In this issue of Blood, O’Brien and colleagues1 demonstrate impaired growth of circulating red-cell progenitor cells ex vivo from patients with Diamond-Blackfan anemia (DBA) with ribosomal protein (RP) or GATA1 mutations. Distinct transcriptional profiles in erythroid cells from patients with RP and GATA1 mutations indicate potential mechanisms underlying dysregulation of translation.1
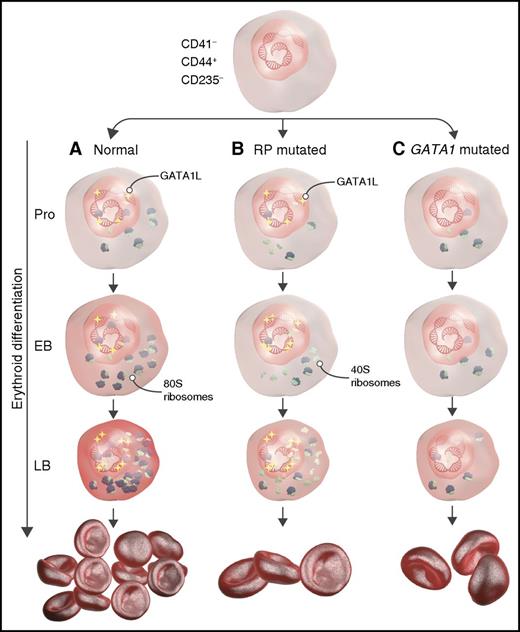
Schematic representation describing the role of RP and GATA1 mutations during erythropoiesis in DBA. (A) Under normal conditions, long-form GATA1 (GATA1L) induces genes that promote translation and heme synthesis, resulting in adequate red-cell production. (B) In RP-mutant cells, GATA1L induces genes responsible for heme synthesis and translation, but defective ribosomal assembly leads to aberrant erythropoiesis and anemia. (C) Mutated GATA1 results in decreased expression of genes encoding the translational apparatus and heme synthesis despite normal ribosome assembly, resulting in anemia. EB, early basophilic erythroblasts; LB, late basophilic erythroblasts; Pro, proerythroblasts. The figure has been adapted from Figure 7 in the article by O’Brien et al that begins on page 3111.
Schematic representation describing the role of RP and GATA1 mutations during erythropoiesis in DBA. (A) Under normal conditions, long-form GATA1 (GATA1L) induces genes that promote translation and heme synthesis, resulting in adequate red-cell production. (B) In RP-mutant cells, GATA1L induces genes responsible for heme synthesis and translation, but defective ribosomal assembly leads to aberrant erythropoiesis and anemia. (C) Mutated GATA1 results in decreased expression of genes encoding the translational apparatus and heme synthesis despite normal ribosome assembly, resulting in anemia. EB, early basophilic erythroblasts; LB, late basophilic erythroblasts; Pro, proerythroblasts. The figure has been adapted from Figure 7 in the article by O’Brien et al that begins on page 3111.
DBA is an inherited bone marrow failure syndrome that is characterized by defects in erythropoiesis, congenital abnormalities, and predisposition to cancer. Treatment of the anemia relies on steroids and red-cell transfusions.2 In 1999, the first ribosomal protein subunit, RPS19, was identified in association with DBA.3 Since then, as many as 72% of patients with DBA have been found to have heterozygous pathogenic mutations in RP genes.4 Another mutation more recently identified through whole-exome sequencing is the erythroid-specific transcription factor GATA1, resulting in altered translation.5,6 The remaining patients with DBA do not have any identifiable mutations.
Several model systems have been developed to recapitulate the phenotype of DBA, including mice, zebrafish, and inducible pluripotent stem cells.7-9 Heterozygous mice with RPS19 mutations do not have an anemia phenotype, whereas RPS19 knockout in mice is lethal. Inducible pluripotent stem cells can be difficult to differentiate to the erythroid lineage. Zebrafish provide insights into development, but there are limited reagents available to study specific pathways. Lymphocyte cell lines from patients with DBA are Epstein-Barr virus transformed, which could confound results of mechanistic experiments and make interpretation of results difficult. Obtaining bone marrow from patients with DBA can be cumbersome and requires anesthesia for a potentially unnecessary procedure, because the diagnosis can be made through gene testing of peripheral blood cells and assaying erythrocyte adenine deaminase activity.
One of the challenges in studying the molecular pathogenesis of DBA has been the limited numbers of cells obtained from bone marrow of patients with DBA. This paper describes a 2-phase ex vivo culture method to characterize erythroid differentiation, transcriptional profiles, and steroid responses of 11 patients with DBA with various RP and GATA1 mutations.1 Peripheral blood CD34+ hematopoietic stem and progenitor cells allow for repeated analysis and are easily accessible in comparison with bone marrow CD34+ cells. The authors nicely show increased erythroid cell proliferation and differentiation using their assay with peripheral blood CD34+ cells from steroid-treated patients with DBA. The system also allows for analysis of transcriptional profiling that characterizes the differences in patients with RP and GATA1 mutations.
Intriguingly, this study demonstrates that RP and GATA1 mutations result not only in defective translation but also unique and overlapping transcriptional programs. The authors define CD41−/CD44−/CD235− cells as immature cells, CD41−/CD44+/CD235− cells as erythroid precursors, and CD41−/CD44+/CD235+ cells as basophilic erythroblasts at day 14 of normal and DBA cultures. Upregulation of genes identified in normal control CD41−/CD44+/CD235+ cells included genes that regulate heme metabolism, cell cycle, and targets of transcription factors GATA1 and E2F. CD41−/CD44+/CD235+ cells also demonstrated a significant increase in genes involved with the translational machinery (eg, RNA Pol I/III, small nucleolar RNA, and ribosomal genes). In contrast, genes upregulated in CD41−/CD44+/CD235− cells seem to be related to growth-factor signaling. Although complex and still preliminary, these results suggest a possible feedback loop between translation and transcription regulation that has yet to be characterized.
Further analysis of transcriptional profiles of cells from RP- vs GATA1-mutant patients with DBA and normal CD41−/CD44+/CD235− populations showed differences in their gene set enrichment analyses. Interestingly, although GATA1- and RP-mutant cells express different genes, the end result is defective translation affecting production of red cells (see figure). The caveats of these studies are whether the samples are being analyzed at the same time points during erythroid differentiation and whether the populations are homogeneous. The key factor to elucidating the specific events mediating bone marrow failure will be to fill the gaps in knowledge of how the changes in gene expression are linked to translation. The identification of key steps in this regulation will lead to potential avenues to treat DBA.
Conflict-of-interest disclosure: The author declares no competing financial interests.