Key Points
Mutation profiling has a high predictive value for identifying individuals with, or at high risk of developing, a myeloid neoplasm.
Patients with clonal cytopenia have a significantly higher risk of developing a myeloid neoplasm than those with no evidence of clonality.
Unexplained blood cytopenias, in particular anemia, are often found in older persons. The relationship between these cytopenias and myeloid neoplasms like myelodysplastic syndromes is currently poorly defined. We studied a prospective cohort of patients with unexplained cytopenia with the aim to estimate the predictive value of somatic mutations for identifying subjects with, or at risk of, developing a myeloid neoplasm. The study included a learning cohort of 683 consecutive patients investigated for unexplained cytopenia, and a validation cohort of 190 patients referred for suspected myeloid neoplasm. Using granulocyte DNA, we looked for somatic mutations in 40 genes that are recurrently mutated in myeloid malignancies. Overall, 435/683 patients carried a somatic mutation in at least 1 of these genes. Carrying a somatic mutation with a variant allele frequency ≥0.10, or carrying 2 or more mutations, had a positive predictive value for diagnosis of myeloid neoplasm equal to 0.86 and 0.88, respectively. Spliceosome gene mutations and comutation patterns involving TET2, DNMT3A, or ASXL1 had positive predictive values for myeloid neoplasm ranging from 0.86 to 1.0. Within subjects with inconclusive diagnostic findings, carrying 1 or more somatic mutations was associated with a high probability of developing a myeloid neoplasm during follow-up (hazard ratio = 13.9, P < .001). The predictive values of mutation analysis were confirmed in the independent validation cohort. The findings of this study indicate that mutation analysis on peripheral blood granulocytes may significantly improve the current diagnostic approach to unexplained cytopenia and more generally the diagnostic accuracy of myeloid neoplasms.
Introduction
The term unexplained cytopenia is used to define a condition characterized by peripheral blood cytopenia whose origin is not attributable to causes detectable with conventional tests or to any concomitant diseases. The most common cytopenia is anemia, which represents a major clinical problem in elderly individuals.1 Overall, the prevalence of anemia in the general population increases sharply after the age of 50, reaching 20% or more in individuals aged 85 years or older.2,3 A significant association has been reported between anemia and risk for all-cause hospitalization and mortality.4,5 In up to one-third of cases, the etiology of anemia remains unexplained, and several potential mechanisms have been proposed, including myelodysplasia.1,-3,6
The myelodysplastic syndromes (MDS) are clonal disorders of hematopoiesis characterized by peripheral blood cytopenia, dysplasia in 1 or more myeloid cell lines, and a propensity to evolve into acute myeloid leukemia.7,8 The incidence of these disorders increases to up 50 per 100 000 persons per year over the age of 60, making MDS one of the most common hematologic malignancies in Western countries. Recent data suggest that this incidence may be four- to fivefold underestimated, because many MDS patients may remain underdiagnosed as anemia or cytopenia of the elderly.9,10
The diagnostic approach to MDS includes morphological studies of peripheral blood and bone marrow aspirate smears; bone marrow biopsy to assess marrow cellularity and topography; and cytogenetics to identify nonrandom chromosomal abnormalities. Additional investigations are also recommended, including flow cytometry immunophenotyping and fluorescence in situ hybridization.11 A patient is diagnosed as having MDS, when he or she has unexplained cytopenia and meets at least 1 of the following criteria: morphologic evidence of dysplasia, and/or MDS-specific cytogenetic abnormalities.12,-14 Excluding or establishing the diagnosis of myeloid neoplasm with myelodysplasia may be challenging in the absence of robust morphological criteria or a cytogenetic abnormality.15 The term of idiopathic cytopenia of undetermined significance (ICUS) has been introduced to describe a condition that does not fit into the diagnostic criteria of MDS.16
Studies of massive parallel DNA sequencing have shown that the vast majority of MDS patients carry 1 or more somatic oncogenic mutations in genes involved in RNA splicing, DNA methylation, chromatin modification, transcription regulation, DNA repair, or signal transduction.17,,,,,-23 Recent genetic studies have shown that somatic mutations in some of these genes are commonly acquired in hematopoietic cells during aging, and that apparently normal subjects with these mutations have increased risk of hematologic malignancies and increased all-cause mortality.24,,-27 This condition has been defined as age-related clonal hematopoiesis or clonal hematopoiesis of indeterminate potential.15,28,29
In this work, we performed a mutation analysis of genes that are recurrently mutated in myeloid malignancies in a prospective cohort of individuals with unexplained cytopenia undergoing a comprehensive diagnostic workup, with the aim to estimate the predictive value of somatic mutations for identifying individuals with a myeloid neoplasm or with an increased risk of developing these malignancies.
Patients and methods
Patients’ characteristics and clinical procedures
This study included 873 patients investigated at the Department of Hematology, IRCCS Policlinico San Matteo Foundation and University of Pavia, Pavia, Italy, between 2003 and 2015. These patients were recruited in 2 distinct cohorts: (1) a learning cohort of 683 consecutive patients investigated for unexplained cytopenia; and (2) a validation cohort of 190 patients referred to the same institution for a second opinion for suspected myeloid neoplasm with myelodysplasia. Diagnostic procedures aimed at distinguishing myeloid neoplasms from reactive causes of cytopenia or other hematopoietic disorders, including bone marrow examination, were in accord to recent recommendations.11 Peripheral blood and bone marrow specimens were analyzed by 2 independent hematopathologists who were blinded to clinical data, and the diagnosis of myeloid neoplasm was formulated according to the criteria of the World Health Organization (WHO) classification of myeloid neoplasms and its revision.12,-14 The category of ICUS was defined as previously reported, including cytopenia(s) in 1 or more lineages that did not meet the criteria of MDS and could not be explained by any other hematologic or nonhematologic disease.16 Patients were prospectively included in the study at the time of their first visit at the Department of Hematology and regularly followed up to establish the diagnosis and to monitor the disease as clinically required. Clinical features of patients included in the study and diagnosis according to conventional diagnostic workup are reported in Table 1 and supplemental Tables 1-3, available on the Blood Web site. The diagnosis of patients in the learning cohort was myeloid neoplasm in 409 cases, other cytopenia (including iatrogenic, immune, or secondary cytopenia) in 120 cases, whereas in 154 patients a provisional diagnosis of ICUS was adopted. In the validation cohort, the final diagnosis was of myeloid neoplasm in 138 patients; in 38 patients, the suspect of myeloid malignancy was excluded, whereas in 14 patients a diagnosis of ICUS was provisionally adopted.
Clinical and hematological features of the patients included in the study and results of the conventional diagnostic workup
| Variable . | Value . |
|---|---|
| Learning cohort | |
| No. of patients | 683 |
| Age, median (range), y | 66 (18-92) |
| Sex, M/F | 388/295 |
| Hemoglobin, median (range), g/dL | 11.2 (5-16.6) |
| Mean cell volume, median (range), fL | 94 (60-125) |
| White blood cell count, median (range), ×109/L | 4.19 (0.19-15.82) |
| Absolute neutrophil count, median (range), ×109/L | 1.9 (0.03-12.62) |
| Platelet count, median (range), ×109/L | 127 (1-496) |
| Diagnosis according to the outcome of conventional diagnostic workup | |
| Myeloid neoplasm | 409 |
| MDS | 233 |
| Myelodysplastic/myeloproliferative neoplasm | 86 |
| Myeloproliferative neoplasm | 35 |
| Acute myeloid leukemia | 55 |
| Other cytopenias | 120 |
| Aplastic anemia | 30 |
| Lymphoid neoplasm | 28 |
| Other (iatrogenic, immune, and secondary cytopenia) | 62 |
| ICUS | 154 |
| Validation cohort | |
| No. of patients | 190 |
| Age, median (range), y | 62 (19-88) |
| Sex, M/F | 105/85 |
| Time from first evaluation to harvest, median (range), d | 124 (1-436) |
| Diagnosis according to the outcome of conventional diagnostic workup | |
| Myeloid neoplasm | 138 |
| ICUS | 14 |
| Other cytopenias | 38 |
| Variable . | Value . |
|---|---|
| Learning cohort | |
| No. of patients | 683 |
| Age, median (range), y | 66 (18-92) |
| Sex, M/F | 388/295 |
| Hemoglobin, median (range), g/dL | 11.2 (5-16.6) |
| Mean cell volume, median (range), fL | 94 (60-125) |
| White blood cell count, median (range), ×109/L | 4.19 (0.19-15.82) |
| Absolute neutrophil count, median (range), ×109/L | 1.9 (0.03-12.62) |
| Platelet count, median (range), ×109/L | 127 (1-496) |
| Diagnosis according to the outcome of conventional diagnostic workup | |
| Myeloid neoplasm | 409 |
| MDS | 233 |
| Myelodysplastic/myeloproliferative neoplasm | 86 |
| Myeloproliferative neoplasm | 35 |
| Acute myeloid leukemia | 55 |
| Other cytopenias | 120 |
| Aplastic anemia | 30 |
| Lymphoid neoplasm | 28 |
| Other (iatrogenic, immune, and secondary cytopenia) | 62 |
| ICUS | 154 |
| Validation cohort | |
| No. of patients | 190 |
| Age, median (range), y | 62 (19-88) |
| Sex, M/F | 105/85 |
| Time from first evaluation to harvest, median (range), d | 124 (1-436) |
| Diagnosis according to the outcome of conventional diagnostic workup | |
| Myeloid neoplasm | 138 |
| ICUS | 14 |
| Other cytopenias | 38 |
F, female; M, male.
This study was approved by the Ethics Committee of the IRCCS Policlinico San Matteo Foundation, Pavia, Italy. The procedures followed were in accordance with the Declaration of Helsinki of 1975, as revised in 2000, and samples were obtained after patients provided written informed consent.
Mutation analysis
A core panel of 40 genes selected on the basis of prior implication in the pathogenesis of myeloid disease was analyzed in the whole study cohort on peripheral blood granulocytes. Details of the sequencing analysis are provided in the “Methods” of the supplemental material and supplemental Table 4. Functionally annotated variants were then filtered based on the information retrieved from public databases, and the expected germ line allele frequency. The remaining variants were finally tagged as oncogenic (supplemental Table 5), based on the information derived from the literature, the Catalog of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic), and in silico prediction effect, as previously described.22
Statistical analysis
All the statistical analyses were performed using Stata SE 12.1 (StataCorp LP, College Station, TX; http://www.stata.com) software. Full details of the statistical analysis are reported in “Methods” of the supplemental material.
Results
We initially performed a mutation analysis, utilizing granulocyte DNA collected at the time of the initial diagnostic workup, in the prospective cohort of 683 patients reported in Table 1. We then studied the relationships between initial mutation pattern and outcome of the diagnostic process.
Predictive value of mutation status and hematopoietic clone size in the differential diagnosis of unexplained cytopenia
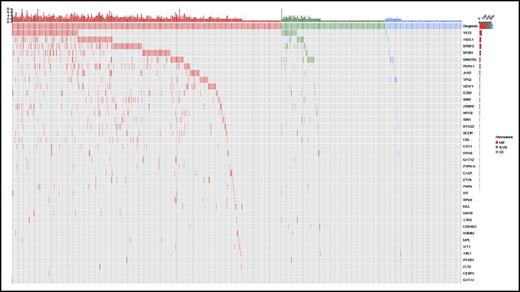
Overall, 435/683 (64%) patients carried a somatic mutation in at least 1 of the 40 genes studied: the most frequently mutated genes and their distribution are reported in Figure 1; supplemental Figure 1; and supplemental Tables 5 and 6. A significantly higher number of mutations per subject was observed in patients with myeloid neoplasms compared with those with ICUS or other cytopenias (P values < .001), as well as in patients with ICUS compared with those with other cytopenias (P = .002) (supplemental Figure 2). In addition, significant differences in variant allele frequency (VAF) were observed between these conditions (P values < .001) (supplemental Figure 2).
Patterns of the mutations identified in the cohort of 683 patients with unexplained cytopenia. Distribution of somatic lesions in the analyzed genes according to the final diagnosis resulting from standard workup. Red color indicates a diagnosis of myeloid neoplasm; green indicates a diagnosis of ICUS, and blue indicates a diagnosis of other cytopenia. Each column represents an individual patient sample. MN, myeloid neoplasm; OC, other cytopenia.
Patterns of the mutations identified in the cohort of 683 patients with unexplained cytopenia. Distribution of somatic lesions in the analyzed genes according to the final diagnosis resulting from standard workup. Red color indicates a diagnosis of myeloid neoplasm; green indicates a diagnosis of ICUS, and blue indicates a diagnosis of other cytopenia. Each column represents an individual patient sample. MN, myeloid neoplasm; OC, other cytopenia.
The absence of mutations in the 40 genes studied had a negative predictive value for myeloid neoplasm of 0.76 (95% confidence interval [CI] 0.70-0.81) (supplemental Table 7). Twenty-four unmutated patients carried cytogenetic abnormalities by standard karyotyping of bone marrow cells, including abnormalities providing presumptive evidence of MDS according to WHO criteria in 14 cases. When mutation status was combined with standard karyotyping, the negative predictive value of the absence of genetic lesions increased to 0.84 (CI 0.79-0.89).
Conversely, detecting 1 or more somatic mutations had a positive predictive value for myeloid neoplasm of 0.81 (95% CI 0.76-0.84), whereas having 2 or more mutations had a positive predictive value of 0.88 (95% CI 0.84-0.92) (supplemental Table 7).
We then performed receiver operating characteristic analyses to explore the best cutoff value of VAF to identify myeloid neoplasm. These analyses were performed on VAF values not corrected for copy number variation. VAF showed a highly significant discrimination ability (area under the curve, 0.83; 95% CI 0.80-0.86; P < .001) (supplemental Figure 3), a VAF of 0.087 being associated with the highest percentage of correct classification (positive predictive value 0.86; negative predictive value 0.77; sensitivity 0.84; specificity 0.80) (supplemental Table 8).
Positive predictive value for myeloid neoplasm of mutation patterns in the differential diagnosis of unexplained cytopenia
We calculated the predictive value for diagnosis of myeloid neoplasm of the most frequently mutated genes. Spliceosome genes (SF3B1, SRSF2, U2AF1), JAK2, and RUNX1 had the highest predictive value for myeloid neoplasm irrespective of co-occurring mutations, with positive predictive values ranging from 0.88 to 0.97 (Figure 2; supplemental Table 9). In order to account for interactions with comutated genes, we calculated positive predictive values of mutated genes when occurring as isolated mutations or in the context of comutation patterns (Figure 2; supplemental Table 9).
Positive predictive value for myeloid neoplasm of the most frequently mutated genes. (A) Distribution and positive predictive value of the most frequently mutated genes. The upper bars indicate positive predictive values for myeloid neoplasm: blue indicates the predictive value according to standard diagnostic approach, whereas lavender depicts the predictive value when accounting for ICUS with mutation pattern highly specific for myeloid neoplasm as true positive cases. The lower bars report the distribution of mutated genes according to the final diagnosis of standard workup (pink, myeloid neoplasm; orange, ICUS; red, other cytopenia). (B) Distribution and positive predictive value of mutations in TET2, ASXL1, and DNMT3A as isolated mutations or comutation patterns (bar colors as in panel A). (C) Positive predictive value of the most frequent mutated genes associated with TET2, ASXL1, and DNMT3A mutations (bar colors as in panels A and B).
Positive predictive value for myeloid neoplasm of the most frequently mutated genes. (A) Distribution and positive predictive value of the most frequently mutated genes. The upper bars indicate positive predictive values for myeloid neoplasm: blue indicates the predictive value according to standard diagnostic approach, whereas lavender depicts the predictive value when accounting for ICUS with mutation pattern highly specific for myeloid neoplasm as true positive cases. The lower bars report the distribution of mutated genes according to the final diagnosis of standard workup (pink, myeloid neoplasm; orange, ICUS; red, other cytopenia). (B) Distribution and positive predictive value of mutations in TET2, ASXL1, and DNMT3A as isolated mutations or comutation patterns (bar colors as in panel A). (C) Positive predictive value of the most frequent mutated genes associated with TET2, ASXL1, and DNMT3A mutations (bar colors as in panels A and B).
Positive predictive values of mutations in spliceosome genes were comparable when these genetic lesions occurred as alone or with co-occurring mutations, although mutations in SRSF2, U2AF1, and ZRSR2 occurred significantly less frequently as isolated mutation than those in SF3B1 (P values ranging from <.001 to .064). Comparable results were observed for JAK2 mutation. Conversely, the positive predictive value of mutations in TET2, DNMT3A, or ASXL1 occurring as single lesions ranged from 0.29 to 0.57, whereas that of the same mutations combined with other genetic lesions was significantly higher (0.81-0.89; P values ranging from .002 to <.001) (Figure 2; supplemental Table 9). A similar trend was observed for mutations in TP53, whose predictive value increased from 0.71 as isolated mutation to 0.90 in comutation patterns, mainly in association with TET2 or ASXL1 mutation. When focusing on the most common comutation patterns of TET2, DNMT3A, and ASXL1 (supplemental Figure 4), mutations of RUNX1, EZH2, CBL, TP53, NRAS, CUX1, or IDH2 showed positive predictive values for diagnosis of myeloid neoplasm ranging from 0.86 to 1.0 (Figure 2). Overall, mutations in spliceosome genes and comutation patterns involving TET2, DNMT3A, and ASXL1 accounted for 73% of mutated myeloid neoplasms.
In a multivariable logistic regression analysis including the number of mutations per subject and the most frequently mutated genes, having 2 or more mutations (odds ratio [OR] = 4.69; P < .001) or carrying SF3B1 mutation (OR = 4.83; P = .003) were independent predictors for MDS or another myeloid neoplasm. Conversely, isolated DNMT3A or TET2 mutations were independently associated with diagnoses other than myeloid neoplasm (OR = .21; P = .001; and OR = .43; P = .023, respectively) (supplemental Table 10). When accounting for interaction between genes, TET2 mutation did not retain independent value.
The predictive values of mutation analysis were tested in an independent cohort of 190 patients referred to our institution for suspected myeloid neoplasm with myelodysplasia (Table 1). The positive predictive values of the number of mutations and VAF as well as of mutation patterns were fully confirmed (supplemental material).
Influence of the set of analyzed genes on the predictive value of mutation status
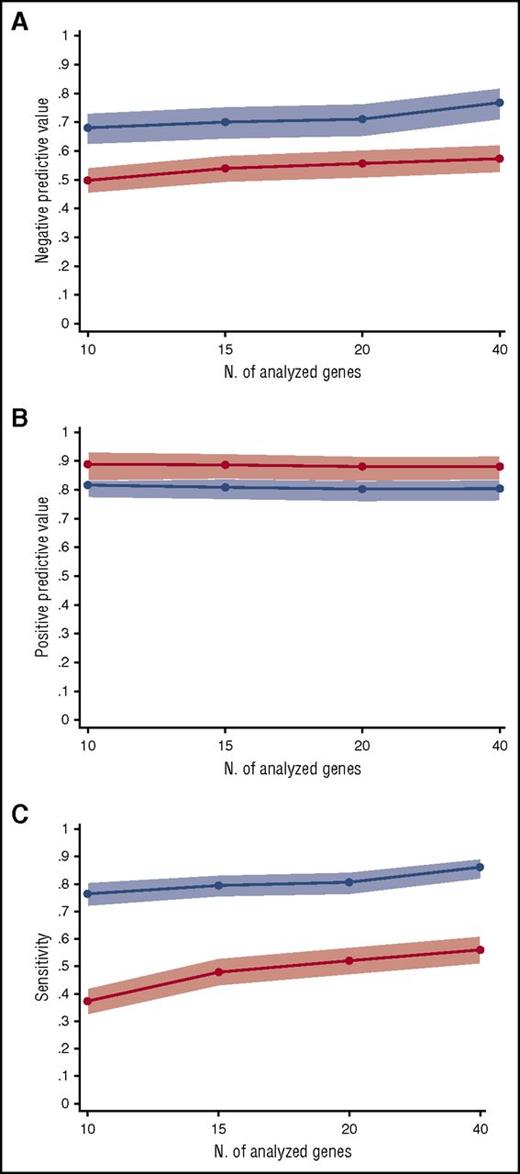
We tested the influence of the number of analyzed genes on the negative and positive predictive values of mutation status. To this purpose, 3 sets of genes, including 10, 15, and 20 genes, were generated, based on the frequency of mutation observed in this study and the information retrieved by previous studies in myeloid neoplasms (supplemental Table 12).22,23 The positive predictive value for myeloid neoplasm of having 1 or more somatic mutations was comparable between panels, ranging from 0.80 to 0.82. Similar results were observed for the positive predictive value of having 2 or more somatic mutations (range 0.88-0.89). Conversely, negative predictive value and sensitivity tend to increase with the number of genes analyzed (negative predictive value from 0.68 to 0.76 and sensitivity from 0.77 to 0.86 for 10-gene to 40-gene panels) (Figure 3; supplemental Table 7).
Predictive values and sensitivity according to the number of analyzed genes. (A) Negative predictive value for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels (supplemental Table 12). The blue line represents the negative predictive value of an unmutated status, whereas the red line indicates the negative predictive value of having no or 1 somatic mutation in the set of analyzed genes. Light blue and red areas indicate 95% CIs. (B) Positive predictive value for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels. The blue line represents the positive predictive value of having 1 or more somatic mutations in the set of analyzed genes, whereas the red line indicates the positive predictive value of having 2 or more somatic mutations. (C) Sensitivity for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels. The blue line represents the sensitivity of having 1 or more somatic mutations in the set of analyzed genes, whereas the red line indicates the sensitivity of having 2 or more somatic mutations.
Predictive values and sensitivity according to the number of analyzed genes. (A) Negative predictive value for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels (supplemental Table 12). The blue line represents the negative predictive value of an unmutated status, whereas the red line indicates the negative predictive value of having no or 1 somatic mutation in the set of analyzed genes. Light blue and red areas indicate 95% CIs. (B) Positive predictive value for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels. The blue line represents the positive predictive value of having 1 or more somatic mutations in the set of analyzed genes, whereas the red line indicates the positive predictive value of having 2 or more somatic mutations. (C) Sensitivity for diagnosis of myeloid neoplasms of 40-gene, 20-gene, 15-gene, and 10-gene panels. The blue line represents the sensitivity of having 1 or more somatic mutations in the set of analyzed genes, whereas the red line indicates the sensitivity of having 2 or more somatic mutations.
Diagnostic value of mutation status in patients with cytopenia of undetermined significance
One hundred fifty-four patients in the learning cohort received a provisional diagnosis of ICUS after the standard diagnostic workup (supplemental Table 2), and 38 of them (25%) developed a myeloid neoplasm (Figure 4). Among patients with a diagnosis of ICUS, 56 of 154 (36%) carried 1 or more mutations (range 1-7) (Figure 1; supplemental Figure 6; supplemental Table 6): this condition has been previously defined as clonal cytopenia of undetermined significance (CCUS).30,31 Patients with CCUS showed a significantly higher probability of developing myeloid neoplasm compared with those without evidence of clonality (hazard ratio [HR] = 13.9; 95% CI 5.40-35.91; 5- and 10-year cumulative probabilities of progression: 82% vs 9% and 95% vs 9%, respectively; P < .001) (Figure 4).
Probability of progression to myeloid neoplasm of patients receiving a provisional diagnosis of ICUS, according to mutation status and pattern. (A) Cumulative probability of developing a myeloid neoplasm in patients receiving a diagnosis of ICUS. (B) Distribution and cumulative incidence of progression to myeloid neoplasms according to the number of somatic mutations per patient. (C) Cumulative probability of progression to myeloid neoplasm according to mutation status. The red curve represents the probability of progression of patients with ICUS carrying 1 or more mutations in the set of genes analyzed (CCUS), whereas the blue curve reports the probability of progression of patients with ICUS without evidence of clonal hematopoiesis. (D) Cumulative probability of progression to myeloid neoplasm according to mutation pattern. The green curve represents the probability of progression of patients with CCUS showing mutation patterns highly predictive for myeloid neoplasm; the red curve represents the probability of progression of patients with low predictive mutation pattern, whereas the blue curve represents the probability of progression of patients with unmutated status.
Probability of progression to myeloid neoplasm of patients receiving a provisional diagnosis of ICUS, according to mutation status and pattern. (A) Cumulative probability of developing a myeloid neoplasm in patients receiving a diagnosis of ICUS. (B) Distribution and cumulative incidence of progression to myeloid neoplasms according to the number of somatic mutations per patient. (C) Cumulative probability of progression to myeloid neoplasm according to mutation status. The red curve represents the probability of progression of patients with ICUS carrying 1 or more mutations in the set of genes analyzed (CCUS), whereas the blue curve reports the probability of progression of patients with ICUS without evidence of clonal hematopoiesis. (D) Cumulative probability of progression to myeloid neoplasm according to mutation pattern. The green curve represents the probability of progression of patients with CCUS showing mutation patterns highly predictive for myeloid neoplasm; the red curve represents the probability of progression of patients with low predictive mutation pattern, whereas the blue curve represents the probability of progression of patients with unmutated status.
We then classified patients according to mutation patterns with low or high predictive value for myeloid neoplasm (number of patients: 24 and 32, respectively); highly predictive patterns included spliceosome gene mutations and mutations in TET2, ASXL1, or DNMT3A with additional mutations. Clinical and hematological features of patients receiving a diagnosis of ICUS grouped according to mutation profile are reported in Table 2. Patients with highly predictive mutation patterns had a significantly higher risk of developing a myeloid neoplasm compared with those with low-predictive mutation patterns (HR = 4.12; 95% CI 1.75-9.69; P = .001) or an unmutated status (HR = 25.26; 95% CI 9.27-68.86; P < .001) (Figure 4). In uni- and multivariable logistic regression analyses, no significant predictive effect of mutation pattern was observed on the evolution into different WHO categories of myeloid neoplasm.
Clinical and hematological features of patients receiving a diagnosis of ICUS grouped according to mutation status compared with features of patients receiving a diagnosis of MDS
| Variable . | ICUS . | MDS . | P value . | ||
|---|---|---|---|---|---|
| ICUS unmutated . | CCUS low predictive pattern . | CCUS high predictive pattern . | |||
| No. of patients | 97 | 24 | 32 | 233 | |
| Age, median (range), y | 53 (18-88) | 68 (18-86) | 68 (41-86) | 69 (27-91) | <.001 |
| Sex, M/F | 44/53 | 12/9 | 25/11 | 142/91 | .012 |
| Hemoglobin, median (range), g/dL | 12.7 (8.6-15.7) | 12.9 (9.7-15.4) | 11.9 (8.2-14.8) | 9.9 (5-15.6) | <.001 |
| Mean cell volume, median (range), fL | 92 (78-110) | 92 (82-102) | 92 (78-118) | 98 (63-124) | <.001 |
| White blood cell count, median (range), ×109/L | 3.72 (0.96-12.4) | 3.72 (1.75-9.5) | 3.94 (0.56-12.2) | 4.26 (0.7-15.82) | .72 |
| Absolute neutrophil count, median (range), ×109/L | 1.85 (0.68-9.2) | 1.88 (0.61-5.67) | 1.70 (0.61-6.22) | 2.4 (0.18-12.62) | .89 |
| Platelet count, median (range), ×109/L | 151 (20-496) | 117 (71-206) | 137 (23-355) | 129 (6-495) | .46 |
| Variable . | ICUS . | MDS . | P value . | ||
|---|---|---|---|---|---|
| ICUS unmutated . | CCUS low predictive pattern . | CCUS high predictive pattern . | |||
| No. of patients | 97 | 24 | 32 | 233 | |
| Age, median (range), y | 53 (18-88) | 68 (18-86) | 68 (41-86) | 69 (27-91) | <.001 |
| Sex, M/F | 44/53 | 12/9 | 25/11 | 142/91 | .012 |
| Hemoglobin, median (range), g/dL | 12.7 (8.6-15.7) | 12.9 (9.7-15.4) | 11.9 (8.2-14.8) | 9.9 (5-15.6) | <.001 |
| Mean cell volume, median (range), fL | 92 (78-110) | 92 (82-102) | 92 (78-118) | 98 (63-124) | <.001 |
| White blood cell count, median (range), ×109/L | 3.72 (0.96-12.4) | 3.72 (1.75-9.5) | 3.94 (0.56-12.2) | 4.26 (0.7-15.82) | .72 |
| Absolute neutrophil count, median (range), ×109/L | 1.85 (0.68-9.2) | 1.88 (0.61-5.67) | 1.70 (0.61-6.22) | 2.4 (0.18-12.62) | .89 |
| Platelet count, median (range), ×109/L | 151 (20-496) | 117 (71-206) | 137 (23-355) | 129 (6-495) | .46 |
Implications of mutation status for the diagnosis of myeloid neoplasms
We first tested the hypothesis that mutation status could improve the diagnosis of MDS. We identified 34 patients receiving a diagnosis of MDS based on mild dysplasia (ie, a percentage of dysplastic cells <25%); in 19 of these patients, no evidence of clonality was obtained using standard cytogenetic analysis or targeted gene sequencing at the time of diagnosis. To further support this finding, we performed X-chromosome inactivation pattern analysis in 7 informative female patients, and in all cases, a balanced inactivation pattern was found (median granulocyte corrected allelic ratio = 1.75, range 1.0-2.98), consistent with a polyclonal hematopoiesis. When examining retrospectively the clinical course of these unmutated patients, after a median follow-up of 64 months (range 8-145), none of them showed signs of disease progression. These data suggest that a diagnosis of MDS might not have been appropriate despite that current WHO criteria were fulfilled (supplemental Figure 7). When treating these cases as true negative cases, an unmutated status had a negative predictive value for myeloid neoplasm of 0.84 (95% CI 0.79-0.88) (0.92, 95% CI 0.88-0.95, when combining the results of mutation and standard cytogenetic analysis).
We then explored whether mutation status could provide presumptive evidence of MDS in the absence of definitive morphological evidence. To this purpose, we calculated the specificity for diagnosis of myeloid neoplasm with myelodysplasia (ie, MDS, myelodysplastic/myeloproliferative neoplasm, or acute myeloid leukemia with myelodysplasia-related changes) of the most frequent mutation patterns (supplemental Figure 8; supplemental Tables 13 and 14). We found that mutations in SF3B1 showed a specificity for myeloid neoplasm with myelodysplasia of 0.97, either as isolated mutation or in combination. Notably, 3 patients receiving an initial diagnosis of CCUS with SF3B1 mutation and VAF ranging from 0.05 to 0.11 showed morphological evidence of ring sideroblasts (median value 2%; range 1% to 3%) and eventually developed an overt MDS. Additional mutations were found to have high specificity, including ZRSR2 mutation and comutation patterns involving TET2, ASXL1, or DNMT3A, mutations in RUNX1, EZH2, CBL, BCOR, CUX1, TP53, or IDH1/IDH2 being the most frequent co-occurring mutations conferring a specificity >0.90. Mutations in SRSF2 and U2AF1 were observed in a fraction of patients with primary myelofibrosis associated with JAK2 mutation. When excluding this specific comutation pattern, SRSF2 and U2AF1 mutations showed a 100% specificity for myeloid neoplasm with myelodysplasia.
We then compared clinical features and outcomes of patients with CCUS and highly specific mutation patterns with those of patients with myeloid neoplasm with myelodysplasia without excess blasts or with specific mutation patterns: no significant differences were observed in overall survival and risk of disease progression in multivariable analysis (Figure 5; supplemental Tables 15 and 16). These data suggest that these mutation patterns may provide presumptive evidence of MDS even in the absence of definitive morphological criteria (supplemental Figure 7). When patients with CCUS and highly specific mutation patterns were considered as true positive cases, the positive predictive value of 2 or more mutations increased to 0.99 (95% CI 0.97-1.0); the positive predictive values of mutant spliceosome genes and comutation patterns involving TET2, ASXL1, or DNMT3A increased to 0.97-1.0 (Figure 2).
Overall survival of patients with CCUS and highly specific mutation pattern and of patients with myeloid neoplasm with myelodysplasia. Overall survival of patients with CCUS and highly specific mutation patterns (blue curve) and of patients with myeloid neoplasm with myelodysplasia without excess blasts (red curve) (P = .55). Comparable results were obtained when analyzing survival of patients with CCUS and highly specific mutation pattern and of patients with myeloid neoplasm with myelodysplasia and similar mutation pattern (P = .56).
Overall survival of patients with CCUS and highly specific mutation pattern and of patients with myeloid neoplasm with myelodysplasia. Overall survival of patients with CCUS and highly specific mutation patterns (blue curve) and of patients with myeloid neoplasm with myelodysplasia without excess blasts (red curve) (P = .55). Comparable results were obtained when analyzing survival of patients with CCUS and highly specific mutation pattern and of patients with myeloid neoplasm with myelodysplasia and similar mutation pattern (P = .56).
Discussion
In this study on a well-annotated cohort of unbiased consecutive patients with unexplained cytopenia undergoing a comprehensive diagnostic workup, including bone marrow examination, we found that mutation profiling on peripheral blood cells has a high predictive value for identifying individuals with, or at high risk of developing, a myeloid neoplasm. These findings suggest that mutation analysis may be a valuable complement to the current diagnostic workup of unexplained cytopenia.
Both the number of somatic mutations per subject and the size of the mutant clone, as defined by the variant allele frequency, had significant predictive values for myeloid neoplasm. In fact, we found that having 2 or more mutations had a positive predictive value of 0.88, whereas a threshold value of variant allele frequency of ∼0.10 was associated with the highest percentage of correct classification. It must be acknowledged that this value of allele burden, obtained from isolated peripheral blood granulocytes, may be subjected to variation if a different cell source is used (eg, whole peripheral blood cells).
However, considerable differences were found between different mutant genes. In agreement with previous biologic and clinical observations,18,19,32,,,,-37 mutations in spliceosome genes showed the highest predictive value. SF3B1 mutation occurred as isolated mutation in a high proportion of patients receiving a diagnosis of myeloid neoplasm.22,33,34 Notably, the positive predictive value of this gene mutation was comparable when occurring as isolated lesion or in comutation patterns. Other mutated splicing factors (SRSF2, U2AF1, ZRSR2) occurred more frequently in combination with other mutations; however, as for SF3B1, their positive predictive value was equally high when these mutations were detected as isolated or in combination patterns. Although additional studies are required to trace the natural history of hematopoietic clones carrying these mutations, the detection of these mutations in patients with unexplained cytopenia should be considered highly predictive of myeloid neoplasm. By contrast, the predictive value of isolated mutations in genes like TET2, DNMT3A, and ASXL1, which are frequently mutated in age-related clonal hematopoiesis,24,-26,38 was lower, in accord with previous observations that these genes require additional genetic events to give rise to a myeloid neoplasm.15,24,25
Recent studies have shown that a substantial portion of patients with cytopenia of undetermined significance carries MDS-associated somatic mutations, and the term clonal cytopenia of undetermined significance has been proposed for describing this condition.30,31 In the present study, individuals with clonal cytopenia had a probability of developing a myeloid neoplasm, which was 14 times higher than that of subjects with no evidence of clonal disease. Furthermore, subjects with spliceosome gene mutations or mutations in TET2, ASXL1, or DNMT3A, combined with additional mutations, had a 5-year cumulative probability of developing a myeloid neoplasm equal to 95%, thus representing a very high-risk group. Although our study did not include sequential mutation analysis, in patients receiving a provisional diagnosis of CCUS, serial analysis is likely to allow detection of changes in mutation pattern and variant allele frequencies and may potentially improve current clinical monitoring.
We further refined these mutation patterns to identify those highly specific for myeloid neoplasms with myelodysplasia. Highly specific patterns included mutations of splicing factors, as well as comutation patterns involving TET2, ASXL1, or DNMT3A, and RUNX1, EZH2, CBL, BCOR, CUX1, TP53, or IDH1/IDH2. Notably, when comparing clinical features and outcomes of patients with CCUS and highly specific mutation patterns with those of patients either with myeloid neoplasm with myelodysplasia without excess blasts or with similar mutation patterns, no significant differences were observed in survival and risk of disease progression. These data indicate that the underlying genetic lesions may provide presumptive evidence of myeloid neoplasm with myelodysplasia even in the absence of definitive morphological features, as previously accepted for selected cytogenetic abnormalities.7,14 These results support the concept that cases with inconclusive dysplasia but highly specific mutation patterns might be classified as bona fide MDS, as those with MDS-defining cytogenetic abnormalities.
A portion of patients receiving a final diagnosis of myeloid neoplasm did not carry any mutation in the set of analyzed genes. Some of these patients had MDS with del(5q), a condition in which the chromosomal deletion alone represents the founding and disease-defining genetic lesion.39 We previously showed that del(5q) can be detected by means of massive parallel sequencing22 ; more generally, optimization of the sequencing design by targeting germline single nucleotide polymorphism to detect clinically relevant copy number alterations, enabling simultaneous detection of both gene mutations and cytogenetic abnormalities in a single assay, is likely to further improve the sensitivity of mutation analysis.40 We also identified a fraction of patients receiving a diagnosis of MDS based on mild dysplasia with no evidence of clonality. This information, combined with a stable clinical course without any sign of disease progression, suggests that a diagnosis of MDS might not have been appropriate despite satisfying the current WHO criteria.
In order to enhance the transferability of targeted sequencing in the clinical practice, we estimated the predictive value of mutation analysis using different sets of 10 to 40 genes. Notably, the positive predictive values for myeloid neoplasm were comparable between panels. Conversely, the negative predictive value tended to increase with the number of genes analyzed. In perspective, however, the number of genes sequenced is not expected to represent a limiting factor in terms of feasibility, and the analysis of less frequently mutated genes not included in the adopted gene set may further improve the predictive value of mutation analysis.
In conclusion, the results of this study suggest that mutation analysis on peripheral blood cells may significantly improve the current diagnostic approach to subjects with unexplained cytopenia. More effective, noninvasive diagnostic procedures are in turn expected to improve compliance to diagnostic tests, in particular, in the frail population of elderly individuals, and overall, in the diagnostic accuracy of myeloid neoplasms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from Associazione Italiana per la Ricerca sul Cancro (IG 15356) (L.M.) and Special Program Molecular Clinical Oncology 5 per Mille, project 1005 (M.C.).
Authorship
Contribution: L.M. and M.C. designed the study, performed statistical analysis, and wrote the manuscript; A.G., E.M., S.C., S.Z., D.P., E.P., and S.O. analyzed sequencing data; E. Rizzo and V.V.F. performed bioinformatics and statistical analysis; E.T., I.A., C.E., E. Bono, A.B., G.T., E. Rumi, and E. Boveri collected clinical data; C.C., D.T., E.P., and S.O. contributed to study design and interpretation of the data.
Conflict-of-interest disclosure: E. Rizzo is an employee of enGenome s.r.l. The remaining authors declare no competing financial interests.
Correspondence: Luca Malcovati, Department of Hematology Oncology, Fondazione IRCCS Policlinico San Matteo, Viale Golgi 19, 27100 Pavia, Italy; e-mail: luca.malcovati@unipv.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal