Key Points
MALT1 protease activity stabilizes MYC.
The MALT1-MYC network might represent a therapeutic target for MCL patients.
Abstract
Mantle cell lymphoma (MCL) is a mature B-cell lymphoma characterized by poor clinical outcome. Recent studies revealed the importance of B-cell receptor (BCR) signaling in maintaining MCL survival. However, it remains unclear which role MALT1, an essential component of the CARD11-BCL10-MALT1 complex that links BCR signaling to the NF-κB pathway, plays in the biology of MCL. Here we show that a subset of MCLs is addicted to MALT1, as its inhibition by either RNA or pharmacologic interference induced cytotoxicity both in vitro and in vivo. Gene expression profiling following MALT1 inhibition demonstrated that MALT1 controls an MYC-driven gene expression network predominantly through increasing MYC protein stability. Thus, our analyses identify a previously unappreciated regulatory mechanism of MYC expression. Investigating primary mouse splenocytes, we could demonstrate that MALT1-induced MYC regulation is not restricted to MCL, but represents a common mechanism. MYC itself is pivotal for MCL survival because its downregulation and pharmacologic inhibition induced cytotoxicity in all MCL models. Collectively, these results provide a strong mechanistic rationale to investigate the therapeutic efficacy of targeting the MALT1-MYC axis in MCL patients.
Introduction
Mantle cell lymphoma (MCL) is characterized by an aggressive clinical course and short overall survival.1 Different cytomorphological variants can be distinguished and, in particular, blastic variants are associated with poor overall survival.2-4 Besides clinical factors summarized in the MCL international prognostic index, high cell proliferation has been identified as a major prognostic factor associated with adverse outcome.5-7
Pathogenetically, MCL is characterized by cyclin D1 overexpression resulting from the chromosomal translocation t(11;14)(q13;q32).8 In addition, various secondary genetic aberrations activating different pathways have been elucidated.9,10 Recently, constitutive activation of B-cell receptor (BCR) signaling and downstream activation of the NF-κB pathway have been identified to be critical for survival of MCL subsets.11,12 Upon BCR stimulation, MALT1 and BCL10 are recruited to CARD11, resulting in the formation of the CARD11-BCL10-MALT1 complex and NF-κB activation.13,14 Additionally, the protease activity of MALT1 is enhanced, leading to cleavage of NF-κB inhibitors such as A20 and RelB.15,16 Other known MALT1 substrates include BCL10, CYLD, Regnase-1, Roquin-1, Roquin-2, and HOIL1.17-25
Preliminary data suggested that MALT1 is constitutively activated in subsets of MCL.11 However, its precise role in the pathogenesis of MCL remains unknown. Thus, we investigated the role of MALT1 in the biology of MCL in the current study.
Methods
Patient samples, immunohistochemistry, and fluorescence in situ hybridization (FISH)
CD20+ MCL cells were separated from peripheral blood mononuclear cells (PBMCs; patient samples 1 and 2) or cell suspensions from lymph nodes (patient samples 3-5) of MCL patients by CD20 magnetic-activated cell sorting (Miltenyi Biotec, Bergisch Gladbach, Germany).
Cell culture, retroviral constructs, and transductions
The experiments were performed as described.28-30 Protocols are available in the supplemental Material and Methods. The sequences of the used small hairpin RNAs (shRNAs) are summarized in supplemental Table 1.
Viability assay, analysis of cell cycle, apoptosis, and proliferation
Isolation and stimulation of mouse splenocytes
Protocols are available in the supplemental Material and Methods.
In vivo xenograft mouse studies
The in vivo xenograft mouse studies were done as described.33 Protocols are available in the supplemental Material and Methods.
Gene expression profiling
Quantitative PCR
Quantitative polymerase chain reaction (PCR) was performed as described using predesigned assays (Applied Biosystems, Carlsbad, CA).29
Western blotting and analysis of MYC stability
Protocols are available in the supplemental Material and Methods.
Results
MALT1 is expressed and activated in MCL
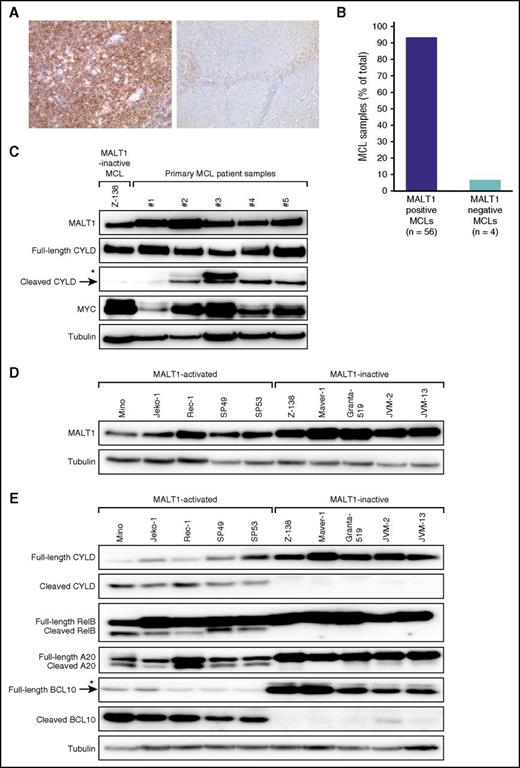
To assess if MALT1 is expressed in MCL, we determined its expression in 60 primary samples by immunohistochemistry. To establish the immunohistochemical assay, we stained 5 reactive lymph node and tonsil specimens. MALT1 was expressed in both B- and T-cell areas of the lymph node, albeit to varying degrees. The germinal center (GC) B cells were strongly positive, and staining was accentuated in the dark zone of the GC (supplemental Figure 1). Fifty-six of 60 (93%) MCL cases stained positive for MALT1 and showed a diffuse cytoplasmic expression that was detectable in virtually all MCL cells (Figure 1A-B). We compared this expression pattern with other aggressive lymphomas by staining 81 primary diffuse large B-cell lymphoma (DLBCL) samples. We determined the molecular DLBCL subtype by applying the Hans algorithm36 and identified 34 germinal center B-cell like (GCB) and 47 non-GCB DLBCLs. All 34 GCB as well as 46 of the 47 (98%) non-GCB DLBCLs expressed MALT1, suggesting that different B-cell lymphoma subtypes express MALT1.
MALT1 expression and activity in MCL. (A) Immunohistochemical MALT1 staining of a MALT1-positive MCL case (left; original magnification ×200) and a MALT1-negative MCL case (right; original magnification ×100). Images were captured using a Leitz DMRB microscope (Leica Microsystems, Wetzlar, Germany) equipped with Fluotar objective lenses (10×/0.30 numeric aperture, 20×/0.50 numeric aperture) and a KY-F75U digital camera (Victor, Yokohama, Japan) and were processed with the Diskus Program 4.20 (Hilgers Technical, Königswinter, Germany) that converts and exports images in .jpg file format. (B) MALT1 expression in MCLs determined by immunohistochemistry. (C) Western blot analysis of MALT1, full-length and cleaved forms of CYLD, and MYC. MALT1 was highly expressed in CD20+ cells isolated from either PBMCs (patient samples 1 and 2) or lymph nodes (patient samples 3-5) of 5 primary MCL patient samples. Cleaved CYLD indicating MALT1 proteolytic activity was detectable in 4 of 5 patient samples. MYC expression was higher in these 4 samples with activated MALT1. The MCL cell line Z-138 was used as a positive control for MALT1 expression and as a negative control for CYLD cleavage. *Nonspecific band, which was not observed in any MALT1-activated MCL cell lines (Figure 1E). (D) Western blot analysis of MALT1 expression in MCL cell lines. MALT1 protein expression was detectable in all MCL cell lines. (E) Western blot analysis of different MALT1 targets. Cleaved forms of CYLD, RelB, A20, and BCL10 were detectable in Mino, Jeko-1, Rec-1, SP49, and SP53 cells. *Nonspecific band.
MALT1 expression and activity in MCL. (A) Immunohistochemical MALT1 staining of a MALT1-positive MCL case (left; original magnification ×200) and a MALT1-negative MCL case (right; original magnification ×100). Images were captured using a Leitz DMRB microscope (Leica Microsystems, Wetzlar, Germany) equipped with Fluotar objective lenses (10×/0.30 numeric aperture, 20×/0.50 numeric aperture) and a KY-F75U digital camera (Victor, Yokohama, Japan) and were processed with the Diskus Program 4.20 (Hilgers Technical, Königswinter, Germany) that converts and exports images in .jpg file format. (B) MALT1 expression in MCLs determined by immunohistochemistry. (C) Western blot analysis of MALT1, full-length and cleaved forms of CYLD, and MYC. MALT1 was highly expressed in CD20+ cells isolated from either PBMCs (patient samples 1 and 2) or lymph nodes (patient samples 3-5) of 5 primary MCL patient samples. Cleaved CYLD indicating MALT1 proteolytic activity was detectable in 4 of 5 patient samples. MYC expression was higher in these 4 samples with activated MALT1. The MCL cell line Z-138 was used as a positive control for MALT1 expression and as a negative control for CYLD cleavage. *Nonspecific band, which was not observed in any MALT1-activated MCL cell lines (Figure 1E). (D) Western blot analysis of MALT1 expression in MCL cell lines. MALT1 protein expression was detectable in all MCL cell lines. (E) Western blot analysis of different MALT1 targets. Cleaved forms of CYLD, RelB, A20, and BCL10 were detectable in Mino, Jeko-1, Rec-1, SP49, and SP53 cells. *Nonspecific band.
MALT1 functions as a protease; therefore, its proteolytic activity is determining its biologic function. To determine MALT1 activity in primary MCL samples, we prepared cell lysates from CD20+ MCL cells that were isolated from either PBMCs or cell suspensions from affected lymph nodes. MALT1 expression was found in all 5 primary MCL samples, whereas CYLD cleavage as a direct marker of MALT1 proteolytic activity22 was detectable in 4 out of 5 samples (Figure 1C).
To investigate MALT1 activity in additional MCLs, we analyzed 10 established cell lines. MALT1 was expressed in all lines assessed by western blotting (Figure 1D). Five cell lines (Mino, Jeko-1, Rec-1, SP49, and SP53) had detectable levels of cleaved forms of CYLD, RelB, A20, and BCL10, indicating constitutive MALT1 activity. In contrast, the other cell lines did not show cleavage of MALT1 targets, suggesting absent MALT1 proteolytic activity (Figure 1E). Collectively, these data implicate that MALT1 is constitutively active in a substantial number of MCLs and that MCLs can be divided into 2 distinct subgroups based on their MALT1 activation status.
Activation of MALT1 in MCL is caused by constitutive BCR signaling
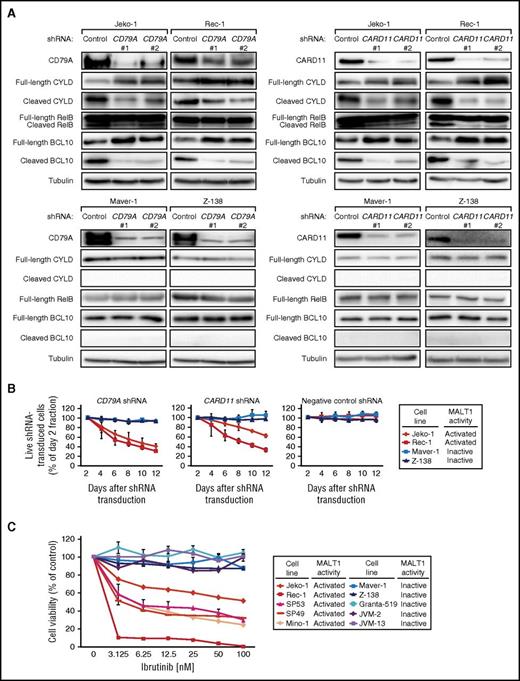
Next, we investigated the mechanisms leading to MALT1 activation in MCL. Because BCR signaling is an activator of MALT1, we knocked down CD79A and CARD11 as central components of the BCR cascade to investigate the effects on MALT1 activity. Cleavage of the MALT1 targets CYLD, RelB, and BCL10 was significantly decreased in 2 MALT1-activated cell lines (Jeko-1 and Rec-1), but unaffected in 2 MALT1-inactive MCL cell lines (Maver-1 and Z-138) after CD79A and CARD11 knockdown using specific shRNAs, respectively (Figure 2A). These results suggest that MALT1 is activated through constitutive BCR signaling in MCL. Interestingly, shRNA-mediated knockdown of CD79A, CARD11, or the CARD11-BCL10-MALT1 complex component BCL10 induced toxicity in Jeko-1 and Rec-1 cells, but not in Maver-1 and Z-138 cells, indicating dependency on BCR signaling only in MALT1-activated MCLs (Figure 2B; supplemental Figure 2A-B). These results were further confirmed by experiments showing that all MALT1-activated MCL cell lines were sensitive to the Bruton’s tyrosine kinase (BTK) inhibitor ibrutinib, whereas all MALT1-inactive models did not respond (Figure 2C).
Activation of MALT1 is caused by constitutive BCR signaling in MCL. (A) Western blot analysis of CD79A, CARD11, CYLD, RelB, and BCL10 following shRNA-mediated knockdown of CD79A and CARD11, respectively. Cleavage of CYLD, RelB, and BCL10 was significantly downregulated following CD79A or CARD11 knockdown, respectively, in MALT1-activated cell lines (Jeko-1 and Rec-1), whereas none of these cleaved forms was detectable in MALT1-inactive cell lines (Maver-1 and Z-138) and the expression levels of the corresponding full-length forms were not affected. (B) shRNA-mediated knockdown of CD79A and CARD11 was toxic to the MALT1-activated cell lines Jeko-1 and Rec-1. In contrast, the MALT1-inactive cell lines Maver-1 and Z-138 were unaffected by CD79A and CARD11 knockdown. A previously described, nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± standard deviations (SDs) of at least 3 independent experiments. (C) Cell viability of MCL cell lines after incubation with the BTK inhibitor ibrutinib. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs.
Activation of MALT1 is caused by constitutive BCR signaling in MCL. (A) Western blot analysis of CD79A, CARD11, CYLD, RelB, and BCL10 following shRNA-mediated knockdown of CD79A and CARD11, respectively. Cleavage of CYLD, RelB, and BCL10 was significantly downregulated following CD79A or CARD11 knockdown, respectively, in MALT1-activated cell lines (Jeko-1 and Rec-1), whereas none of these cleaved forms was detectable in MALT1-inactive cell lines (Maver-1 and Z-138) and the expression levels of the corresponding full-length forms were not affected. (B) shRNA-mediated knockdown of CD79A and CARD11 was toxic to the MALT1-activated cell lines Jeko-1 and Rec-1. In contrast, the MALT1-inactive cell lines Maver-1 and Z-138 were unaffected by CD79A and CARD11 knockdown. A previously described, nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± standard deviations (SDs) of at least 3 independent experiments. (C) Cell viability of MCL cell lines after incubation with the BTK inhibitor ibrutinib. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs.
Downregulation of MALT1 is toxic to MALT1-activated MCLs in vitro and in vivo
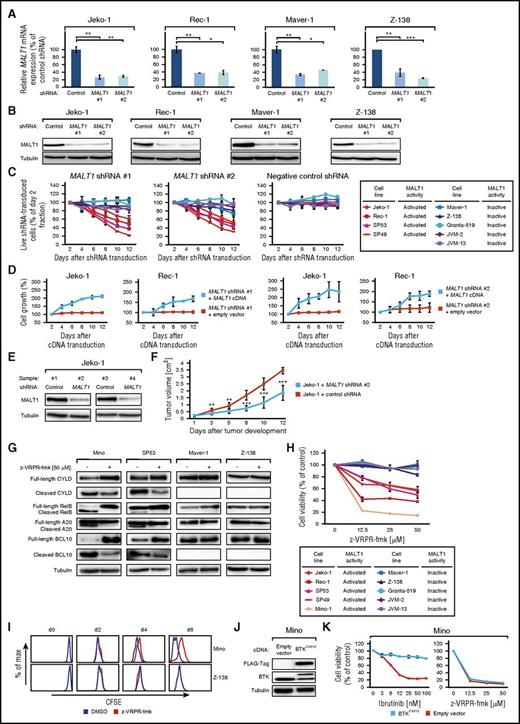
To elucidate the functional significance of MALT1 in MCL, we knocked down its expression using different MALT1-specific shRNAs. Both shRNAs significantly decreased MALT1 messenger RNA (mRNA) and protein levels after 48 hours (Figure 3A-B). Transduction of MALT1 shRNAs induced cytotoxicity in MALT1-activated MCLs, whereas it did not affect survival of any of the MALT1-inactive models (Figure 3C). To demonstrate that MALT1 shRNA-mediated toxicity was specifically caused by MALT1 knockdown, we performed a rescue experiment by transducing Jeko-1 and Rec-1 cells with a vector carrying the MALT1 complementary DNA (cDNA) that is not targetable by both MALT1 shRNAs. Indeed, exogenous MALT1 expression restored growth of both MALT1-activated MCL models, indicating the specificity of our approach (Figure 3D).
Subsets of MCLs are addicted to MALT1. (A) Effect of MALT1 shRNA #1 and #2 on MALT1 mRNA level in MALT1-activated (Jeko-1 and Rec-1) and MALT1-inactive (Maver-1 and Z-138) MCLs 48 hours after shRNA induction measured by quantitative PCR. MALT1 mRNA levels were normalized to expression of GAPDH. Error bars indicate SDs. (B) Effect of MALT1 shRNA #1 and #2 on MALT1 protein in MALT1-activated (Jeko-1 and Rec-1) and MALT1-inactive (Maver-1 and Z-138) MCLs 48 hours after shRNA induction measured by western blotting. (C) Effect of MALT1 knockdown by 2 independent shRNAs on viability of MCL cell lines. A previously described, nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± SDs of at least 3 independent experiments. (D) Rescue of Jeko-1 and Rec-1 cells from MALT1 shRNA-induced toxicity by exogenous expression of a MALT1 cDNA. Data are shown as means ± SDs of at least 3 independent experiments. (E) Western blot analysis of MALT1 knockdown in Jeko-1 mouse xenograft tumor biopsies from cells transduced with MALT1 shRNA #2 compared with control shRNA-transduced cells (shRNA against MSMO1). (F) Tumor growth curve of Jeko-1 xenograft mouse models that inducibly express MALT1 shRNA #2 (blue) or a control shRNA against MSMO1 (red). MALT1 knockdown significantly reduced in vivo tumor growth (P = 1.9 × 10−5, MALT1 shRNA vs control shRNA on day 12; 1-tailed 2-sample t test). Error bars indicate SDs. (G) Western blot analysis of MCL cell lines, treated with z-VRPR-fmk for 48 hours, for cleavage of CYLD, RelB, A20, and BCL10 in MALT1-activated MCL cell lines (Mino and SP53) vs MALT1-inactive MCLs (Maver-1 and Z-138). (H) Cell viability of MCL cell lines after incubation with the MALT1 inhibitor z-VRPR-fmk. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. (I) Carboxyfluorescein diacetate succinimidyl ester staining after treatment with z-VRPR-fmk or DMSO was measured on day 0 and after 2, 4, and 6 days. In Z-138 cells, no difference in cell proliferation was detectable (P = .4 on day 6). In contrast, Mino cells showed reduced proliferation after treatment with z-VRPR-fmk (P < 10−15 on day 6). Representative results from at least 3 independent replicates are shown. (J) Western blotting for FLAG and BTK following transduction of Mino cells with either a BTKC481S cDNA or an empty vector. (K) Determination of cell viability of Mino cells expressing either an empty vector (red) or a BTKC481S cDNA (blue) following treatment with ibrutinib or z-VRPR-fmk. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. *P < .05, **P < .01, ***P < .001.
Subsets of MCLs are addicted to MALT1. (A) Effect of MALT1 shRNA #1 and #2 on MALT1 mRNA level in MALT1-activated (Jeko-1 and Rec-1) and MALT1-inactive (Maver-1 and Z-138) MCLs 48 hours after shRNA induction measured by quantitative PCR. MALT1 mRNA levels were normalized to expression of GAPDH. Error bars indicate SDs. (B) Effect of MALT1 shRNA #1 and #2 on MALT1 protein in MALT1-activated (Jeko-1 and Rec-1) and MALT1-inactive (Maver-1 and Z-138) MCLs 48 hours after shRNA induction measured by western blotting. (C) Effect of MALT1 knockdown by 2 independent shRNAs on viability of MCL cell lines. A previously described, nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± SDs of at least 3 independent experiments. (D) Rescue of Jeko-1 and Rec-1 cells from MALT1 shRNA-induced toxicity by exogenous expression of a MALT1 cDNA. Data are shown as means ± SDs of at least 3 independent experiments. (E) Western blot analysis of MALT1 knockdown in Jeko-1 mouse xenograft tumor biopsies from cells transduced with MALT1 shRNA #2 compared with control shRNA-transduced cells (shRNA against MSMO1). (F) Tumor growth curve of Jeko-1 xenograft mouse models that inducibly express MALT1 shRNA #2 (blue) or a control shRNA against MSMO1 (red). MALT1 knockdown significantly reduced in vivo tumor growth (P = 1.9 × 10−5, MALT1 shRNA vs control shRNA on day 12; 1-tailed 2-sample t test). Error bars indicate SDs. (G) Western blot analysis of MCL cell lines, treated with z-VRPR-fmk for 48 hours, for cleavage of CYLD, RelB, A20, and BCL10 in MALT1-activated MCL cell lines (Mino and SP53) vs MALT1-inactive MCLs (Maver-1 and Z-138). (H) Cell viability of MCL cell lines after incubation with the MALT1 inhibitor z-VRPR-fmk. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. (I) Carboxyfluorescein diacetate succinimidyl ester staining after treatment with z-VRPR-fmk or DMSO was measured on day 0 and after 2, 4, and 6 days. In Z-138 cells, no difference in cell proliferation was detectable (P = .4 on day 6). In contrast, Mino cells showed reduced proliferation after treatment with z-VRPR-fmk (P < 10−15 on day 6). Representative results from at least 3 independent replicates are shown. (J) Western blotting for FLAG and BTK following transduction of Mino cells with either a BTKC481S cDNA or an empty vector. (K) Determination of cell viability of Mino cells expressing either an empty vector (red) or a BTKC481S cDNA (blue) following treatment with ibrutinib or z-VRPR-fmk. Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. *P < .05, **P < .01, ***P < .001.
We next determined if MALT1 dependency in MALT1-activated MCLs translates into an in vivo setting. To this end, we created MCL xenograft mouse models using Jeko-1 cells transduced with vectors encoding either MALT1 shRNA #2 or a negative control shRNA. shRNA-mediated MALT1 knockdown was detectable by western blotting in different samples from euthanized mice (Figure 3E). MALT1 knockdown significantly inhibited tumor growth over 12 days (P = 1.9 × 10−5 for MALT1 shRNA vs control shRNA on day 12; 1-tailed 2-sample t test; Figure 3F), suggesting that MALT1 promotes lymphoma growth in MALT1-activated MCLs.
Next, we asked if signaling through MALT1 can be used therapeutically in MCL. Thus, we treated our MCL lines with the specific MALT1 inhibitor z-VRPR-fmk.23 To confirm that z-VRPR-fmk indeed exerts its effect through inhibiting MALT1’s proteolytic activity, expression of the MALT1 targets CYLD, RelB, A20, and BCL10 were studied 48 hours after incubation with z-VRPR-fmk by immunoblotting. We detected a significant downregulation of their cleaved forms in MALT1-activated MCLs (Mino and SP53). In contrast, no changes in their expression levels were observed in the MALT1-inactive MCLs (Maver-1 and Z-138) (Figure 3G). These results confirm that z-VRPR-fmk inhibits the proteolytic function of MALT1. Subsequently, we determined cell viability 7 days after z-VRPR-fmk treatment. In line with our MALT1 knockdown data, pharmacologic inhibition of MALT1 significantly reduced cell viability of MALT1-activated MCLs, whereas survival of MALT1-inactive MCLs was not affected (Figure 3H).
To obtain insights into the nature of the growth inhibitory effects of MALT1 inhibition through z-VRPR-fmk, we measured cell proliferation and the rate of apoptosis and performed cell cycle analyses. We treated MALT1-activated (Mino) and MALT1-inactive (Z-138) MCLs with z-VRPR-fmk or DMSO. To assess proliferation, cellular divisions were determined by measuring carboxyfluorescein diacetate succinimidyl ester dilutions in viable cells by flow cytometry. z-VRPR-fmk significantly downregulated proliferation of Mino cells (P < 10−15 on day 6), whereas Z-138 cells were not affected (P = .4 on day 6; Figure 3I). In addition, we quantified the number of cell divisions following treatment with z-VRPR-fmk. These analyses revealed that Mino cells divided 1.8 ± 0.17 or 0.9 ± 0.1 ×/day after DMSO or z-VRPR-fmk treatment, respectively, whereas Z-138 cells divided 2.6 ± 0.03 ×/day after DMSO treatment and 2.5 ± 0.04 ×/day following MALT1 inhibition. In contrast, neither changes in apoptosis (data not shown) nor cell cycle (supplemental Figure 3) were detectable following MALT1 inhibition, indicating that z-VRPR-fmk exerts its growth inhibitory effect in MALT1-activated MCLs predominantly through reduction of cell proliferation (Figure 3I).
Inhibition of MALT1 overcomes ibrutinib resistance
The BTK inhibitor ibrutinib is effective in the treatment of relapsed/refractory MCL patients.37 A recent study identified that the BTKC481S mutation confers resistance to ibrutinib in MCL.38 To investigate whether inhibition of MALT1 is able to overcome BTKC481S-induced ibrutinib resistance, we expressed a BTKC481S cDNA or an empty vector in all 5 MALT1-activated and 2 MALT1-inactive MCL lines (Figure 3J; supplemental Figure 4A,C,E,G,I,K) and subsequently treated these cells with ibrutinib or z-VRPR-fmk. Introduction of the BTKC481S mutation rescued all MALT1-activated lines from the toxic effect of ibrutinib (Figure 3K; supplemental Figure 4B,D,F,H). In contrast, in MALT1-inactive MCLs, transduction of the BTKC481S mutant or an empty vector did not alter sensitivity to ibrutinib or z-VRPR-fmk (Figures 2C and 3H; supplemental Figure 4J,L). These data indicate that inhibition of MALT1 might be effective in ibrutinib-resistant MCLs.
MALT1 regulates MYC expression in MCL
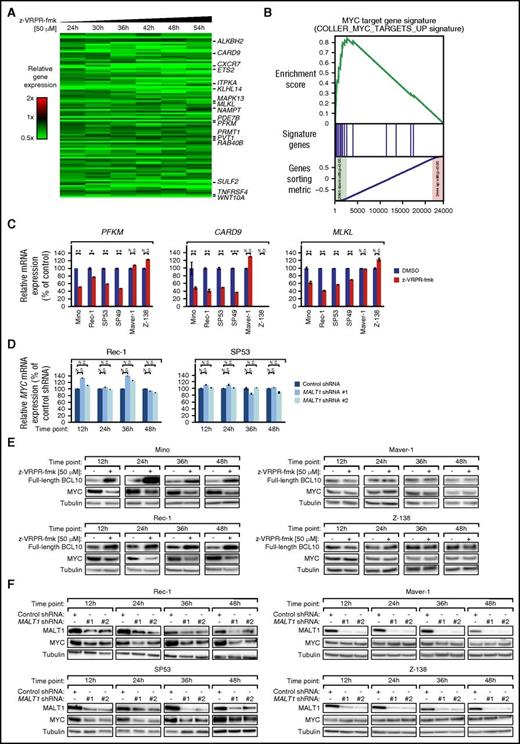
To understand which biologic processes are regulated by MALT1 in MCL, we profiled gene expression changes after 24, 30, 36, 42, 48, and 54 hours of z-VRPR-fmk treatment in Mino cells. We identified 93 genes that were significantly downregulated (P ≤ 1 × 10−5; paired t tests over all time points) and 126 genes significantly upregulated (P ≤ 1 × 10−5) following pharmacologic MALT1 inhibition (Figure 4A; supplemental Figure 5A; supplemental Table 2).
MALT1 regulates the gene expression network of MYC in MCL. (A) Gene expression profiling following pharmacologic inhibition of the proteolytic MALT1 activity using z-VRPR-fmk vs DMSO in Mino cells. Changes of gene expression were profiled at the indicated time points following treatment with z-VRPR-fmk. Each time point depicted the mean of log2-transformed expression ratios for 2 replicates. Gene expression changes were depicted according to the color scale shown. Genes that are involved in critical biological processes are highlighted. (B) Gene set enrichment analysis of a previously described MYC gene expression signature. The MYC signature was significantly enriched with genes that are downregulated following pharmacologic MALT1 inhibition using z-VRPR-fmk in Mino cells. (C) Expression levels of MALT1 target genes in MALT1-activated and MALT1-inactive MCL cell lines determined by quantitative PCR. mRNA levels of PFKM, CARD9, and MLKL were normalized to expression of GAPDH. Error bars indicate SDs. (D) MYC mRNA levels in Rec-1 and SP53 cells following shRNA-mediated knockdown of MALT1 as measured by quantitative PCR. MYC mRNA levels were normalized to expression of GAPDH. Error bars indicate SDs. (E) Treatment with z-VRPR-fmk downregulated MYC protein in the MALT1-activated MCL cell lines Mino and Rec-1. In contrast, in the MALT1-inactive cell lines Maver-1 and Z-138, MYC was not affected by inhibition of MALT1 activity. Accumulation of full-length BCL10 in MALT1-activated MCL models after treatment with z-VRPR-fmk was used as a surrogate marker of MALT1 inhibition. (F) MALT1 shRNA #1 and #2 downregulated MYC protein in MALT1-activated MCLs (Rec-1 and SP53), but not in MALT1-inactive MCLs (Maver-1 and Z-138) at the indicated time points after shRNA induction as measured by western blotting. N.D., not detectable; N.S., not significant. *P < .05, **P < .01, ***P < .001.
MALT1 regulates the gene expression network of MYC in MCL. (A) Gene expression profiling following pharmacologic inhibition of the proteolytic MALT1 activity using z-VRPR-fmk vs DMSO in Mino cells. Changes of gene expression were profiled at the indicated time points following treatment with z-VRPR-fmk. Each time point depicted the mean of log2-transformed expression ratios for 2 replicates. Gene expression changes were depicted according to the color scale shown. Genes that are involved in critical biological processes are highlighted. (B) Gene set enrichment analysis of a previously described MYC gene expression signature. The MYC signature was significantly enriched with genes that are downregulated following pharmacologic MALT1 inhibition using z-VRPR-fmk in Mino cells. (C) Expression levels of MALT1 target genes in MALT1-activated and MALT1-inactive MCL cell lines determined by quantitative PCR. mRNA levels of PFKM, CARD9, and MLKL were normalized to expression of GAPDH. Error bars indicate SDs. (D) MYC mRNA levels in Rec-1 and SP53 cells following shRNA-mediated knockdown of MALT1 as measured by quantitative PCR. MYC mRNA levels were normalized to expression of GAPDH. Error bars indicate SDs. (E) Treatment with z-VRPR-fmk downregulated MYC protein in the MALT1-activated MCL cell lines Mino and Rec-1. In contrast, in the MALT1-inactive cell lines Maver-1 and Z-138, MYC was not affected by inhibition of MALT1 activity. Accumulation of full-length BCL10 in MALT1-activated MCL models after treatment with z-VRPR-fmk was used as a surrogate marker of MALT1 inhibition. (F) MALT1 shRNA #1 and #2 downregulated MYC protein in MALT1-activated MCLs (Rec-1 and SP53), but not in MALT1-inactive MCLs (Maver-1 and Z-138) at the indicated time points after shRNA induction as measured by western blotting. N.D., not detectable; N.S., not significant. *P < .05, **P < .01, ***P < .001.
To analyze the gene expression data in an unbiased manner, we performed a gene set enrichment analysis using a previously described gene expression signatures database consisting of 13 593 signatures (supplemental Table 3). Our analysis revealed that the second most enriched downregulated signature was a previously described MYC target gene set (enrichment score = 0.841; P ≤ .001; Figure 4B; supplemental Figure 5B; supplemental Table 3). In addition, various other independent MYC signatures were significantly enriched with downregulated genes and among the top downregulated signatures, suggesting that MYC expression and its gene expression network is regulated by MALT1 (supplemental Tables 3 and 4).
To confirm that MYC deregulation by MALT1 is a general mechanism in MCL, we further performed gene expression profiling in Rec-1 cells (supplemental Figure 6A-B; supplemental Table 5). These analyses confirmed that various previously identified MYC target sets were downregulated following z-VRPR-fmk treatment (supplemental Figure 6C-D; supplemental Tables 6 and 7). Additionally, the identified Mino target gene signature was significantly downregulated in Rec-1 following MALT1 inhibition (supplemental Figure 6E), suggesting that very similar target genes are affected by MALT1 inhibition in Mino and Rec-1 cells. Finally, to confirm that the detected Mino target gene signature is also downregulated in other MCL models, we performed real-time PCR for 11 selected genes following MALT1 inhibition in both MALT1-activated and MALT1-inactive cell lines. Real-time PCR confirmed downregulation of 10 out of 11 target genes in all MALT1-activated but not MALT-inactive models (Figure 4C; supplemental Figure 7). These results suggest that the MYC target gene network seems to be controlled by MALT1 in MCL. In contrast, previously identified NF-κB target gene signatures were not affected by MALT1 inhibition in both Mino and Rec-1 cells.
Because of the strong impact of MALT1 inhibition on the MYC expression profile, we asked whether MYC itself is regulated by MALT1. MYC mRNA expression was moderately suppressed by MALT1 inhibition (log2[ratio] = −0.33 in Mino and log2[ratio] = −0.23 in Rec-1). This result was confirmed by shRNA-mediated MALT1 knockdown that did not substantially alter MYC mRNA levels measured by quantitative PCR (Figure 4D). To elucidate if MYC is regulated posttranscriptionally by MALT1, we evaluated MYC protein expression after z-VRPR-fmk treatment. Immunoblotting revealed that MYC levels were reduced in MALT1-activated but not in MALT1-inactive MCLs following MALT1 inhibition (Figure 4E).
To corroborate these findings, we next investigated MYC expression levels following shRNA-mediated MALT1 knockdown in MALT1-activated and MALT1-inactive cells by western blotting. MALT1 silencing induced a substantial decrease in MYC protein levels in MALT1-activated, but not MALT1-inactive, MCLs (Figure 4F). Because MALT1 is activated by BCR signaling in MCL, we investigated whether inhibition of BCR signaling by ibrutinib alters MYC expression levels. To this end, we treated MALT1-activated (Mino and Rec-1) and MALT1-inactive (Maver-1 and Z-138) cell lines with 5 and 10 nM ibrutinib for 12, 24, 36, and 48 hours. Ibrutinib treatment significantly decreased MYC expression in Mino and Rec-1 cells, whereas MYC levels in Maver-1 and Z-138 were unaffected (supplemental Figure 8). Collectively, these data indicate that BCR-driven MALT1 activity regulates MYC expression.
MALT1 regulates MYC expression in primary mouse splenocytes
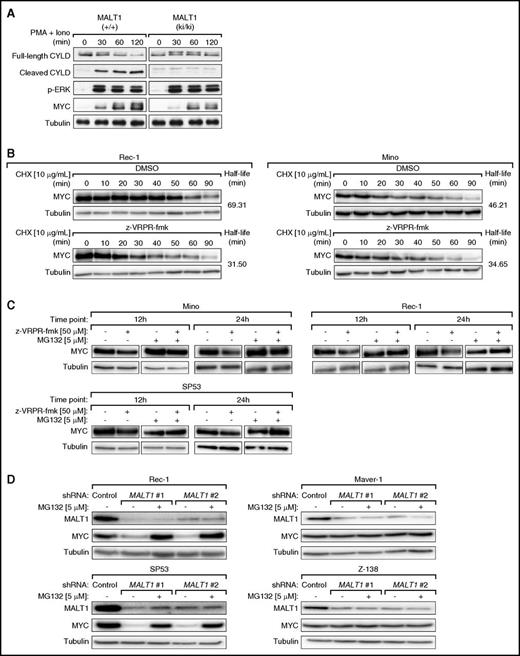
To elucidate if MALT1-induced regulation of MYC protein expression is relevant in other settings than MCL, we isolated primary mouse splenocytes expressing either wild-type MALT1 (+/+) or a catalytically inactive MALT1C472A mutant (ki/ki)39 and subsequently stimulated the splenocytes with phorbol myristate acetate and ionomycin for 30, 60, and 120 minutes (Figure 5A). Stimulation of MALT1 (+/+) and MALT1 (ki/ki) splenocytes was equally strong, as determined by monitoring the induction of extracellular signal-regulated kinase phosphorylation (Figure 5A). In contrast, CYLD cleavage, which served as a marker of MALT1 activity, was only detectable in MALT1 (+/+) splenocytes. Likewise, the stimulation-induced expression of MYC was considerably stronger in MALT1 (+/+) compared with MALT1 (ki/ki) splenocytes. Collectively, these findings suggest that MALT1 activity promotes MYC expression in activated primary lymphocytes (Figure 5A).
MYC is stabilized by MALT1 function. (A) Primary mouse splenocytes expressing either wild-type MALT1 (+/+) or a catalytically inactive MALT1 mutant (ki/ki) were stimulated with phorbol myristate acetate (PMA) and ionomycin for the indicated time points. Stimulation efficiency and MALT1 activation were assessed by western blotting using anti-p-extracellular signal-regulated kinase (p-ERK) and anti-CYLD antibodies, respectively. (B) Rec-1 and Mino cells were first treated with z-VRPR-fmk or DMSO for 24 hours and subsequently with cycloheximide (CHX). MYC protein expression was assessed by western blot using samples collected at the indicated time points. In both cell lines, MALT1 inhibition resulted in a reduced half-life of MYC protein. (C) Mino, Rec-1, and SP53 cells were treated with z-VRPR-fmk or DMSO and subsequently with MG132 or DMSO. MYC protein levels were increased by MG132 treatment as evaluated by western blotting. (D) In Rec-1, SP53, Maver-1, and Z-138 cells, either a control shRNA against MSMO1 or 1 of the 2 MALT1 shRNAs were induced with doxycycline for 24 hours. Subsequently, cells were treated with MG132 or DMSO. MYC protein levels were increased by MG132 treatment in MALT1-activated MCLs (Rec-1 and SP53), but not in MALT1-inactive MCLs (Maver-1 and Z-138) as evaluated by western blotting.
MYC is stabilized by MALT1 function. (A) Primary mouse splenocytes expressing either wild-type MALT1 (+/+) or a catalytically inactive MALT1 mutant (ki/ki) were stimulated with phorbol myristate acetate (PMA) and ionomycin for the indicated time points. Stimulation efficiency and MALT1 activation were assessed by western blotting using anti-p-extracellular signal-regulated kinase (p-ERK) and anti-CYLD antibodies, respectively. (B) Rec-1 and Mino cells were first treated with z-VRPR-fmk or DMSO for 24 hours and subsequently with cycloheximide (CHX). MYC protein expression was assessed by western blot using samples collected at the indicated time points. In both cell lines, MALT1 inhibition resulted in a reduced half-life of MYC protein. (C) Mino, Rec-1, and SP53 cells were treated with z-VRPR-fmk or DMSO and subsequently with MG132 or DMSO. MYC protein levels were increased by MG132 treatment as evaluated by western blotting. (D) In Rec-1, SP53, Maver-1, and Z-138 cells, either a control shRNA against MSMO1 or 1 of the 2 MALT1 shRNAs were induced with doxycycline for 24 hours. Subsequently, cells were treated with MG132 or DMSO. MYC protein levels were increased by MG132 treatment in MALT1-activated MCLs (Rec-1 and SP53), but not in MALT1-inactive MCLs (Maver-1 and Z-138) as evaluated by western blotting.
MALT1 stabilizes MYC expression
Next, we investigated whether MALT1 promotes MYC expression by controlling MYC protein stability. We treated Rec-1 and Mino cells with 50 μM z-VRPR-fmk or DMSO for 24 hours, followed by incubation with 10 μg/mL of the protein synthesis inhibitor cycloheximide. Immunoblotting revealed that MYC levels in Rec-1 cells declined faster following z-VRPR-fmk treatment (half-life, 31.5 minutes) compared with DMSO (69.31 minutes). This was confirmed in Mino cells, in which the half-life of MYC decreased from 46.21 minutes in DMSO-treated cells compared with 34.65 minutes in MALT1-inhibited cells (Figure 5B; supplemental Figure 9A).
Next, to decipher if MALT1 affected proteasomal degradation of MYC, we assessed MYC levels in Mino, Rec-1, and SP53 cells that were incubated with z-VRPR-fmk or DMSO for 12 hours and 24 hours, respectively, followed by treatment with the proteasome inhibitor MG132 for 2 hours. Under these conditions, we detected a marked increase of MYC expression in z-VRPR-fmk and MG132-treated cells (Figure 5C; supplemental Figure 9B). To confirm our MALT1 inhibitor data, we transduced MALT1-activated (Rec-1 and SP53) and MALT1-inactive (Maver-1 and Z-138) MCLs with our 2 MALT1 shRNAs and treated these cells with either DMSO or MG132. MG132 treatment significantly increased MYC expression levels following shRNA-mediated MALT1 knockdown in MALT1-activated MCLs (Figure 5D). Collectively, these results indicate that MALT1 increases MYC stability posttranslationally by preventing its proteasomal degradation.
MCLs depend on MYC signaling
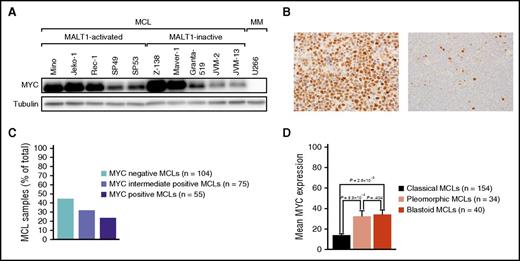
Our analyses implicated that MALT1 regulates MYC expression. To validate these findings, we determined MYC expression in our cell lines and in MCL patient samples. All MCL cell lines expressed MYC protein by western blotting irrespective of their MALT1 activation status (Figure 6A; supplemental Figure 10), whereas the 4 MALT1-activated primary MCLs showed higher MYC expression levels compared with the MALT1-inactive specimen (Figure 1C).
MYC expression in MCL. (A) Western blot analysis of MYC expression in 10 MCL cell lines and in the MM cell line U266. All MCL cell lines had detectable MYC expression compared with the negative control U266. (B) Immunohistochemical MYC staining of an MYC-positive MCL case (left: original magnification ×400) and an MYC-negative MCL case (right: original magnification ×400). Images were captured using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP73 camera and were processed with Olympus cellSens software. (C) Frequency of MYC expression in MCL. (D) Mean MYC expression in cytological MCL variants. MYC expression was significantly higher in pleomorphic and blastoid variants compared with classical MCLs. Error bars indicate the standard error of the mean.
MYC expression in MCL. (A) Western blot analysis of MYC expression in 10 MCL cell lines and in the MM cell line U266. All MCL cell lines had detectable MYC expression compared with the negative control U266. (B) Immunohistochemical MYC staining of an MYC-positive MCL case (left: original magnification ×400) and an MYC-negative MCL case (right: original magnification ×400). Images were captured using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP73 camera and were processed with Olympus cellSens software. (C) Frequency of MYC expression in MCL. (D) Mean MYC expression in cytological MCL variants. MYC expression was significantly higher in pleomorphic and blastoid variants compared with classical MCLs. Error bars indicate the standard error of the mean.
Next, we determined MYC expression in 234 primary MCL samples. A total of 104 (44.4%) samples did not show MYC expression. In contrast, 75 (32.1%) samples displayed an intermediate and 55 (23.5%) samples a high MYC positivity (Figure 6B-C). To compare the MYC staining pattern in MCL to other lymphomas, we stained 93 primary DLBCLs (10 MYC rearranged) and 7 Burkitt lymphomas (BLs) (all MYC rearranged). In BLs, nuclear positivity was strong in >90% of cells. Eight of 10 MYC-rearranged DLBCLs expressed MYC, whereas 52 of 83 primary nonrearranged DLBCLs were MYC-positive. In general, primary DLBCLs and MCLs were similar with respect to their staining intensity and more variable compared with the BL cases (supplemental Figure 11).
To investigate if MYC expression correlates with the cytological subtype, we compared MYC expression in classical (n = 154), pleomorphic (n = 34), and blastoid (n = 40) MCLs (Figure 6D). The cytological subtype was not available for 6 samples. MYC expression was significantly higher in pleomorphic (P = 9.9 × 10−4) and blastoid variants (P = 2.6 × 10−5) compared with classical MCLs. There was no difference in MYC expression between pleomorphic and blastoid MCLs (P = .4; Figure 6D).
To rule out that genetic aberrations involving the MYC locus are causative for increased MYC expression, we performed FISH in 80 MCLs with available MYC expression data: 55 (69%) of these cases were classical, 6 (8%) pleomorphic, and 13 (16%) had blastoid MCLs (the cytological subtype was not available for 6 cases). Forty (50%) cases did not express MYC, 28 (35%) had intermediate MYC expression, whereas 12 (15%) samples had high MYC expression. Of the 80 cases, only 1 (1.3%) harbored a MYC translocation, whereas none of the cases showed a high-level MYC amplification, indicating that these genetic aberrations are extremely rare in MCL.
To elucidate the functional role of MYC expression in MCL, we transduced MCL cell lines with specific MYC shRNAs, which induced MYC downregulation 48 hours after induction (supplemental Figure 12A). MYC knockdown was lethal to all MCL models (Figure 7A). To confirm the specificity of our approach, an exogenous MYC cDNA (which is not targeted by MYC shRNA #2) was introduced in Jeko-1, Rec-1, and SP53 cells before transduction with the MYC shRNA. Indeed, exogenous MYC expression rescued all MCL cells from shRNA-mediated toxicity (Figure 7B).
MCLs depend on MYC signaling. (A) shRNA-mediated MYC knockdown induced cytotoxicity in MCL cell lines. A previously described nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± SDs of at least 3 independent experiments. (B) Expression of a MYC cDNA rescued Jeko-1, Rec-1, and SP53 cells transduced with MYC shRNA #2 (targeting the 3′UTR of MYC) from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (C) Expression of a MYC cDNA partially rescued Jeko-1, Rec-1, and SP53 cells transduced with MALT1 shRNA #1 from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (D) Expression of an MYC cDNA partially rescued Jeko-1, Rec-1, SP53, and Mino cells treated with z-VRPR-fmk from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (E) shRNA-mediated knockdown of MYC significantly downregulated cell proliferation. Data are shown as means ± SDs of at least 2 independent experiments. (F) Correlation of Ki-67 and MYC expression determined by immunohistochemistry. Error bars indicate the standard error of the mean. (G) Viability of MCL cell lines following MYC inhibition using the small molecule inhibitor 10058-F4 that inhibits MYC-MAX heterodimerization. Baseline MYC expression was assessed by western blotting (Figure 6A). Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. **P < .01, ***P < .001.
MCLs depend on MYC signaling. (A) shRNA-mediated MYC knockdown induced cytotoxicity in MCL cell lines. A previously described nontoxic shRNA against MSMO1 did not induce toxicity in any cell line. Data are shown as means ± SDs of at least 3 independent experiments. (B) Expression of a MYC cDNA rescued Jeko-1, Rec-1, and SP53 cells transduced with MYC shRNA #2 (targeting the 3′UTR of MYC) from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (C) Expression of a MYC cDNA partially rescued Jeko-1, Rec-1, and SP53 cells transduced with MALT1 shRNA #1 from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (D) Expression of an MYC cDNA partially rescued Jeko-1, Rec-1, SP53, and Mino cells treated with z-VRPR-fmk from toxicity. Data are shown as means ± SDs of at least 2 independent experiments. (E) shRNA-mediated knockdown of MYC significantly downregulated cell proliferation. Data are shown as means ± SDs of at least 2 independent experiments. (F) Correlation of Ki-67 and MYC expression determined by immunohistochemistry. Error bars indicate the standard error of the mean. (G) Viability of MCL cell lines following MYC inhibition using the small molecule inhibitor 10058-F4 that inhibits MYC-MAX heterodimerization. Baseline MYC expression was assessed by western blotting (Figure 6A). Representative results from at least 3 independent replicates are shown. Error bars indicate SDs. **P < .01, ***P < .001.
To evaluate the degree to which MYC downregulation contributes to the impaired viability of MALT1-silenced cells, we performed a rescue experiment introducing an MYC cDNA or an empty vector into MALT1 shRNA-transduced Jeko-1, Rec-1, and SP53 cells. We detected a partial MYC-induced rescue in all 3 cell lines, suggesting that MYC knockdown, at least partially, contributes to the lethal effect of MALT1 silencing in these cells (Figure 7C; supplemental Figure 12B). To confirm these results, we treated Jeko-1, Rec-1, SP53, and Mino cells that expressed either MYC cDNA or an empty vector with z-VRPR-fmk. In all cell lines, we could detect a substantial MYC-induced rescue confirming that MYC downregulation is at least partially causative for the toxic effects of MALT1 inhibition in MALT1-activated MCLs (Figure 7D). To investigate whether this rescue effect is specific to z-VRPR-fmk, we determined the cell viability of these exogenous MYC harboring cells after doxorubicin treatment. No resistance against doxorubicin was conferred by MYC cDNA expression, indicating the specificity of our findings (supplemental Figure 12C).
To obtain insights into the nature of the growth inhibitory effects of MYC knockdown, we analyzed whether cell proliferation was negatively affected by MYC silencing. To this end, SNARF-1 staining was performed in Rec-1 and SP53 cells expressing MYC shRNA #1/#2. Cell divisions were compared after 2 days between cells with and without MYC knockdown. In both cell lines, the proliferation rate decreased substantially after MYC silencing (Figure 7E), indicating that MYC controls MCL proliferation.
To validate our in vitro findings, we stained our cohort of primary MCL samples for Ki-67 to assess proliferation. Indeed, overall Ki-67 and MYC expression correlated (r = .63; P = 5 × 10−27; supplemental Figure 12D). Furthermore, samples with intermediate MYC expression had higher Ki-67 levels compared with MYC-negative MCLs (P = 1.1 × 10−6; Figure 7F), whereas MYC-positive MCLs had the highest Ki-67 levels (P = 4.6 × 10−24 vs MYC-negative MCLs and P = 1.4 × 10−12 vs MYC-intermediate MCLs; Figure 7F). This suggests that MYC regulates MCL proliferation in vivo.
Finally, we investigated if inhibiting MYC signaling can be exploited therapeutically. Cell viability was measured 3 days after treating MCL lines with the small molecule inhibitor 10058-F4, which inhibits MYC-MAX heterodimerization. U266 cells that do not express MYC were used as a negative control. Irrespective of MALT1 activity, 3 cell lines (Mino, Rec-1, and Z-138) were highly sensitive to 10058-F4. In contrast, U266 cells were virtually unaffected, whereas Maver-1 cells showed an intermediate sensitivity (Figure 7G). Taken together, these data suggest that MYC represents a promising target for future therapies in MCL patients.
Discussion
We detected a novel role of MALT1 in the biology of MCL. A substantial fraction of MCLs exhibit constitutive MALT1 activity, and these MCLs are addicted to MALT1 function. In contrast, some MCLs do not show constitutive MALT1 proteolytic activity, and these lymphomas do not depend on MALT1. Thus, our results indicate that MCLs can be divided into 2 distinct subgroups based on their MALT1 activation status.
MALT1 activity seems to be caused by constitutive BCR signaling. Recent work showed that activity of BCR signaling is correlated with increased MCL proliferation.12 It seems conceivable that this could be caused by MALT1-induced upregulation of the oncogenic transcription factor MYC. MYC is crucial for the regulation of cell proliferation.40 In line with this, pharmacologic MALT1 inhibition or shRNA-mediated MYC knockdown significantly decreased MCL proliferation. Moreover, Ki-67 expression as a marker for cell proliferation was significantly higher in primary MCL samples with MYC expression. MYC expression was detectable in more than 55% of primary MCLs, indicating that MYC is frequently expressed in MCL. In our series of primary samples, common genetic aberrations such as MYC locus translocations or high-level amplifications that can cause upregulation of MYC were extremely rare, confirming the results of previous studies.41,42 However, given that MYC expression and MALT1 activation status did not correlate in all cell lines, additional molecular mechanisms regulate MYC expression in MCL besides MALT1 activity.
Recent work in chronic lymphocytic leukemia has linked BCR signaling to upregulation of MYC expression, as anti-immunoglobulin M–induced BCR signaling increased translation of MYC mRNA in primary chronic lymphocytic leukemia cells.43 However, the exact molecular mechanisms how BCR signaling and MYC expression are linked were not elucidated. Our data using primary splenocytes suggest that MALT1-driven MYC expression is not restricted to MCL, but seems to be a common mechanism of MYC regulation. MALT1 seems to regulate MYC expression through different mechanisms. We detected a very moderate MYC downregulation on mRNA levels following MALT1 knockdown or pharmacologic inhibition of MALT1. However, the predominant mechanism of MYC regulation involves control of MYC stability.
Interestingly, our gene expression data following MALT1 inhibition failed to show downregulation of previously identified NF-κB gene sets, including signatures defined in MCL.12 This finding is surprising given the role of MALT1 in activating NF-κB signaling. The protease activity of MALT1 is dispensable for initial NF-κB activation and instead promotes NF-κB signaling through cleavage of the negative NF-κB regulators RelB and A20.15,16,44 The mechanisms why downregulation of NF-κB was not detectable by our transcriptome analyses are unclear and should be addressed in future studies.
Finally, our work revealed that the MALT1-MYC network could be exploited therapeutically in MCL patients. Despite improvements in therapy, MCL remains an incurable disease.1 MALT1 inhibition was highly effective in all MALT1-activated models. These data warrant future clinical trials with MALT1 inhibitors in MCL patients with constitutive MALT1 activity. Moreover, MALT1 inhibition was able to overcome BTKC481S-induced ibrutinib resistance and might represent a novel therapeutic option for patients failing ibrutinib therapy. Similarly, MYC inhibition was lethal to MYC expressing models. These data suggest that MYC inhibition offers a promising target and a novel therapeutic strategy to overcome therapy resistance in MCL patients.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE81552).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank André Weilemann for technical support.
This work was supported by research grants from the Deutsche Forschungsgemeinschaft (G.L.); the Else Kröner-Fresenius-Stiftung (G.L.); the Brigitte und Konstanze Wegener-Stiftung (G.L.); the Deutsche Krebshilfe (G.L.); the Swiss National Science Foundation (Sinergia grant) (G.L.); the Deutsche Forschungsgemeinschaft EXC 1003 Cells in Motion–Cluster of Excellence, Münster, Germany (G.L.); the Ministry of Health of the Czech Republic (grant AZV 15-27757A) (P.K.); the e:BIO project Molecular Mechanisms in Malignant Lymphomas with MYC Deregulation (MMML-MYC-Sys) (R.S. and W.K.); the German Ministry for Education and Research (grants 0316166B and 0316166I) (R.S. and W.K.); the Robert Bosch Foundation, Stuttgart, Germany (A.M.S., E. Höring, and G.O.); and the Wilhelm Sander-Stiftung (grant 2012.075.2) (D.K.).
Authorship
Contribution: B.D. designed research, performed experiments, analyzed data, and wrote the manuscript; M.G. performed bioinformatic and biophysical analyses; M.J., P.K., E. Höring, J.M., G.S., S.M.A., E. Hoster, N.V., A.M.S., T.E., W.X., K.E., N.D., and H.M. performed and analyzed experiments; W.E.B., M.T., M.D., K.J., P.L., A.R., R.S., A.T., W.K., I.A., D.K., G.O., and M.T. analyzed data; and G.L. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no conflict of interest.
Correspondence: Georg Lenz, Translational Oncology, University Hospital Münster, Albert-Schweitzer-Campus 1, 48149 Münster, Germany; e-mail: georg.lenz@ukmuenster.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal