated MDS-RS patients (n = 9) and healthy NBM (n = 4). (C) Mean (SEM) erythroid and myeloid colony formation from purified CMPs, GMPs, and MEPs from 8 MDS-RS patients and 4 NBMs. (D) Representative images of an erythroid colony (BFU-E) from an MDS-RS patient and a healthy NBM control. Original magnification for light microscopy ×10. (E) LTC-CFC activity in HSCs, CMPs, GMPs, and MEPs from MDS-RS patients (n = 8) and healthy NBM controls (n = 5). Results are mean (SEM) values for all patients and healthy controls, in each case derived from the mean of 3 technical replicates for each cell population and patient. **P < .005.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/7/10.1182_blood-2017-03-776070/4/m_blood776070f1.jpeg?Expires=1769094250&Signature=CyDeOArumlUedfkUSG21olxsXJ4c49e43obG9mlEwjMqdGUqNUlP1hCfm~xOe-6d0xHxvQbZXm-w-77q6Nq638d6P28DbnHgEuYfgus-LrCxX89i3XIUlNNtmP15buhNCOHkrS6tA-nog~uT4~5qbgv5NGOGqVkKcs6MxsA7nw9tw20uQ4-fMT0lex-oDjFz5IjSUQqM7dWRp2qZ6xncHlGeNrd~1UN8~voenvpHKAz4K198PDFskZt-eIYs8ELNGHVD~z50JEsoMaQeSMmILH3mtuXjrBeeadoYwBCUnJIRGtx7ieWCwXQ481h9nxP~HcVrYxRfVd7APU0rUmDetg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal