Key Points
Pediatric patients with de novo AML on average acquire 1.14 somatic CNAs in a study sample of 446 patients.
The presence of CNAs is significantly associated with survival in standard-risk patients.
Abstract
Childhood acute myeloid leukemia (AML) is frequently characterized by chromosomal instability. Approximately 50% of patients have disease relapse, and novel prognostic markers are needed to improve risk stratification. We performed genome-wide genotyping in 446 pediatric patients with de novo AML enrolled in Children’s Oncology Group (COG) studies AAML0531, AAML03P1, and CCG2961. Affymetrix and Illumina Omni 2.5 platforms were used to evaluate copy-number alterations (CNAs) and determine their associations with treatment outcome. Data from Affymetrix and Illumina studies were jointly analyzed with ASCAT and GISTIC software. An average of 1.14 somatically acquired CNAs per patient were observed. Novel reoccurring altered genomic regions were identified, and the presence of CNAs was found to be associated with decreased 3-year overall survival (OS), event-free survival (EFS), and relapse risk from the end of induction 1 (hazard ratio [HR], 1.7; 95% confidence interval [CI], 1.2-2.4; HR, 1.4; 95% CI, 1.0-1.8; and HR, 1.4; 95% CI, 1.0-2.0, respectively). Analyses by risk group demonstrated decreased OS and EFS in the standard-risk group only (HR, 1.9; 95% CI, 1.1-3.3 and HR, 1.7; 95% CI, 1.1-2.6, respectively). Additional studies are required to test the prognostic significance of CNA presence in disease relapse in patients with AML. COG studies AAML0531, AAML03P1, and CCG2961 were registered at www.clinicaltrials.gov as #NCT01407757, #NCT00070174, and #NCT00003790, respectively.
Introduction
Acute leukemia is the most frequent childhood cancer, and acute myeloid leukemia (AML) comprises approximately 25% of pediatric acute leukemias. Therapy for pediatric AML is intensive, with approximately 25% of patients receiving allogeneic donor stem-cell transplantation in first remission. Despite receiving intensive therapy, approximately 50% of patients with AML have disease relapse.1 Currently, the Children’s Oncology Group (COG) stratifies patients into risk groups according to cytogenetic and molecular features of the response of AML blasts to treatment by flow cytometry detection of minimal residual disease after the first course of chemotherapy.2,3 Extensive research efforts by the COG and other pediatric cooperative groups have been undertaken to refine risk stratification by identifying additional prognostic markers.
Conventional karyotyping has revealed the frequent occurrence of large chromosomal gains or losses, most commonly deletion 5q, monosomy 7, and trisomies of chromosomes 8, 11, and 13 in patients with AML. Genomic profiling of DNA copy-number alterations (CNAs) and loss of heterozygosity (LOH),4,5 as well as the complete sequencing of AML genomes,6,7 indicates that the genetic alterations in AML are fewer than those in other malignancies. However, there is variability in the number of CNAs. Kühn et al4 identified an average of 2.38 somatic CNAs per patient, whereas Radtke et al5 reported 1.28 CNAs per patient in AML. However, the latest data from The Cancer Genome Atlas Pan-Cancer data set report an average of 0.4 focal CNAs in 200 patients genotyped on the Affymetrix 6.0 Array platform.8 The discrepancy might be explained by the distinguishing of broad from focal events, but DNA quality, CNA calling algorithms, breakpoint accuracy, and rigor in manually reviewing CNA calls may also play a role.9,10
Two prior studies examined CNAs in pediatric AML. In a study of 111 children with de novo AML genotyped by using the Affymetrix 100K and 500K single-nucleotide polymorphism (SNP) arrays (combined resolution of 615 000), Radtke et al5 identified 5 reoccurring gains and 12 focal lesions. In a study of 43 t(8;21) and 39 inv(16) pediatric patients with core binding factor (CBF) AML genotyped by using the Affymetrix Genome-Wide Human SNP Array 6.0, Kühn et al4 reported 11 reoccurring focal losses and 4 gains. Neither study found an association between CNA and relapse risk (RR).
In this study, we sought to expand our knowledge of the genomic architecture of CNAs in de novo pediatric AML. We determined CNA presence and its prognostic significance regarding survival in a large cohort of patients with newly diagnosed AML from 3 COG trials.
Methods
Study cohort
A total of 528 matched tumor remission samples were obtained from 460 patients with de novo AML enrolled in COG trials CCG-2961,11 AAML-03P1,12 and AAML-0531.13 CCG-2961 evaluated idarubicin and fludarabine versus an intensive-timing 5-drug combination therapy in induction and interleukin-2 as postconsolidation therapy with related-donor SCT for patients with an available donor (supplemental Figure 1, available on the Blood Web site). AAML03P1 evaluated the feasibility and efficacy of combining gemtuzumab ozogamicin with standard chemotherapy used in the Medical Research Council backbone. AAML0531 randomly assigned patients to standard chemotherapy with or without gemtuzumab ozogamicin (supplemental Figure 2). Written informed consent was obtained from all study participants. Informed consent was obtained in accordance with the Declaration of Helsinki. Institutional review boards of all participating institutions approved the clinical protocols.
Cytogenetic analysis, molecular characterization, and risk stratification
Cytogenetic analysis and fluorescence in situ hybridization were performed by using standard G-banding/fluorescence in situ hybridization techniques, and results were centrally reviewed. Molecular analyses for FLT3-ITD, NPM1, and CEBPα mutations were performed as previously described.14-16 Patients were stratified into cytogenetic risk groups according to the COG risk classification used in the AAML0531 trial.13 Patients with CBF, which included t(8;21), inv(16)/(16;16), and mutated NPM1 and/or CEBPα, were classified as favorable risk. Patients with high allelic ratio FLT3-ITD, monosomy 7, monosomy 5, or del(5q) were classified as poor risk. All remaining patients for whom sufficient data were available for classification were categorized as standard risk. Minimal residual disease data were not used for risk stratification, because they were not uniformly available for all patients. Thirteen patients were excluded from analysis because cytogenetic and molecular testing was not successful.
Genome-wide genotyping
Genomic DNA was isolated from frozen bone marrow aspirate specimens from patients who provided consent for biological studies and for whom specimens were available at diagnosis and at the end of induction 1. DNA from paired tumor remission samples from 274 patients was genotyped with the HumanOmni 2.5-Quad BeadChip platform (Illumina) in 3 array runs at the Children’s Hospital of Philadelphia. Raw image intensity data normalization, genotype clustering, and calling of individual sample genotypes were performed by using GenomeStudio software (version 2011.1) and Genotyping Module (version 1.9.4; Illumina). Signal intensities were also adjusted for patterns of genomic wave by using PennCNV.17 A total of 254 paired tumor remission samples were genotyped with the Affymetrix Genome-Wide Human SNP Array 6.0 at the Seattle Children’s Hospital, University of Washington (Affymetrix, Santa Clara, CA). Raw Affymetrix CEL files were converted to allele-specific signals and genotype calls by using Affymetrix Power Tools 9 and the BirdSeed algorithm (Broad Institute) to calculate log R ratio and B-allele frequency values, and all markers were updated to hg19, GRCh37.
Copy-number assessment
A matched allele-specific copy-number analysis of tumors (ASCAT) was performed by using ASCAT 2.2 in R software (R Foundation for Statistical Computing, Vienna, Austria).18 All samples passed the quality-control criteria, and ASCAT profiles of patients genotyped on both platforms were manually reviewed as described previously.19 Raw segmentation of log R ratio and B-allele frequency profiles obtained from the ASCAT algorithm were used to create the manually reviewed CNA profile by using visual inspection.
The Genomic Identification of Significant Targets in Cancer (GISTIC) method was used to aggregate data from different tumors to differentiate between driver and passenger aberrations by combining prevalence and amplitude.20 All markers on Illumina and Affymetrix were merged into 1 single-reference SNP annotation file. GISTIC was performed by using the Web-based interface of the Broad Institute (http://genepattern.broadinstitute.org), with CNA thresholds set to ±0.3, a minimum of 10 markers, and a default q-value threshold of 0.25.
Statistical analysis
The Kruskal-Wallis test was used to compare differences in continuous variables, and the χ2 test was used to compare categorical variables between patients with and without somatic CNAs. Bivariate association testing with 3-year overall survival (OS), event-free survival (EFS), and RR was performed by the Kaplan-Meier analysis. OS was defined as the time from study entry for patients in complete remission (CR) to death. EFS was defined as the time from study entry to death, failure to achieve remission during induction therapy, or relapse. Disease-free survival (DFS) was defined as the time from the end of induction 1 for patients in CR to relapse or death. RR was defined as the time from the end of induction 1 for patients in CR to relapse or death resulting from progressive disease, wherein deaths resulting from causes other than progressive disease were censored. Patients lost to follow-up were censored at their date of last known contact. The significance of candidate predictor variables was tested with the log-rank statistic for OS, EFS, and DFS. Cox proportional hazard models were also used to estimate hazard ratios (HRs) with 95% confidence intervals (CIs) for univariate and multivariate analyses. Multivariate analyses included the covariates of age, sex, and race/ethnicity. The significance level for P values was set at .05 using SAS software (SAS Institute, Inc., Cary, NC).
Results
Of 528 genotyped patient samples, 82 were excluded from analysis as follows: 45 duplicates, 24 genotyping failures, and 13 with unavailable cytogenetic data. Thus, 446 patient samples were included in the final analysis. Table 1 compares demographics, risk group classification, cytogenetics, and clinical characteristics between patients with and without CNAs by study enrollment (CCG2916, AAML03P1, or AAML0531). A total of 363 patients were enrolled from AAML0531, 60 patients from AAML03P1, and 23 from CCG2961. Patients with CNAs did not differ by age, sex, race, initial white blood cell count, platelet count, or bone marrow blast percentage. However, Hispanics were significantly more likely than non-Hispanics to have CNAs (P = .03). Patient samples that showed chromosomal gains and losses as detected by the SNP array analysis also revealed cytogenetic abnormalities in the karyotyping analysis.
Cohort characteristics
| Characteristic . | No. (%) . | P . | |
|---|---|---|---|
| No CNAs (n = 191) . | 1+ CNAs (n = 255) . | ||
| Study | |||
| AAML0531 | 156 (81.7) | 207 (81.2) | .89 |
| AAML03P1 | 26 (13.6) | 34 (13.3) | .93 |
| CCG2961 | 9 (4.7) | 14 (5.5) | .71 |
| Age, y | |||
| <1 | 13 (6.8) | 13 (5.1) | .45 |
| –1-9 | 64 (33.5) | 113 (44.3) | .02 |
| ≥10 | 114 (59.7) | 129 (50.6) | .06 |
| Sex | |||
| Male | 111 (58.1) | 130 (51.0) | .14 |
| Female | 80 (41.9) | 125 (49.0) | |
| Race | |||
| White | 164 (90.1) | 218 (88.6) | .62 |
| Black | 7 (3.8) | 19 (7.7) | .10 |
| Other | 11 (5.9) | 7 (2.9) | .25 |
| Ethnicity | |||
| Hispanic | 32 (17.7) | 67 (26.6) | .03 |
| Non-Hispanic | 149 (82.3) | 185 (73.4) | |
| Risk group | |||
| Favorable | 95 (50.7) | 102 (41.1) | .16 |
| Standard | 64 (34.2) | 102 (46.4) | .07 |
| Poor | 28 (15) | 31 (12.5) | .42 |
| Cytogenetics | |||
| Normal | 74 (40.7) | 24 (9.8) | <.001 |
| t(8;21) | 14 (7.7) | 50 (20.3) | <.001 |
| Inv(16) | 34 (18.7) | 33 (13.4) | .138 |
| 11q23 | 42 (23.1) | 48 (19.5) | .371 |
| t(6;9) | 3 (1.6) | 4 (1.6) | 1.000 |
| Monosomy 7 | 0 (0) | 5 (2) | .075 |
| Del(7q) | 0 (0) | 2 (0.8) | .510 |
| −5/5q- | 1 (0.5) | 3 (1.2) | .640 |
| Plus 8 | 0 (0) | 28 (11.4) | <.001 |
| Other | 14 (7.7) | 49 (19.9) | <.001 |
| WBC count, × 109/L | .337 | ||
| Median | 56.6 | 46.2 | |
| Range | 1.3-473 | 1.3-405 | |
| Platelet count, × 103/mL | .592 | ||
| Median | 39.5 | 41 | |
| Range | 2-253 | 4-1177 | |
| BM blasts, % | .789 | ||
| Median | 77 | 77 | |
| Range | 7-100 | 6-99 | |
| Characteristic . | No. (%) . | P . | |
|---|---|---|---|
| No CNAs (n = 191) . | 1+ CNAs (n = 255) . | ||
| Study | |||
| AAML0531 | 156 (81.7) | 207 (81.2) | .89 |
| AAML03P1 | 26 (13.6) | 34 (13.3) | .93 |
| CCG2961 | 9 (4.7) | 14 (5.5) | .71 |
| Age, y | |||
| <1 | 13 (6.8) | 13 (5.1) | .45 |
| –1-9 | 64 (33.5) | 113 (44.3) | .02 |
| ≥10 | 114 (59.7) | 129 (50.6) | .06 |
| Sex | |||
| Male | 111 (58.1) | 130 (51.0) | .14 |
| Female | 80 (41.9) | 125 (49.0) | |
| Race | |||
| White | 164 (90.1) | 218 (88.6) | .62 |
| Black | 7 (3.8) | 19 (7.7) | .10 |
| Other | 11 (5.9) | 7 (2.9) | .25 |
| Ethnicity | |||
| Hispanic | 32 (17.7) | 67 (26.6) | .03 |
| Non-Hispanic | 149 (82.3) | 185 (73.4) | |
| Risk group | |||
| Favorable | 95 (50.7) | 102 (41.1) | .16 |
| Standard | 64 (34.2) | 102 (46.4) | .07 |
| Poor | 28 (15) | 31 (12.5) | .42 |
| Cytogenetics | |||
| Normal | 74 (40.7) | 24 (9.8) | <.001 |
| t(8;21) | 14 (7.7) | 50 (20.3) | <.001 |
| Inv(16) | 34 (18.7) | 33 (13.4) | .138 |
| 11q23 | 42 (23.1) | 48 (19.5) | .371 |
| t(6;9) | 3 (1.6) | 4 (1.6) | 1.000 |
| Monosomy 7 | 0 (0) | 5 (2) | .075 |
| Del(7q) | 0 (0) | 2 (0.8) | .510 |
| −5/5q- | 1 (0.5) | 3 (1.2) | .640 |
| Plus 8 | 0 (0) | 28 (11.4) | <.001 |
| Other | 14 (7.7) | 49 (19.9) | <.001 |
| WBC count, × 109/L | .337 | ||
| Median | 56.6 | 46.2 | |
| Range | 1.3-473 | 1.3-405 | |
| Platelet count, × 103/mL | .592 | ||
| Median | 39.5 | 41 | |
| Range | 2-253 | 4-1177 | |
| BM blasts, % | .789 | ||
| Median | 77 | 77 | |
| Range | 7-100 | 6-99 | |
Missing values in race and ethnicity have been omitted.
BM, bone marrow; WBC, white blood cell count.
Patient samples harbored an average of 1.14 somatically acquired CNAs with a mean of 1.1, 1.3, and 0.8 in the favorable-, standard-, and poor-risk groups, respectively (Figures 1 and 2). Although the average number of CNAs in this cohort was less than those previously reported,4,5 it is consistent with results from The Cancer Genome Atlas network.8 Specifically, these included frequent genomic losses in 7q36.1 (MLL3, EZH2), 16p13.11 (MYH11), 9q21.32, 11p13 (WT1), 2q37.1 (SEPT2), 10p12.31 (MLLT10), 11q23.3 (MLL), 16q22.1 (CBFB), and 1p36.3 (RUNX3). Genomic gains in 17q24 (thymidine kinase 1 [TK1]), 1q32, 3q28 (LPP), 11q23 (MLL), 6q27 (DLL1), 2q32.1, and 4q35 (SORBS2) were frequently observed.
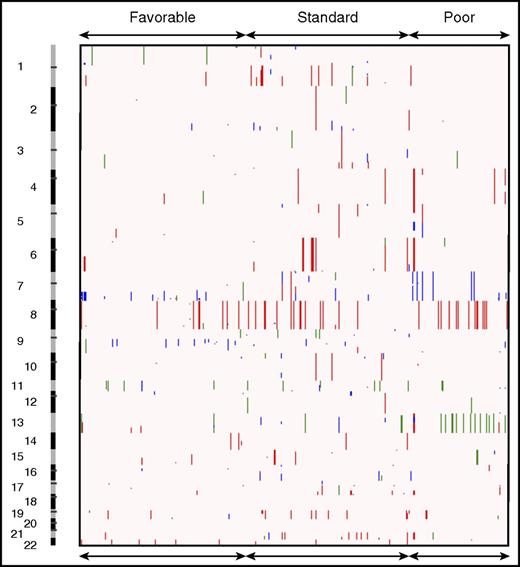
CNAs in pediatric AML. Broad copy number landscape of pediatric AML, shown as a heatmap of CNAs as seen in the COG cancer cohort. Deletions are shown in blue, amplifications in red, and copy-neutral (CN) LOHs in green.
CNAs in pediatric AML. Broad copy number landscape of pediatric AML, shown as a heatmap of CNAs as seen in the COG cancer cohort. Deletions are shown in blue, amplifications in red, and copy-neutral (CN) LOHs in green.
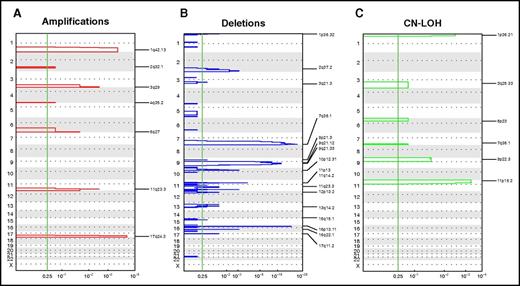
Focal recurrent aberrations. Significant focal CNAs in the COG pediatric AML cohort. GISTIC analysis revealed significantly recurring regions of focal CNAs, stratified according to amplifications (A), deletions (B), and CN LOHs (C). The vertical green line shows a false-discovery rate Q P > .25, which is considered significant.
Focal recurrent aberrations. Significant focal CNAs in the COG pediatric AML cohort. GISTIC analysis revealed significantly recurring regions of focal CNAs, stratified according to amplifications (A), deletions (B), and CN LOHs (C). The vertical green line shows a false-discovery rate Q P > .25, which is considered significant.
Table 2 compares reoccurring CNAs in pediatric AML reported in previous studies with those seen in our cohort by using GISTIC analysis. Patient samples in our study harbored most of the CNA losses reported by Kühn et al4 and Radtke et al.5 In addition, we also identified novel recurring gains and losses in 1p36.3, 2q37.1, and 10p12.31. Of interest, most of the recurring focal gains (1q32, 2q32.1, 3q28, 4q35.2, 6q27, 11q23, and 17q24) identified in our study have not been previously reported. However, a focal loss on chromosome 8 (ie, 8q24.21) reported by Kühn et al and Radtke et al was not seen in our study. This gain was identified in those prior studies because the presence of whole-chromosome gains and losses were included in the GISTIC analysis, whereas our GISTIC analysis excluded whole-arm and whole-chromosome gains and losses.
Comparison of focal losses and gains among 3 different SNP array studies
| Region . | Genes, n . | Patients, n . | Study . | ||
|---|---|---|---|---|---|
| Vujkovic . | Radtke . | Kühn . | |||
| 1p36.3 | 6 (*RUNX3, *NRAS) | 4 | Yes | No | No |
| 2q37.1 | 19 | 6 | Yes | No | No |
| 5p15.33 | 3 | — | No | No | Yes |
| 7p21.3 | 4 | — | No | No | Yes |
| 7q33-34 | 18 | — | No | Yes | No |
| 7q36.1 | 22 | 21 | Yes | Yes | Yes |
| 8q21.3 | RUNX1T1 | — | No | Yes | Yes |
| 9p21.3 | 12 (*CDKN2A) | — | No | No | Yes |
| 9q21.32 | 13 (*XPA) | 16 | Yes | Yes | No |
| 9q22.33 | 9 | — | No | No | Yes |
| 10p12.31 | 6 | 5 | Yes | No | No |
| 10q24.32-q24.33 | 9 | — | No | Yes | No |
| 11p13 | 2 (*WT1) | 7 | Yes | Yes | No |
| 11p14.1 | 7 | — | No | No | Yes |
| 11q23.3 | 4 (*MLL) | 3 | Yes | No | Yes |
| 12p13.31 | 31 | — | No | No | Yes |
| 16p11.2 | 33 | — | No | Yes | No |
| 16p13.11 | 5 (*MYH11) | 3 | Yes | Yes | Yes |
| 16q22.1 | 17 (*CBFB) | 7 | Yes | Yes | Yes |
| 17q11.2 | 12 | — | No | Yes | No |
| 18p11.21 | 0 | — | No | No | Yes |
| 21q22.12 | RUNX1 | — | No | Yes | No |
| Region . | Genes, n . | Patients, n . | Study . | ||
|---|---|---|---|---|---|
| Vujkovic . | Radtke . | Kühn . | |||
| 1p36.3 | 6 (*RUNX3, *NRAS) | 4 | Yes | No | No |
| 2q37.1 | 19 | 6 | Yes | No | No |
| 5p15.33 | 3 | — | No | No | Yes |
| 7p21.3 | 4 | — | No | No | Yes |
| 7q33-34 | 18 | — | No | Yes | No |
| 7q36.1 | 22 | 21 | Yes | Yes | Yes |
| 8q21.3 | RUNX1T1 | — | No | Yes | Yes |
| 9p21.3 | 12 (*CDKN2A) | — | No | No | Yes |
| 9q21.32 | 13 (*XPA) | 16 | Yes | Yes | No |
| 9q22.33 | 9 | — | No | No | Yes |
| 10p12.31 | 6 | 5 | Yes | No | No |
| 10q24.32-q24.33 | 9 | — | No | Yes | No |
| 11p13 | 2 (*WT1) | 7 | Yes | Yes | No |
| 11p14.1 | 7 | — | No | No | Yes |
| 11q23.3 | 4 (*MLL) | 3 | Yes | No | Yes |
| 12p13.31 | 31 | — | No | No | Yes |
| 16p11.2 | 33 | — | No | Yes | No |
| 16p13.11 | 5 (*MYH11) | 3 | Yes | Yes | Yes |
| 16q22.1 | 17 (*CBFB) | 7 | Yes | Yes | Yes |
| 17q11.2 | 12 | — | No | Yes | No |
| 18p11.21 | 0 | — | No | No | Yes |
| 21q22.12 | RUNX1 | — | No | Yes | No |
—, Not applicable.
In addition to detecting genomic gains and losses at a lower resolution than that detected by conventional karyotyping, SNP arrays can also detect regions of CN LOH, also known as acquired uniparental disomy. We identified several recurring regions of CN LOH in the AML genome in 14% of patients (n = 64), with 28% involving chromosome 13 and the others involving the arms of 11p (23%), 1p (11%), 9p (8%), 7q (6%), 19q (6%), and 3q (5%). Furthermore, focal CN LOH regions were confined to 11p15.5 (NUP98, PICALM, WT1), 1p36.3 (RUNX3, NRAS), 9p22.3 (MLLT3), 3q25.3, 6p23, and 7q35 (MLL3).
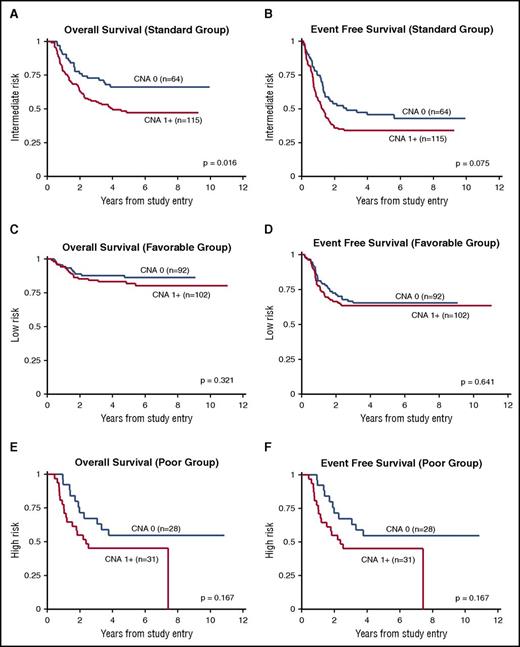
The univariate association between CNA presence and OS from study entry was statistically significant for all patients (P = .007; Table 3). This association remained significant in multivariate models that controlled for age, sex, and race/ethnicity (HR, 1.7; 95% CI, 1.2-2.4; P = .005; Table 4; Figure 3). Similarly, the univariate and multivariate associations between CNA presence and EFS and RR were significant (EFS: HR, 1.4; 95% CI, 1.0-1.8; P = .029; RR: HR, 1.4; 95% CI, 1.0-2.0; P = .043). In a subgroup analysis by risk group, CNA presence was significantly associated with survival only in standard-risk patients. Specifically, associations with OS and EFS were significant in the multivariate models, as shown in Figure 4 and Table 4.
Univariate association of CNA presence
| CNAs, n . | Patients, n . | OS ± 2 × SE, %* . | P . | EFS ± 2 × SE, %* . | P . | DFS ± 2 × SE, %† . | P . |
|---|---|---|---|---|---|---|---|
| Overall | .007 | .064 | .050 | ||||
| 0 | 191 | 73 ± 7 | 54 ± 7 | 58 ± 8 | |||
| 1+ | 255 | 62 ± 6 | 46 ± 6 | 50 ± 7 | |||
| Favorable risk | .321 | .641 | .805 | ||||
| 0 | 92 | 86 ± 7 | 65 ± 10 | 65 ± 11 | |||
| 1+ | 102 | 82 ± 8 | 64 ± 10 | 64 ± 10 | |||
| Standard risk | .016 | .075 | .102 | ||||
| 0 | 64 | 66 ± 12 | 46 ± 13 | 48 ± 15 | |||
| 1+ | 115 | 47 ± 10 | 34 ± 9 | 37 ± 10 | |||
| Poor risk | .167 | .409 | .214 | ||||
| 0 | 28 | 55 ± 20 | 37 ± 19 | 51 ± 23 | |||
| 1+ | 31 | 45 ± 18 | 32 ± 17 | 43 ± 22 |
| CNAs, n . | Patients, n . | OS ± 2 × SE, %* . | P . | EFS ± 2 × SE, %* . | P . | DFS ± 2 × SE, %† . | P . |
|---|---|---|---|---|---|---|---|
| Overall | .007 | .064 | .050 | ||||
| 0 | 191 | 73 ± 7 | 54 ± 7 | 58 ± 8 | |||
| 1+ | 255 | 62 ± 6 | 46 ± 6 | 50 ± 7 | |||
| Favorable risk | .321 | .641 | .805 | ||||
| 0 | 92 | 86 ± 7 | 65 ± 10 | 65 ± 11 | |||
| 1+ | 102 | 82 ± 8 | 64 ± 10 | 64 ± 10 | |||
| Standard risk | .016 | .075 | .102 | ||||
| 0 | 64 | 66 ± 12 | 46 ± 13 | 48 ± 15 | |||
| 1+ | 115 | 47 ± 10 | 34 ± 9 | 37 ± 10 | |||
| Poor risk | .167 | .409 | .214 | ||||
| 0 | 28 | 55 ± 20 | 37 ± 19 | 51 ± 23 | |||
| 1+ | 31 | 45 ± 18 | 32 ± 17 | 43 ± 22 |
COG cohort (n = 466). Mantel-Cox log-rank P value is used to denote significance. CNA presence is defined as the presence of any CNA in the tumor genome (dichotomous). The reference category is no CNA presence (CNA = 0).
OS and EFS from study entry.
DFS after induction 1.
Multivariate Cox proportional hazards models of the association of CNA presence and clinical variables with survival
| CNAs, n . | OS* . | EFS* . | RR† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients, n . | HR . | 95% CI . | P . | Patients, n . | HR . | 95% CI . | P . | Patients, n . | HR . | 95% CI . | P . | |
| Overall | ||||||||||||
| 0 | 181 | 1 | — | — | 181 | 1 | — | — | 146 | 1 | — | — |
| 1+ | 252 | 1.7 | 1.2-2.4 | .005 | 251 | 1.4 | 1.0-1.8 | .029 | 203 | 1.4 | 1.0-2.0 | .043 |
| Favorable risk | ||||||||||||
| 0 | 87 | 1 | — | — | 87 | 1 | — | — | 78 | 1 | — | — |
| 1+ | 100 | 1.5 | 0.7-3.1 | .286 | 100 | 1.1 | 0.7-1.8 | .709 | 87 | 1.0 | 0.6-1.8 | .885 |
| Standard risk | ||||||||||||
| 0 | 60 | 1 | — | — | 60 | 1 | — | — | 44 | 1 | — | — |
| 1+ | 114 | 1.9 | 1.1-3.3 | .021 | 114 | 1.7 | 1.1-2.6 | .020 | 88 | 1.5 | 0.9-2.6 | .105 |
| Poor risk | ||||||||||||
| 0 | 27 | 1 | — | — | 27 | 1 | — | — | 20 | 1 | — | — |
| 1+ | 31 | 1.7 | 0.8-3.6 | .193 | 31 | 1.4 | 0.7-2.6 | .374 | 21 | 1.6 | 0.7-4.0 | .278 |
| CNAs, n . | OS* . | EFS* . | RR† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients, n . | HR . | 95% CI . | P . | Patients, n . | HR . | 95% CI . | P . | Patients, n . | HR . | 95% CI . | P . | |
| Overall | ||||||||||||
| 0 | 181 | 1 | — | — | 181 | 1 | — | — | 146 | 1 | — | — |
| 1+ | 252 | 1.7 | 1.2-2.4 | .005 | 251 | 1.4 | 1.0-1.8 | .029 | 203 | 1.4 | 1.0-2.0 | .043 |
| Favorable risk | ||||||||||||
| 0 | 87 | 1 | — | — | 87 | 1 | — | — | 78 | 1 | — | — |
| 1+ | 100 | 1.5 | 0.7-3.1 | .286 | 100 | 1.1 | 0.7-1.8 | .709 | 87 | 1.0 | 0.6-1.8 | .885 |
| Standard risk | ||||||||||||
| 0 | 60 | 1 | — | — | 60 | 1 | — | — | 44 | 1 | — | — |
| 1+ | 114 | 1.9 | 1.1-3.3 | .021 | 114 | 1.7 | 1.1-2.6 | .020 | 88 | 1.5 | 0.9-2.6 | .105 |
| Poor risk | ||||||||||||
| 0 | 27 | 1 | — | — | 27 | 1 | — | — | 20 | 1 | — | — |
| 1+ | 31 | 1.7 | 0.8-3.6 | .193 | 31 | 1.4 | 0.7-2.6 | .374 | 21 | 1.6 | 0.7-4.0 | .278 |
Adjusted for the following potential covariates: age, sex, and ethnicity.
—, Not applicable.
OS and EFS from study entry.
RR from end of induction 1.
Stratified survival analysis according to risk group. (A,C,E) OS and (B,D,F) EFS for standard- (A-B), favorable- (C-D), and poor-risk groups (E-F).
Stratified survival analysis according to risk group. (A,C,E) OS and (B,D,F) EFS for standard- (A-B), favorable- (C-D), and poor-risk groups (E-F).
Discussion
In this large pediatric de novo AML study cohort, we observed a significant association between CNA presence and 3-year OS, EFS and RR. However, the significant association between CNA status and treatment outcome was seen only for patients in the standard-risk group.
To our knowledge, this is the first study reporting an association between CNA presence and treatment outcome in pediatric AML. There are several explanations for the association between CNA presence and treatment outcome found in our study that has not been seen in prior studies.4,5 First, the sample size of our study was substantially larger than those of the 2 prior studies in pediatric AML. Thus, the higher statistical power of our study may have enabled the observation of a significant association. Second, our subgroup analysis showed no significant association between CNA presence and treatment outcome in the favorable-risk group, which is consistent with the data reported by Kühn et al4 on patients with CBF AML. Because the association between CNA presence and treatment outcome has not been previously reported, additional studies are required to confirm this association, particularly in standard-risk patients.
We identified novel focal losses in 1p36.3, which harbors the genes RUNX3 and NRAS; 2q37.1 with SEPT2; and breakpoint-associated 10p12.31 with MLLT10. In a previous study of children with AML, AML with high RUNX3 expression was associated with reduced EFS.21 However, in a study of 2502 patients, Bacher et al22 showed that mutations in NRAS, which lies in the same region of 1p36.3, were not associated with OS, EFS, or DFS. Similarly, Berman et al23 showed in a large pediatric COG cohort that NRAS mutations were not associated with CR, RR, or 5-year OS or EFS. The lesion in 10p12.31 is a breakpoint-associated CNA resulting from the PICALM-MLLT10 fusion gene generated by the t(10;11)(p12;q14) translocation or the MLL(KMT2A)-MLLT10 by the t(10;11)(p12;q23) translocation, respectively. Similarly, the focal CNA at 2q37.1 likely affects the SEPT2 fusion partner of the MLL t(2;11)(q37;q23) translocation.24
We also observed novel focal gains in 1q32, 2q32.1, 3q28, 4q35.2, 6q27, 11q23, and 17q24 and explored the probable genes altered at each breakpoint. There are no known AML-associated genes within 1q32 and 2q32.1. The observed gain on 2q38 might involve the LPP-MLL fusion t(3;11) (q28;q23) translocation,25 and that on 4q35 might involve the t(4;11)(q35.1;q23) translocation, resulting in an ArgBP2-MLL fusion.26 The identified region of 6q27 contains DLL1, 11q23 contains MLL (KMT2A), and 17q24 contains TK1. The 11q23 amplification is likely a breakpoint-associated amplification. DLL1 is a NOTCH ligand that belongs to a family of single-pass transmembrane receptor proteins, the role of which in AML is poorly understood. DLL1 expression is high in acute promyelocytic leukemia.27 Two studies showed that DLL4 and NOTCH1 expression was significantly higher in patients with AML than in controls, indicating that activation of NOTCH signaling may promote AML development and be associated with an unfavorable prognosis.28,29 Also at this breakpoint is MLLT4 (AF6), the partner of MLL in t(6;11)(q27;q23) translocation. Lastly, the 17q24 region identified with a focal gain contains TK1, an enzyme involved in nucleic acid synthesis. TK1 seems to play a primary role in regulating intracellular thymidine pools throughout the cell cycle and is an important marker of tumor proliferation. Serum TK1 levels are elevated in patients with AML, and this is an accurate indicator of treatment outcome and stage of disease in patients with acute lymphoblastic leukemia or AML.30
In our study, focal CN LOH regions were confined to 11p15.5 (NUP98), 1p36.3 (RUNX3), 9p22.3 (MLLT3), 3q25.3, 6p23, and 7q35 (MLL3). Broad CN LOH analysis showed that complete LOH of chromosome 13 frequently occurred in the high-risk group, potentially as a result of FLT-ITD. Our focal analysis suggests that novel biological mechanisms drive these CNAs and that EZH2, WT1, and RUNX3 may be studied further for possible association with CNAs. Gronseth et al31 recently demonstrated in a study of 112 patients with AML that CN LOH was associated with a shorter duration of CR and worse OS. However, their observation was largely explained by the 13q CH LOH, which is acquired as result of a FLT3-ITD mutation. In our study, a majority of the patients with a 13q CN LOH were classified as poor risk because of the cooccurrence of FLT3-ITD mutations. Furthermore, the presence of a CNA within the poor-risk group indicated a tendency toward worse OS (HR, 1.7; 95% CI, 0.8-3.6); however, this association did not reach significance. The etiology underlying the development of acquired CN LOH remains unknown, although it is thought that interchromosomal homologous recombination activity may be involved. The recent investigation by Gaymes et al32 of homologous recombination events in mutated FLT3 revealed that FLT3 and JAK2 mutations lead to the production of reactive oxygen species and confer interchromosomal homologous recombination, leading to subsequent acquisition of CN LOH. The authors also observed that common breakpoints in the TK locus contribute to the propagation of CN LOH. It remains to be elucidated whether the use of antioxidants could prevent the accumulation of mutations and the CN LOH acquisition to slow the rate of disease progression in AML.
Radtke et al5 reported a substantially higher average number of CNAs by using the Affymetrix 100K and 500K SNP arrays (2.38 CNAs per patient) than those reported by Kühn et al4 by using the Affymetrix Array 6.0 containing 1.8 million genetic markers (1.28 CNAs per patient) and in our study by using the Illumina Omni 2.5M SNP/Affymetrix 6.0 Array (1.14 CNAs per patient). Our results are consistent with the average of 0.4 focal CNAs reported in specimens from 200 patients genotyped on the Affymetrix 6.0 Array in The Cancer Genome Atlas Pan-Cancer data set.8 The finding of a low frequency of CNAs in the 2 previous studies in pediatric AML performed on lower-resolution array seems counterintuitive. The lower-density array may have yielded a higher number of false-positive CNA calls, which might have contributed to the observed null association between CNA presence and survival.4,5
There were other differences between the analytic pipelines used by our group and those used in the Radtke et al5 and Kühn et al4 studies. Our data analysis was performed on ASCAT, which incorporates methods to account for stromal contamination, tumor heterogeneity, and aneuploidy but does not use a fixed probe number to partition individual probes into segments. This approach also enabled the integration of data from the Illumina and Affymetrix platforms, resulting in a substantially larger sample size.19 This large sample size allowed us to perform GISTIC analysis on focal CNAs only, and our results are therefore independent of the presence of whole-chromosome gains and losses. The analytic pipeline we used likely resulted in the high number of focal CNAs identified in our data set.
Our study has several limitations. SNP array analysis was performed on bone marrow aspirates at diagnosis and remission, with those at remission serving as controls. Residual CNAs in samples from remission might have prevented the identification of CNAs in the matched tumor remission sample analysis, because nonhematopoietic germline DNA was not available. Furthermore, although the DNA samples passed quality-control standards for inferring genotype calls, a manual inspection process was still required to limit the number of false-positive CNA calls. Further work will be required to integrate these results with rapidly evolving next-generation sequencing technologies, which are now being validated for the routine care of pediatric patients with AML at major centers.33
In conclusion, to our knowledge, this is the largest study using SNP genotyping arrays to examine CNAs in pediatric AML. Focal CNAs in AML frequently included losses in 7q36.1, 16p13.11, 9q21.32, 11p13, 2q37.1, 10p12.31, 11q23.3, 16q22.1, and 1p36.3 and gains in 17q24, 1q32, 3q28, 11q23, 6q27, 2q32.1, and 4q35.2; many of these loci have been associated with known structural aberrations in AML. CNAs were associated with survival for patients in the standard-risk group. If replicated in other data sets, CNA presence could be considered for inclusion in AML risk stratification. Future studies will require the integration of CNA data with next-generation sequencing data such as those recently published from the National Cancer Institute Therapeutically Applicable Research to Generate Effect Treatments Initiative.34
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE95691, GSE95496, and GSE95690).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Vani Shanker, senior scientific editor at St Jude Children’s Research Hospital, for editing the manuscript; the COG institutions and their principal investigators for their diligent efforts in completing this trial; and the patients and their families for their participation.
This work was supported by the National Institutes of Health, National Cancer Institute (grant 1R01CA133881) (M.V. and R.A.) and the Alex's Lemonade Stand Foundation (M.V. and R.A.).
Authorship
Contribution: R.A., T.A.A., R.B.G., B.H., S.R., A.S.G., and S.M. designed the research; M.V., E.F.A., R.E.R., E.K.G., Y.D., M.K., Y.-C.W., and L.S. performed the research; M.V. analyzed the data; and all authors contributed to writing the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Corresponding author: Richard Aplenc, Children’s Hospital of Philadelphia, 4018 CTRB, 3501 Civic Center Blvd, Philadelphia, PA 19104; e-mail: aplenc@e-mail.chop.edu.
References
Author notes
S.M. and R.A. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal