Key Points
CDC42 regulates AML cell polarity and division symmetry.
CDC42 suppression in AML cells promotes differentiation and blocks leukemia progression.
Abstract
As a central regulator of cell polarity, the activity of CDC42 GTPase is tightly controlled in maintaining normal hematopoietic stem and progenitor cell (HSC/P) functions. We found that transformation of HSC/P to acute myeloid leukemia (AML) is associated with increased CDC42 expression and activity in leukemia cells. In a mouse model of AML, the loss of Cdc42 abrogates MLL-AF9–induced AML development. Furthermore, genetic ablation of CDC42 in both murine and human MLL-AF9 (MA9) cells decreased survival and induced differentiation of the clonogenic leukemia-initiating cells. We show that MLL-AF9 leukemia cells maintain cell polarity in the context of elevated Cdc42-guanosine triphosphate activity, similar to nonmalignant, young HSC/Ps. The loss of Cdc42 resulted in a shift to depolarized AML cells that is associated with a decrease in the frequency of symmetric and asymmetric cell divisions producing daughter cells capable of self-renewal. Importantly, we demonstrate that inducible CDC42 suppression in primary human AML cells blocks leukemia progression in a xenograft model. Thus, CDC42 loss suppresses AML cell polarity and division asymmetry, and CDC42 constitutes a useful target to alter leukemia-initiating cell fate for differentiation therapy.
Introduction
The Rho GTPase family member, CDC42, plays an integral role in coordinating normal hematopoietic stem/progenitor cell (HSC/P) function in response to input from the bone marrow (BM) niche. Cdc42 signaling has been implicated in HSC/P proliferation, cytoskeleton organization, polarity, adhesion and directional migration within the microenvironment.1,2 An excess level of active, guanosine triphosphate (GTP)-bound Cdc42 in normal HSC/Ps, as found in aged cells or the gain of Cdc42 activity Cdc42GAP−/− cells, also leads to decreased adhesion and migration, less competitive engraftment, and loss of cell polarity, suggesting that Cdc42 activity must be tightly regulated for proper HSC/P function.2,3
The role of CDC42 activity in hematopoietic cell transformation and leukemia progression remains unclear. Interestingly, both murine and human models of AML derived from hematopoietic stem cells (HSCs) expressing the MA9 oncogene show elevated levels of GTP-bound active CDC42.4,5 Furthermore, recent analysis ranks the pathways involved in adherens junction, regulation of actin cytoskeleton, tight junction, focal adhesion, Rho cell motility signaling, and Wnt signaling among the top 10 most highly dysregulated gene expression networks distinguishing AML stem cells from normal HSCs.6 CDC42 is integrally involved in the regulation of these pathways, and indeed CDC42 gene expression and the CDC42 controlled genes involved in cellular functional outcomes are upregulated in leukemia stem cells.6
A proposal was put forward that the transformation of HSC/Ps to AML promotes a symmetric division pattern, which enhances the leukemia-initiating cell (LIC) self-renewal activity and maintains leukemia progression.7-9 This concept is at odds with our understanding of the normal HSC self-renewal mechanism that typically involves an asymmetric division to give rise to a HSC and a committed progeny in the absence of reprogramming factors.10,11 In the present study, we used a CDC42 genetic targeting approach to better define the functional relationship of LIC polarity, division symmetry, and cell fate decision. We show that CDC42 targeting suppresses LIC polarity and division symmetry, blocking LIC survival and AML progression. Our data suggest CDC42 represents a novel target in LIC self-renewal and differentiation.
Methods
Murine MLL-AF9 cell lines
Animal experiments followed protocols approved by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (CCHMC). The generation of C57Bl/6 Cdc42flox/flox (Cdc42 floxed [FL]) mice was described previously.1,12 Crossbreeding to B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J mice generated ROSA26Cre/ERT2;Cdc42FL/FL mice (Cre+;Cdc42FL). Mice heterozygous at the Cdc42 allele (Cre+;FL/WT) or absent the ROSA26Cre/ERT2 allele (Cre-;Cdc42FL) served as control donors. Lineage-negative BM was transduced with pMSCV-MLL-AF9-pgk-EGFP retroviral vector to establish immortalized cell lines in vitro, as previously described.5,13,14 To assay colony-forming unit (CFU) content, 2500-5000 MA9 cells were plated in triplicate in methylcellulose (MethoCult GF M3434, StemCell Technologies). Primary transplants were established by injecting 200 000 EGFP+ cells into lethally irradiated 6- to 8-week-old congenic recipients (BoyJ; The Jackson Laboratory). Floxed Cdc42 sequences were knocked out by injecting 2 mg tamoxifen (TAM; Sigma-Aldrich) intraperitoneally once daily on 2 consecutive days, and repeating every other week, beginning at day 15, with corn oil as the vehicle control. Secondary transplants were established in sublethally irradiated BoyJ mice, and Cdc42 deletion was performed from day 5 posttransplant, as previously described.
Differentiation, survival, and cell cycle analysis
Conditional Cdc42 (Cre+;Cdc42FL) and control (Cre–;Cdc42FL) murine MA9 leukemia was harvested from diseased, primary transplant recipients and treated in vitro with 4-hydroxy TAM, with 0.01% ethanol (EtOH) as the vehicle control. Apoptosis was measured by AnnexinV/7-aminoactinomycin D according to manufacturer’s protocol (BD Biosciences). Cell cycle analysis was performed by fluorescence-activated cell sorting (FACS) after staining with Vybrant DyeCycle Violet (ThermoFisher Scientific). RNA was isolated 72 hours posttreatment with TAM vs EtOH, per the manufacturer’s protocol (RNeasy; Qiagen). RNA was reverse transcribed and complementary DNA measured by quantitative polymerase chain reaction (PCR) using Taqman probe for Egr1. Relative gene expression was calculated using the ΔΔ cycle threshold method, normalizing to Gapdh and vehicle control. Cell lines were transduced with retroviral MSCV-Bcl-xL-Venus vector or empty vector control, as previously described.13 Sorted Venus+ cells were plated in methylcellulose for CFU assay.
Structural and functional cell polarity analysis
Murine AML derived from knockin of the MLL-AF9 allele into the ROSA26CreERt2;Cdc42FL/FL background was treated with TAM to delete Cdc42 knockout(KO) vs ethanol control. For confocal microscopy, 30 000 cells were seeded on RetroNectin (Takara)–coated, 35-mm glass bottom dishes (In Vitro Scientific), and then fixed with 3% paraformaldehyde, pH 7.4, for 10 minutes at room temperature. Cells were stained with primary antibody (rat anti-tubulin, Abcam, ab6160) and secondary antibody (AF647 goat anti-rat, Jackson ImmunoResearch Laboratories, 112-606-003), as previously described.3 After washing, cells were treated with Prolong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (ThermoFisher Scientific, P-36931) and imaged on a Nikon Eclipse Ti microscope, Plano Apo VC 60× oil objective, numerical aperture 1.4, with a Nikon C2+ camera. NIS Elements AR software (Nikon) was used to measure fluorescence intensity along a bisecting vector, and cells were considered polarized when clear asymmetric distribution of tubulin was seen, as previously described.3 Fifty to 100 MA9 cells were individually analyzed per sample. Data were plotted as a percentage of the total number of cells scored per sample.
Functional polarity was assayed by measuring cell adhesion and directional migration. Samples were seeded on RetroNectin in 8 replicate wells, and then nonadherent cells were removed by washing with phosphate-buffered saline. Adherent cells were stained with Vybrant DyeCycle Violet (ThermoFisher Scientific) in phosphate-buffered saline then imaged on the Nikon A1Rsi inverted SP microscope. Cells were counted by using NIS Elements (Nikon). For migration, cells were cultured 2 hours in Iscove modified Dulbecco medium (IMDM) +0.5% bovine serum albumin, and then placed in the upper chamber of a 5.0-μm pore transwell membrane (Corning no. 3241) with 300 ng/mL SDF-1α (Peprotech) in the lower chamber. After a 3-hour incubation, migrated cells collected from the lower chamber were counted. Data represents 2 independent experiments, each consisting of Cre–;Cdc42FL-MA9 control and Cre+;Cdc42FL-MA9 conditional KO cell lines plated in triplicate.
Division symmetry assay
Cre– and Cre+;Cdc42FL-MA9 cells were treated for 48 hours with 4-hydroxytamoxifen (4-OHT) vs EtOH vehicle control, and then plated in methylcellulose. Sixteen hours later, doublets were identified, indicating cells at the completion of the first cell division. Doublets were isolated and then transferred by pipette to a single well of RetroNectin-coated 96-well plates in media (IMDM, 10% fetal bovine serum [FBS], 20 ng/mL recombinant rat stem cell factor [SCF], and 10 ng/mL murine granulocyte-macrophage colony-stimulating factor, murine interleukin-3 [IL-3], and human IL-6). Daughter cells were separated by pipetting gently, and then visualized 4 hours later to confirm 2 distinct single cells (day 0). Wells were visualized over the next 48 hours to confirm subsequent divisions by each daughter cell, indicating viability. Efficiency of transfer of 2 distinct, viable daughter cells to wells that could be subsequently evaluated for division symmetry was similar between groups (data not shown). Evaluable wells were visualized at day 7 and scored as having 2 colonies (symmetric self-renewal [SS]), 1 colony (asymmetric self-renewal [AS]), or 0 colonies (symmetric differentiation [SD]).
Human AML cell lines and xenografts
Human umbilical cord blood was obtained from the Translational Trials Development and Support Laboratory at CCHMC in adherence with a protocol approved by the CCHMC Institutional Review Board. Patient samples were obtained from the CCHMC Cancer and Blood Diseases Institute tissue repository under an Institutional Review Board–approved research protocol with written informed consent in accordance with the Declaration of Helsinki. MA9 and MA9/NRas cell lines were generated by retroviral transduction of umbilical cord blood CD34+ cells, as previously described.5,7,8 Patient AML cells were cultured on OP9-DL1 stroma in IMDM containing 20% BIT 9500 (StemCell Technologies) and 10 ng/mL SCF, thrombopoietin, FLT3 ligand, IL-3, and IL-6 (KTF36 cytokines), as previously described.14 Xenografts were established by IV injection of cells into 6- to 8-week-old sublethally irradiated (280 cGy) NOD/LtSz-scid IL2RG (NSG) or NOD/LtSz-scid IL2RG-SGM3 mice without conditioning, or 24 hours after intraperitoneal injection of 40 mg/kg busulfan (Busulfex, Otsuka).14,15 Femoral or tibial aspirates were sampled to measure engraftment.
Short hairpin RNA–mediated knockdown of CDC42
TRMPVIR tetracycline-inducible short hairpin RNA (shRNA) expression vector was a gift from Johannes Zuber.16 This vector allows constitutive expression of Venus and tetracycline-inducible expression of microRNA-30–based shRNA sequence and dsRed fluorescent marker. Two distinct shRNA sequences were validated for knockdown of CDC42: 170 (5′-ACAGACAATTAAGTGTGTTGTTTAGTGAAGCCA CAGATGTAAACAACACACTTAATTGTCTGC) and 347 (ACAAGAGGATTATG ACAGATTATAGTGAAGCCACAGATGTATAATCTGTCATAATCCTCTTGC-3′). Sequence targeting Renilla-luciferase served as the control vector, as previously described.16 Each microRNA-30–based shRNA sequence was subcloned into the TRMPVIR backbone, and retroviral supernatant was produced. Following transduction, MA9/NRas cells were sorted on Venus+, and then maintained in culture (IMDM, 20% Tet-free FBS, 1% penicillin/streptomycin, and 10 ng/mL KTF36 cytokines. Knockdown was induced with 2 mg/mL doxycycline vs vehicle control. Cultures were analyzed for percent dsRed-positive over time using FACSVantage (BD Biosciences). MA9/NRas TRMPVIR cells were plated in triplicate in Methocult Express (StemCell Technologies) in the presence of 2 mg/mL doxycycline vs vehicle control, and colonies counted at days 7 to 14. For in vivo studies, MA9/NRas TRMPVIR cells were transduced with firefly luciferase–expressing vector and injected into NSG mice receiving doxycycline chow vs regular chow. Disease progression was measured at 4 weeks posttransplant by an IVIS bioluminescence imaging system (Perkin Elmer).
Lentiviral MISSION pLKO.1-shRNA-puro constructs targeting human CDC42 were obtained from Sigma-Aldrich. Efficient knockdown was confirmed with CDC42 shRNA sequence no. 471(5′-CCGGCCCTCTACTATTGAGAAACTTCTCGAGAAGTT TCTCAATAGTAGAGGGTTTTTG-3′). The puromycin cassette was replaced with the Venus insert from pLKO.1-nontargeting-Venus control vector. Lentiviral supernatant was used to transduce AML cells, as previously described.5 A total of 100 000 cells were IV injected into busulfan-conditioned NOD/LtSz-scid IL2RG-SGM3 mice. Leftover cells were maintained in culture and analyzed 72 hours posttransduction to determine the baseline Venus+ percentage. Intrafemoral aspirates were performed 4 weeks posttransplant to analyze engraftment.
Statistical analysis
Statistical analysis was performed with GraphPad Prism software, and significance was determined by using unpaired Student t test except for survival curves, where the log-rank test was used, polarity analysis, where Fisher’s exact test was used, and in vivo engraftment, where the Mann-Whitney U test was used. A value of P < .05 was considered significant.
Results
CDC42 is elevated in human AML
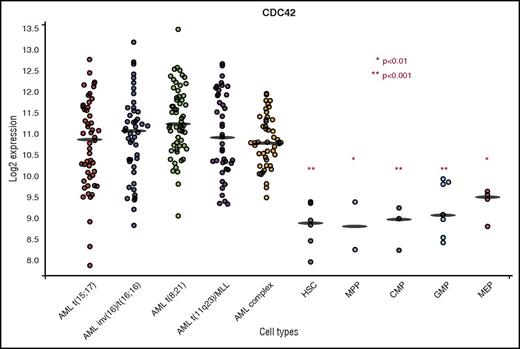
Previous work using both murine and human HSC/Ps expressing MLL-AF9 demonstrated an increase in active, GTP-bound CDC42 in the transformation to AML.4,5 Analysis of the BloodSpot database (www.bloodspot.eu) found increased CDC42 messenger RNA expression in primary human AML compared with normal stem and progenitor controls (Figure 1).17 In the AML subset with MLL rearrangement [t(11q23] CDC42 expression was increased relative to normal HSCs (P < .001), multipotential progenitor (P < .01), common myeloid progenitor (P < .001), granulocyte monocyte progenitor (P < .001), and megakaryocyte-erythroid progenitor (P < .01) populations. Although the present study uses MLL-rearranged leukemia as a model system, BloodSpot analysis showed similar elevation of CDC42 expression in AML across cytogenetic subgroups, including t(15;17), inv(16), t(8;21), and complex karyotype, suggesting that upregulation and increased activity of CDC42 may be common to AML transformation.
BloodSpot analysis showed increased CDC42 messenger RNA expression in primary human AML across cytogenetic subtypes in comparison with normal HSC/P controls. CDC42 expression was increased in t(11q23)/MLL rearranged AML compared with HSC, multipotential progenitor (MPP), common myeloid progenitor (CMP), granulocyte monocyte progenitor (GMP), and megakaryocyte-erythroid progenitor (MEP) subsets. *P < .01; ** P < .001. A similar trend was seen across cytogenetic subtypes of AML, including t(15;17), inv(16), t(8;21), and complex karyotype. BloodSpot is a curated database of publicly available gene expression datasets. AML samples came from the Microarray Innovations in Leukemia study headed by the European Leukemia Network and sponsored by Roche Molecular Systems, Inc. The normal hematopoietic subsets were from multiple studies. Platforms included Affymetrix Human 133U plus 2, Affymetrix Human 133UA, and Affymetrix Human 133UB chips (http://servers.binf.ku.dk/bloodspot/).
BloodSpot analysis showed increased CDC42 messenger RNA expression in primary human AML across cytogenetic subtypes in comparison with normal HSC/P controls. CDC42 expression was increased in t(11q23)/MLL rearranged AML compared with HSC, multipotential progenitor (MPP), common myeloid progenitor (CMP), granulocyte monocyte progenitor (GMP), and megakaryocyte-erythroid progenitor (MEP) subsets. *P < .01; ** P < .001. A similar trend was seen across cytogenetic subtypes of AML, including t(15;17), inv(16), t(8;21), and complex karyotype. BloodSpot is a curated database of publicly available gene expression datasets. AML samples came from the Microarray Innovations in Leukemia study headed by the European Leukemia Network and sponsored by Roche Molecular Systems, Inc. The normal hematopoietic subsets were from multiple studies. Platforms included Affymetrix Human 133U plus 2, Affymetrix Human 133UA, and Affymetrix Human 133UB chips (http://servers.binf.ku.dk/bloodspot/).
CDC42 is required for murine AML initiation
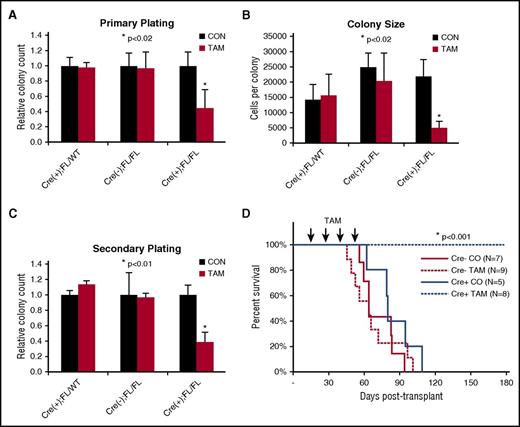
Finding upregulation of CDC42 expression/activity in AML, we sought to determine whether leukemia initiation was dependent on CDC42 integrity. To interrogate CDC42 in LIC self-renewal, MLL-AF9 cell lines were established following retroviral transduction of BM HSC/Ps from tamoxifen-inducible Cdc42 KO mice (Cre+;Cdc42FL), with Cre+;Cdc42FL/WT and Cre-;Cdc42FL donors as controls. Upon TAM treatment, Cdc42KO-MA9 cells had decreased CFU and colony size (Figure 2A-B). This decrease was also seen on secondary plating of Cdc42KO-MA9 cells (Figure 2C). Thus, Cdc42 loss impairs proliferation of neoplastic MA9-transduced progenitor cells. Mice transplanted with untreated MA9 cells with intact Cdc42 alleles were divided to receive injections of TAM vs control. TAM treatment was deferred until day +15 posttransplant to allow initial engraftment prior to Cdc42 deletion, thus avoiding the negative effects of Cdc42 loss on homing and lodging (supplemental Figure 1A-B). The Cdc42KO-MA9 cohort survived >200 days posttransplant, whereas vehicle control mice died of AML with latency similar to Cre-MA9 cell recipients (Figure 2D, P < .005). Expected blood genotypes of transplant donors and recipients were confirmed by PCR analysis (supplemental Figure 2). Thus, the loss of Cdc42 suppresses leukemia-initiating activity.
Conditional deletion of Cdc42 blocks AML initiation. (A) Cre+;Cdc42FL-MA9 cells treated with TAM to delete Cdc42 had reduced CFU cell (CFU-C) in primary plating in methylcellulose compared with heterozygous Cdc42 FL/WT and Cre null (–) controls (CON). Data are representative of 3 to 5 experiments using 2 distinct cell lines per genotype. (B) Colony size, as determined by cell count per colony, was also decreased. (C) CFU-C numbers were still reduced on replating in secondary methylcellulose cultures. (D) TAM-induced deletion of Cdc42 from Cre+;Cdc42FL-MA9 cells suppressed leukemia initiation vs Cre null (–) and corn oil (CO) injected controls (P < .001).
Conditional deletion of Cdc42 blocks AML initiation. (A) Cre+;Cdc42FL-MA9 cells treated with TAM to delete Cdc42 had reduced CFU cell (CFU-C) in primary plating in methylcellulose compared with heterozygous Cdc42 FL/WT and Cre null (–) controls (CON). Data are representative of 3 to 5 experiments using 2 distinct cell lines per genotype. (B) Colony size, as determined by cell count per colony, was also decreased. (C) CFU-C numbers were still reduced on replating in secondary methylcellulose cultures. (D) TAM-induced deletion of Cdc42 from Cre+;Cdc42FL-MA9 cells suppressed leukemia initiation vs Cre null (–) and corn oil (CO) injected controls (P < .001).
CDC42 maintains clonogenic AML cells
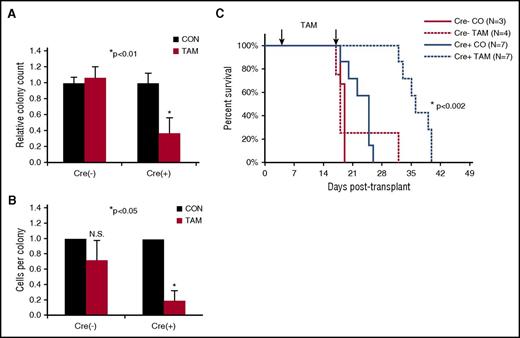
AML cells harvested from diseased mice bearing the Cre+;Cdc42FL-MA9 transplant were plated in methylcellulose in the presence or absence of TAM to test the requirement for Cdc42 in the maintenance of clonogenic LICs in vitro. Cdc42KO-MA9 cells showed decreased colony size and number, demonstrating the requirement for Cdc42 in intrinsic self-renewal of LICs (Figure 3A-B). To test the requirement for Cdc42 in AML maintenance in vivo, secondary transplants were established by using Cre+;Cdc42FL-MA9 cells harvested from control treated mice with Cdc42 alleles still intact. TAM treatment was deferred until transplant day +5 to allow sufficient time for engraftment. In vivo deletion of Cdc42 from fully transformed MA9 leukemia in congenic recipients significantly prolonged disease latency (Figure 3C; P < .005), indicating that Cdc42 is required for maintenance of MA9 leukemia clonality. Mice in the Cdc42KO-MA9 cohort eventually succumbed to a delayed AML resembling Cdc42WT-MA9 leukemia, and PCR genotyping showed complete deletion of floxed Cdc42 alleles in the recovered Cdc42KO-MA9 leukemia tumors (supplemental Figure 2A). Secondary recipients of these Cdc42KO-MA9 cells died with similar latency as Cdc42FL-MA9 recipients (supplemental Figure 2B), suggesting the leukemia eventually compensates for Cdc42 deletion. Interestingly, a compensatory elevation of Rac1 activity was seen in Cdc42KO-MA9 leukemia cells (supplemental Figure 3), suggesting that Cdc42 resistance is associated with Rac1 activation, which has previously been implicated in MA9 leukemia maintenance.5,7 Thus, Cdc42 targeting significantly impedes MA9 leukemia development but may result in a compensatory bypass in the long term. Cdc42 loss from the MLL-AF9 knockin mouse,18 with regulation of MLL-AF9 expression by the endogenous promoter, showed similar effects on clonogenicity and survival (supplemental Figure 4A-B), indicating the requirement for Cdc42 is not limited to an overexpression system.
Loss of Cdc42 disrupts maintenance of the fully transformed MA9 leukemia. MA9 leukemia cells were harvested from diseased animals in the vehicle control group (Cdc42 alleles still intact) and plated in methylcellulose in the presence or absence of 4-OHT. (A) On Cdc42 deletion, MA9 cells had a reduced number of CFU-C compared with controls and (B) fewer cells per colony. (C) Cdc42 deletion from MA9 cells in secondary transplants prolonged the latency of disease (P < .002).
Loss of Cdc42 disrupts maintenance of the fully transformed MA9 leukemia. MA9 leukemia cells were harvested from diseased animals in the vehicle control group (Cdc42 alleles still intact) and plated in methylcellulose in the presence or absence of 4-OHT. (A) On Cdc42 deletion, MA9 cells had a reduced number of CFU-C compared with controls and (B) fewer cells per colony. (C) Cdc42 deletion from MA9 cells in secondary transplants prolonged the latency of disease (P < .002).
Loss of CDC42 promotes AML cell differentiation
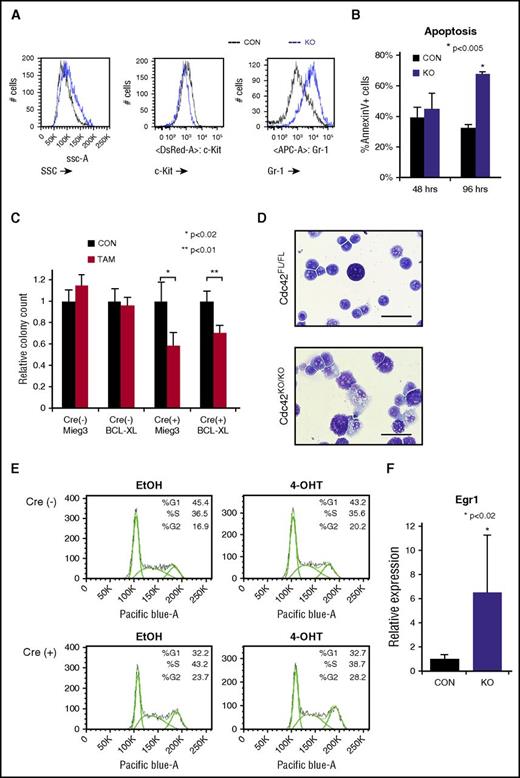
Fully transformed Cre+;Cdc42FL-MA9 cells harvested from diseased animals were cultured ex vivo and treated with TAM to induce deletion of Cdc42. PCR genotyping showed loss of floxed Cdc42 alleles within 36 hours of TAM treatment (supplemental Figure 5A). Cdc42 protein expression was undetectable within 48 to 72 hours of TAM treatment (supplemental Figure 5A). On the loss of Cdc42, MA9 leukemia cultures had higher side scatter and Gr-1 expression and decreased c-Kit expression, indicating differentiation (Figure 4A). Although no significant difference in apoptosis was seen at 48 hours, Cdc42KO-MA9 cells displayed increased apoptosis as time progressed (96 hours), consistent with terminal differentiation (Figure 4B). Previously, we found that Rac2, a small Rho GTPase closely related to Cdc42, sustains MA9 leukemia cell survival through upregulation of Bcl-xL.13 We sought to determine whether Cdc42 similarly impacts the MA9 cell through survival signaling. Cdc42FL-MA9 cells harvested from primary mice were transduced to ectopically express Bcl-xL, with empty vector-transduced cells as a control (supplemental Figure 6). Colony-forming ability was assayed after deletion of Cdc42. Interestingly, overexpression of Bcl-xL was insufficient to rescue colony-forming ability of Cdc42KO-MA9 cells, suggesting Cdc42 loss blocks self-renewal and promotes differentiation, rather than directly inducing apoptosis (Figure 4C). Cdc42KO-MA9 cells showed differentiated cytomorphology (Figure 4D). Cell cycle analysis revealed no significant difference between Cdc42FL-MA9 cells and Cdc42KO-MA9 cells (Figure 4E). Consistent with the differentiation phenotype, Cdc42KO-MA9 cells also had increased expression of the myeloid differentiation gene Egr-1 (Figure 4F), which is known to increase differentiated MLL-AF9 cells following the loss of Ezh2.19 Together, these data suggest that CDC42 targeting leads to LIC differentiation.
Loss of Cdc42 leads to MA9 cell differentiation. (A) Cdc42KO-MA9 leukemia cells had a differentiated immunophenotype and (B) a delayed increase in apoptosis. (C) Overexpression of the prosurvival protein Bcl-xL was not sufficient to rescue the loss of CFU on deletion of Cdc42. (D) Cdc42KO-MA9 cells showed a differentiated cellular morphology on Wright-Giemsa stain compared with Cdc42FL controls. (E) Cdc42KO-MA9 leukemia cells had no significant difference in cell cycle analysis. (F) Relative expression of the myeloid differentiation gene Egr1 was increased in Cdc42KO-MA9 cells normalized to Gapdh. Data are representative of 3 to 5 independent experiments. SSC, side scatter.
Loss of Cdc42 leads to MA9 cell differentiation. (A) Cdc42KO-MA9 leukemia cells had a differentiated immunophenotype and (B) a delayed increase in apoptosis. (C) Overexpression of the prosurvival protein Bcl-xL was not sufficient to rescue the loss of CFU on deletion of Cdc42. (D) Cdc42KO-MA9 cells showed a differentiated cellular morphology on Wright-Giemsa stain compared with Cdc42FL controls. (E) Cdc42KO-MA9 leukemia cells had no significant difference in cell cycle analysis. (F) Relative expression of the myeloid differentiation gene Egr1 was increased in Cdc42KO-MA9 cells normalized to Gapdh. Data are representative of 3 to 5 independent experiments. SSC, side scatter.
Loss of CDC42 disrupts structural and functional polarity of AML cells
Whereas LICs perpetuate the leukemic clone, among its progeny are differentiated blasts with limited capacity for self-renewal. Agents that promote LIC differentiation offer potential therapeutic benefit either alone or in combination with cytoxic chemotherapy, as exemplified by the success of all-trans retinoic acid in the treatment of acute promyelocytic leukemia.
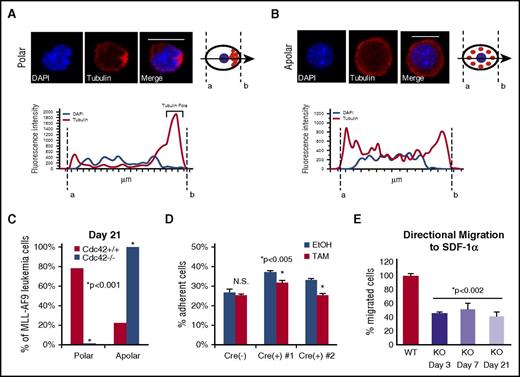
In HSCs, Cdc42 activity increases with age in association with the loss of cell polarity and diminished stem cell function in competitive repopulation assays.3 The interplay between Cdc42 expression and leukemia cell polarity has not been previously demonstrated. We examined the expression and distribution of Cdc42 and tubulin in murine MA9 cells with intact (Cdc42WT-MA9) or conditionally deleted (Cdc42KO-MA9) alleles of Cdc42 by immunofluorescent staining. In a blinded analysis, we measured pixel intensity along a vector bisecting each cell to evaluate polar (Figure 5A) vs apolar (Figure 5B) distribution of tubulin, as previously described.3 Manual scoring of polar and apolar cells showed the majority of cells lose polarity in culture over time, shown at 3 weeks postdeletion of Cdc42 in Figure 5C. In a secondary approach, polarity analysis by imaging flow cytometry of cells in suspension similarly demonstrated increased apolar cells in Cdc42KO-MA9 cultures over time (supplemental Figure 7A-B).
Loss of cell polarity on deletion of Cdc42. (A) MA9 cells were stained with anti-tubulin primary and Rhodamine secondary antibodies (Tubulin) and 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain. Confocal images were analyzed using NIS Elements software (Nikon) to measure fluorescence intensity along a bisecting vector through individual cells. Scale bar, 10 μm. (A) Cells were considered polar when clear asymmetric distribution of tubulin was seen or (B) apolar in the absence of tubulin asymmetry, as previously described.3 Data are plotted as the percentage of the total number of cells scored per sample. (C) A loss of polarity and increase in apolar cell fraction was observed following Cdc42 deletion from MA9 cells. This change became more pronounced as Cdc42KO-MA9 cells were passaged in culture, as shown at 3 weeks postdeletion of Cdc42. Functional polarity was assayed by MA9 cell adhesion and migration. (D) Cre+;Cdc42FL-MA9 cells treated with TAM to induce deletion of Cdc42 had decreased adhesion to fibronectin compared with vehicle (EtOH)–treated controls whereas no difference was seen in Cre null (–) MA9 control cells. (E) Similarly, Cre+;Cdc42FL-MA9 cells treated with TAM had a sustained decrease in migration to SDF-1α compared with controls.
Loss of cell polarity on deletion of Cdc42. (A) MA9 cells were stained with anti-tubulin primary and Rhodamine secondary antibodies (Tubulin) and 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain. Confocal images were analyzed using NIS Elements software (Nikon) to measure fluorescence intensity along a bisecting vector through individual cells. Scale bar, 10 μm. (A) Cells were considered polar when clear asymmetric distribution of tubulin was seen or (B) apolar in the absence of tubulin asymmetry, as previously described.3 Data are plotted as the percentage of the total number of cells scored per sample. (C) A loss of polarity and increase in apolar cell fraction was observed following Cdc42 deletion from MA9 cells. This change became more pronounced as Cdc42KO-MA9 cells were passaged in culture, as shown at 3 weeks postdeletion of Cdc42. Functional polarity was assayed by MA9 cell adhesion and migration. (D) Cre+;Cdc42FL-MA9 cells treated with TAM to induce deletion of Cdc42 had decreased adhesion to fibronectin compared with vehicle (EtOH)–treated controls whereas no difference was seen in Cre null (–) MA9 control cells. (E) Similarly, Cre+;Cdc42FL-MA9 cells treated with TAM had a sustained decrease in migration to SDF-1α compared with controls.
Cdc42 activity plays a central role in cellular functions dependent on cell polarization. Integrin-dependent activation of Cdc42 leads to filopodia and focal complex formation necessary for cell spreading and adhesion.20-22 Directional cell migration to chemotactic signals, such as the chemokine SDF-1α, requires Cdc42 activation at the leading edge of polarized cells.21-23 Cdc42KO-MA9 cells exhibited decreased adhesion compared with wild-type (WT) MA9 cell controls (Figure 5D). Furthermore, Cdc42KO-MA9 cells had a persistent, although not progressive, decrease in directional migration to SDF-1α in transwell assays over time (Figure 5E). These findings suggest that Cdc42 is important for structural and functional regulation related to MA9 cell polarity.
CDC42 loss reduces the frequency of cell divisions, giving rise to clonogenic daughter cells
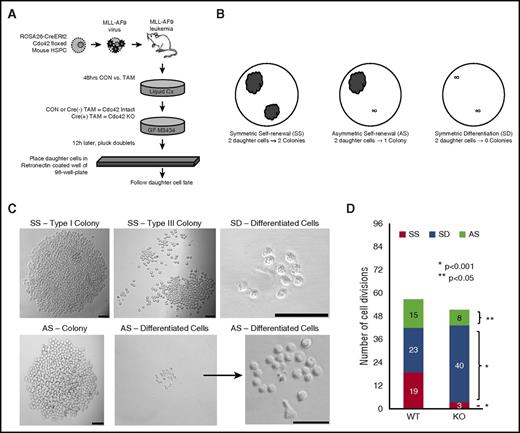
Stem cell polarity can intimately affect division symmetry and cell fate. We next investigated whether the loss of cell polarity from Cdc42 deletion impacts LIC division. Cell division produces 2 daughter cells that may have congruous (symmetric) or disparate (asymmetric) cell fates. Relevant to the LICs, the daughter cells produced may either have the potential to establish a colony (self-renewal) or show limited proliferative potential, resulting in the failure to give rise to a colony (differentiation). We treated Cre+;Cdc42FL-MA9 cells with TAM (KO) vs vehicle control (WT) for 48 hours, and then plated cells in methylcellulose (Figure 6A). To separate daughter cells resulting from single cell divisions, doublets were identified after 16 hours, plucked by pipette, and placed in RetroNectin-coated wells. Only wells in which the successful transfer of 2 distinct daughter cells was confirmed were deemed evaluable for subsequent analysis. Wells were examined 7 days later to determine the fate of each daughter cell with respect to their ability to form a colony. Each cell division could therefore be classified as resulting in SS (each daughter cell giving rise to a colony), SD (neither daughter cell establishing a colony), or AS (one of the daughter cells giving rise to a colony, and the other cell with differentiated fate; Figure 6B). Colonies were defined as having >50 cells and displayed type I and type III colony morphology, as previously described (Figure 6C).24 Asymmetric divisions occurred in wells in which 1 daughter cell produced a typical colony, but the sister cell had a differentiated fate (representative images are show in Figure 6C). Comparing the distribution of each type of cell division, we found that Cdc42KO-MA9 cells had a loss of SS and a marked increase in SD (Figure 6D; P < .001). The Cdc42KO-MA9 cells also had fewer asymmetric divisions (Figure 6D; P < .05). Thus, the loss of cell polarity by Cdc42KO-MA9 cells closely correlates with the loss of SS and AS.
Loss of Cdc42 increased the percentage of cell divisions, resulting in symmetric differentiation. (A) Murine MLL-AF9 leukemia cells were treated with 4-OHT to induce Cre-mediated deletion of Cdc42 (KO), with ethanol-treated cells serving as the vehicle control (WT). After 48 hours, the cells were plated in methylcellulose media (MethoCult GF M3434, StemCell Technologies). Sixteen hours later, doublets were identified, indicating cells at completion of the first cell division. Doublets were isolated and then transferred by pipette to a single well of RetroNectin-coated 96-well plates in IMDM + 10% FBS + 20 ng/mL recombinant rat SCF and 10 ng/mL murine granulocyte-macrophage colony-stimulating factor, murine IL-3, and human IL-6. Daughter cells were separated by gently pipetting up and down, then visualized 4 hours later to confirm 2 distinct single cells adherent to the RetroNectin. Wells were visualized over the next 48 hours to confirm subsequent divisions by each daughter cell, indicating viability. Wells in which 2 distinct, viable daughter cells could not be visualized were excluded from analysis. (B,C) Wells were then visualized after 7 days and scored as having 2 colonies (SS), 1 colony (AS), or 0 colonies (SD). Scale bar, 50 μm. (D) Data are shown as the number of each type of cell division scored per 96 wells plated with WT vs KO MA9 leukemia daughter cell pairs, and the statistical significance of differences between the 2. Data are representative of 6 independent experiments.
Loss of Cdc42 increased the percentage of cell divisions, resulting in symmetric differentiation. (A) Murine MLL-AF9 leukemia cells were treated with 4-OHT to induce Cre-mediated deletion of Cdc42 (KO), with ethanol-treated cells serving as the vehicle control (WT). After 48 hours, the cells were plated in methylcellulose media (MethoCult GF M3434, StemCell Technologies). Sixteen hours later, doublets were identified, indicating cells at completion of the first cell division. Doublets were isolated and then transferred by pipette to a single well of RetroNectin-coated 96-well plates in IMDM + 10% FBS + 20 ng/mL recombinant rat SCF and 10 ng/mL murine granulocyte-macrophage colony-stimulating factor, murine IL-3, and human IL-6. Daughter cells were separated by gently pipetting up and down, then visualized 4 hours later to confirm 2 distinct single cells adherent to the RetroNectin. Wells were visualized over the next 48 hours to confirm subsequent divisions by each daughter cell, indicating viability. Wells in which 2 distinct, viable daughter cells could not be visualized were excluded from analysis. (B,C) Wells were then visualized after 7 days and scored as having 2 colonies (SS), 1 colony (AS), or 0 colonies (SD). Scale bar, 50 μm. (D) Data are shown as the number of each type of cell division scored per 96 wells plated with WT vs KO MA9 leukemia daughter cell pairs, and the statistical significance of differences between the 2. Data are representative of 6 independent experiments.
CDC42 maintains the human AML LIC
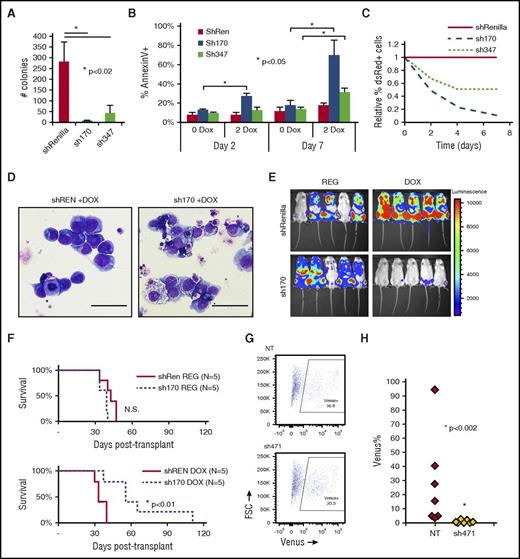
We used Tet-inducible shRNA to knockdown CDC42 in human AML cell lines transformed by MA9 and mutant NRas (MA9/NRas)5,15,16 to test whether CDC42 represents a target for AML differentiation therapy. Immunoblot analysis confirmed efficient knockdown of CDC42 compared with shRNA targeting Renilla-luciferase as a control (supplemental Figure 8A). CDC42-deficient MA9/NRas cells had significantly decreased colony-forming ability (Figure 7A). Similar to the effect seen in murine MA9 cells, reduced CDC42 expression led to late induction of apoptosis, consistent with terminal differentiation (Figure 7B). CDC42 knockdown in human MA9/NRas cells reduced growth in culture (Figure 7C) and also resulted in differentiated cytomorphology (Figure 7D). MA9/NRas cells coexpressing inducible CDC42 shRNA and the firefly luciferase reporter were transplanted into NSG mice, and mice were maintained on doxycycline chow to induce shRNA expression. Bioluminescence imaging showed delayed AML progression in the knockdown group compared with nontargeting shRNA and regular chow controls (Figure 7E). This resulted in prolonged survival (Figure 7F). Thus, CDC42 targeting in human MA9 LIC reproduces the phenotype seen in the mouse model, indicating that targeted inhibition of CDC42 is effective in abolishing AML maintenance.
Human MA9/NRas cells were transduced to allow Tet-inducible shRNA expression marked by dsRed. Knockdown of CDC42 was achieved by 2 distinct shRNA sequences (sh170 and sh347) on treatment with 2 mg/ml doxycycline (2 DOX). Nonspecific shRNA targeting Renilla-luciferase served as the control (shREN). (A) CDC42 knockdown led to decreased CFU-C in methylcellulose and (B) increased apoptosis as measured by AnnexinV+ staining, with greater effect on apoptosis seen later at day 7 in culture. (C) CDC42 knockdown inhibited human MA9/NRas cell growth in liquid culture. (D) Wright-Giemsa stain showed typical blast morphology in shRenilla control cultures treated with doxycycline (DOX; left panel), whereas sh170 DOX cultures showed increased differentiation and vacuolization (right panel). Scale bar, 50 μm. (E) Bioluminescent imaging obtained at 4 weeks posttransplant showed decreased in vivo leukemia progression on CDC42 knockdown. (F) Kaplan-Meier analysis showed prolonged survival from knockdown of CDC42 (P < .01). A primary human patient AML sample bearing a t(X;11) translocation encoding the MLL-SEPT6 fusion was transduced with lentiviral vectors expressing CDC42 shRNA (sh471) vs nontargeting control and the Venus fluorescent protein, then injected into NSG mice. (G) FACS analysis at day 3 showed 16.8% and 23.3% Venus+ cells in the nontargeting control (NT) and sh471 transduction cultures, respectively. (H) BM was harvested and analyzed for Venus+ cells at week 4 posttransplant to reveal the loss of MLL-SEPT6 AML graft on CDC42 knockdown (P < .002). FSC, forward scatter; N.S., not significant.
Human MA9/NRas cells were transduced to allow Tet-inducible shRNA expression marked by dsRed. Knockdown of CDC42 was achieved by 2 distinct shRNA sequences (sh170 and sh347) on treatment with 2 mg/ml doxycycline (2 DOX). Nonspecific shRNA targeting Renilla-luciferase served as the control (shREN). (A) CDC42 knockdown led to decreased CFU-C in methylcellulose and (B) increased apoptosis as measured by AnnexinV+ staining, with greater effect on apoptosis seen later at day 7 in culture. (C) CDC42 knockdown inhibited human MA9/NRas cell growth in liquid culture. (D) Wright-Giemsa stain showed typical blast morphology in shRenilla control cultures treated with doxycycline (DOX; left panel), whereas sh170 DOX cultures showed increased differentiation and vacuolization (right panel). Scale bar, 50 μm. (E) Bioluminescent imaging obtained at 4 weeks posttransplant showed decreased in vivo leukemia progression on CDC42 knockdown. (F) Kaplan-Meier analysis showed prolonged survival from knockdown of CDC42 (P < .01). A primary human patient AML sample bearing a t(X;11) translocation encoding the MLL-SEPT6 fusion was transduced with lentiviral vectors expressing CDC42 shRNA (sh471) vs nontargeting control and the Venus fluorescent protein, then injected into NSG mice. (G) FACS analysis at day 3 showed 16.8% and 23.3% Venus+ cells in the nontargeting control (NT) and sh471 transduction cultures, respectively. (H) BM was harvested and analyzed for Venus+ cells at week 4 posttransplant to reveal the loss of MLL-SEPT6 AML graft on CDC42 knockdown (P < .002). FSC, forward scatter; N.S., not significant.
To validate the role of CDC42 in primary AML cells, we used a patient-derived xenograft model established with primary AML blasts harboring a t(X;11) translocation expressing the MLL-SEPT6 fusion oncogene.14 After confirmation of efficient knockdown (supplemental Figure 8B), lentiviral shRNA vector targeting CDC42 and expressing Venus fluorescent protein was used to transduce patient-derived CD45+/CD33+ AML cells, which were then transplanted into immunodeficient mice, with nontargeting shRNA as control. Initial transduction efficiency was measured by FACS analysis of Venus+ cells at day +3 posttransduction (Figure 7G). AML cells expressing CDC42 shRNA failed to progress in xenografted mice, as shown by the loss of Venus+ cells in BM aspirates at 4 weeks posttransplant, whereas nontargeting shRNA-expressing AML cells readily engrafted and progressed in vivo (Figure 7H). A similar phenotype was seen on CDC42 knockdown in 2 additional patient AML samples (supplemental Figure 9A-B). Thus, consistent with the murine genetic model, CDC42 deficiency in a patient-derived xenograft model blocks leukemia formation, implicating CDC42 in LIC maintenance and progression.
Discussion
Future success in AML therapy aspires to overcome the survival and self-renewal advantages enjoyed by LICs. Nevertheless, specific signaling pathways involved in LIC maintenance that may be therapeutically exploited remain limited. LICs, like their normal HSC counterparts, depend on interactions with the extracellular matrix, soluble factors, and cellular components of the BM niche.25-27 These interactions retain LICs in the niche and may also contribute to resistance of the LIC to chemotherapy and treatment failure.28-30 The Rho GTPase CDC42 plays an integral role in coordinating adhesion, migration, and homing in response to diverse inputs for proper localization of HSC/Ps in the BM niche.1,31 Similarly, we found that the loss of Cdc42 from the LICs blocks AML cell adhesion and migration (Figure 5D-E). CDC42 expression is increased in human AML patient samples across cytogenetic subtypes compared with normal hematopoietic cell subsets, suggesting that this pathway is upregulated broadly in AML, not just in the setting of MLL gene rearrangement (Figure 1). Furthermore, recent analysis ranks pathways involved in adherens junction, regulation of the actin cytoskeleton, tight junction, focal adhesion, Rho cell motility signaling, and Wnt signaling among the top 10 most highly dysregulated gene networks distinguishing human AML stem cells from normal HSCs.6 CDC42 is involved in each of these pathways and is upregulated in CD34+/CD38– leukemia stem cells.6 Recently, the CDC42 effector PAK1 was found overexpressed in AML patients with high H2.0-like homeobox gene expression and constituted part of the H2.0-like homeobox–induced core signature that predicts inferior outcome,32 and inhibition of PAK1 induces differentiation of LICs leading to apoptosis.33 Collectively, CDC42 appears at the nexus of signaling from oncogenes and niche in LIC regulation.
LICs, like HSCs, undergo self-renewal and differentiation.34,35 Central to the balance between self-renewal and differentiation is the mode (symmetric vs asymmetric) of stem cell division.7 In Drosophila melanogaster as well as Caenorhabditis elegans, studies have demonstrated asymmetric stem cell division.36 These stem cells are polarized during division and segregate cell-fate determinants unequally to daughter cells. Previous studies have shown that HSCs maintain their self-renewal predominantly through asymmetric cell divisions, resulting in unequal distribution of proteins to daughter cells and disparate functional fates, resulting in an HSC and a committed progeny in the absence of reprogramming factors.10,11 Published data also support that both stem cell intrinsic and niche signals determine the cell fate of the daughter cells.10,37,38 A logical, yet still unverified, model proposes that the mode of LIC division determines the fate of daughter cells, and thus LIC homeostasis and differentiation, in that symmetric division results in daughter cells with similar potential, whereas asymmetric division generates daughter cells with different potentials. A recently proposed concept is that some LICs adopt a symmetric division mechanism in promoting regeneration and maintenance.7-9 Although molecular mechanisms that regulate the mode of LIC division remain largely unknown, our data indicate that cell polarity and division symmetry of LICs are closely associated, and CDC42, a central regulator of polarity, coordinates LIC polarity and division symmetry to influence differentiation vs self-renewal. We demonstrate that CDC42 deficiency results in the loss of LIC polarity, in association with more SD divisions. Significantly, our results support a model that loss of polarity and reduced division asymmetry hinder, rather than promote, the self-renewal of LICs. Because CDC42-regulated functions, including polarity, division asymmetry, and differentiation, appear conserved in murine and human LICs, our studies of the role of CDC42 in mediating LIC polarity and cell fate add to the cancer stem cell field and imply that CDC42 targeting in AML bears therapeutic value. Small molecules specifically targeting CDC42 activity are under preclinical development3,39,40 and may offer a promising therapy when studies related to in vivo pharmacokinetics, delivery, and toxicity progress further.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Flow Cytometry Core, the Comprehensive Mouse and Cancer Core, and the Confocal Imaging Core at Cincinnati Children’s Hospital Medical Center for their assistance. The authors thank Ashish Kumar for helpful discussion and advice regarding MA9 knockin mouse experiments.
This work was supported in part by National Institutes of Health, National Cancer Institute grants R01 CA 204895 (J.C.M. and Y.Z.), R01 CA193350 (Y.Z.), and R50 CA211404 (M.W.), and by Child Health Research Career Development Award K12 HD028827 (B.M.). B.M. is a St. Baldrick’s Foundation Scholar. Portions of this work were also supported by grants from CancerFree KIDS (B.M.), Hyundai Hope on Wheels (J.C.M. and B.M.), a William Cooper Procter Research Award from Cincinnati Children’s Hospital Research Foundation (B.M.), and Pediatric Scientist Development Program grant K12 HD000850 (B.M.). D.C.M. was supported by the American Society of Hematology HONORS award.
Authorship
Contribution: B.M. and E. O’Brien designed experiments, performed the research, and participated in data analysis and manuscript preparation; D.C.M., M.W., C.L.H., X.D., W.L., and E. Orr participated in the design and performance of some experiments; H.L.G. contributed reagents and participated in research design and analysis; Y.Z. contributed reagents, reviewed the research design and analysis, and prepared the manuscript; and J.C.M. was responsible for the overall design of the study, analysis of data, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yi Zheng, Cincinnati Children’s Hospital Medical Center, 3333 Burnett Ave, Cincinnati, OH 45229; e-mail: yi.zheng@cchmc.org; and James C. Mulloy, Cincinnati Children’s Hospital Medical Center, 3333 Burnett Ave, Cincinnati, OH 45229; e-mail: james.mulloy@cchmc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal