Key Points
Inhibition of RNA Pol I by CX-5461 treats aggressive AML and outperforms standard chemotherapy regimens.
CX-5461 induces p53-dependent apoptosis, p53-independent cell-cycle defects and differentiation, and reduces LICs.
Abstract
Despite the development of novel drugs, the prospects for many patients with acute myeloid leukemia (AML) remain dismal. This study reveals that the selective inhibitor of RNA polymerase I (Pol I) transcription, CX-5461, effectively treats aggressive AML, including mixed-lineage leukemia-driven AML, and outperforms standard chemotherapies. In addition to the previously characterized mechanism of action of CX-5461 (ie, the induction of p53-dependent apoptotic cell death), the inhibition of Pol I transcription also demonstrates potent efficacy in p53null AML in vivo. This significant survival advantage in both p53WT and p53null leukemic mice treated with CX-5461 is associated with activation of the checkpoint kinases 1/2, an aberrant G2/M cell-cycle progression and induction of myeloid differentiation of the leukemic blasts. The ability to target the leukemic-initiating cell population is thought to be essential for lasting therapeutic benefit. Most strikingly, the acute inhibition of Pol I transcription reduces both the leukemic granulocyte-macrophage progenitor and leukemia-initiating cell (LIC) populations, and suppresses their clonogenic capacity. This suggests that dysregulated Pol I transcription is essential for the maintenance of their leukemia-initiating potential. Together, these findings demonstrate the therapeutic utility of this new class of inhibitors to treat highly aggressive AML by targeting LICs.
Introduction
Acute myeloid leukemia (AML) is a clinically heterogeneous disease characterized by a multitude of gene mutations and chromosomal abnormalities, resulting in marked differences in responses and survival following chemotherapy. In particular, AML driven by translocations involving the mixed-lineage leukemia (MLL) KMT2A gene represent an aggressive subtype associated with early relapse following chemotherapy.1 MLL translocations occur in >70% of pediatric and >10% of adult AML, which are associated with an intermediate to unfavorable prognosis depending on the translocation partner and the presence of additional cytogenetic aberrations.2 New approaches targeting epigenetic regulators associated with the MLL-fusion protein complex, eg, bromodomain and extraterminal proteins and DOT1L histone methyltransferase, are currently being investigated in phase 1 clinical trials.3-5 However, it was recently reported that bromodomain and extraterminal protein inhibitors failed to target the leukemia-initiating cell (LIC) population, and thus drug resistance emerged.6 Consequently, there is still an urgent need for new therapies to treat these and other aggressive AML subtypes.
Here, we have tested the therapeutic efficacy of a novel inhibitor of RNA polymerase I (Pol I) transcription, CX-5461,7 in genetically modified mouse models of AML driven by MLL or AML1/ETO fusion proteins, and primary patient-derived xenograft (PDX) models. In both murine and human AML, CX-5461 demonstrated a remarkable single-agent efficacy. Unexpectedly, in addition to the previously characterized mechanism of action of CX-5461 involving activation of p53,8 we observed a p53-independent response involving phosphorylation of checkpoint kinase 1/2 (CHK 1/2) associated with a G2/M cell-cycle defect and induction of myeloid differentiation in leukemic blasts. Analysis of the hematopoietic compartment reveals that CX-5461 reduces the LIC population in p53 wild-type (WT) and null AML, thus decreasing the disease-initiating potential in vivo and their clonogenic capacity. Together, these studies suggest that Pol I transcription inhibition may represent a promising new approach to treat human AML by targeting the LIC independent of functional p53.
Experimental procedures
Animal work was approved by the Animal Ethics Committees at the Peter MacCallum Cancer Centre (E462), Australian National University (E2015/12), SA Pathology/Central Adelaide Local Health Network Animal Ethics Committee (#52/15), and Alfred Medical Research and Education Precinct (E/1563/2015/M). C57Bl/6 mice were purchased (Walter and Eliza Hall Institute or Australian Phenomics Facility) and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice were supplied in house. Human patient samples were obtained following informed patient consent and with ethical approval (project 12/17) from the Peter MacCallum Cancer Centre Tissue Bank, the Royal Adelaide Hospital (#041009), and the Alfred Hospital Human Research Ethics Committee (#459/12). CX-5461 and CX-5447 were provided by Cylene Pharmaceuticals; and KU-5593, VE-821, BMH-21, and actinomycin D (ActD) were purchased from Selleckchem or Sigma.
Animal transplantation and in vivo drug studies
Murine AML were generated by fetal liver transduction.9 For in vivo drug studies, 5 × 105 AML cells were IV injected into C57Bl/6 mice. Disease was monitored by bioluminescence imaging using an IVIS100 (Caliper LifeScience) and white blood cell (WBC) counts were measured using an Advia120 (Bayer Diagnostics). Kaplan-Meier survival studies were performed in disease-bearing mice treated with: CX-5461 or vehicle every 3 days via oral gavage, or cytarabine (5 days) in combination with doxorubicin (3 days) intraperitoneally.
For AML PDX, 1-5 × 106 cells were IV injected into irradiated NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice.10 Age- and sex-matched mice were treated with CX-5461 (40 mg/kg) or vehicle.
Tissue culture
MLL-AML cell lines were maintained in Anne-Kelso Dulbecco’s modified Eagle medium with 10% fetal bovine serum, 100 mM l-asparagine (Sigma), 55 μM β-mercaptoethanol, antibiotic/antimycotic, and glutamax (Gibco). M/E p53WT cell lines were cultured in the above media plus 6 ng/mL recombinant interleukin-3 (PeproTech). Human AML cell lines were obtained from the German Collection of Microorganisms and Cell Cultures and maintained in RPMI 1640 (Gibco) containing 10% to 20% fetal bovine serum, antibiotic/antimycotic, and glutamax.
RNA-fluorescence in situ hybridization (FISH)
Cells were fixed in paraformaldehyde, centrifuged onto charged slides, then hybridized to Cy3-tagged RNA probes complementary to the ribosomal RNA (rRNA) external transcribed spacer and internal transcribed spacer regions, designed using the Stellaris RNA FISH Probe Designer (Biosearch Technologies).
Orthophosphate labeling
2 × 106 AML cells were seeded 24 hours prior to treatment and pulsed with 500 μCi 32P for 30 minutes prior to RNA extraction using the RNeasy Kit (Qiagen). RNA was processed as described.8
Immunophenotyping, cell death, and cell-cycle analysis
Immunophenotype and leukemic granulocyte-macrophage progenitor population (L-GMP) analysis was performed as described6 (see supplemental Table 3, available on the Blood Web site). Propidium iodide (PI) or 4′,6-diamidino-2-phenylindole (DAPI) was added as cell viability stains.
Cell death assays were performed in 96-well plates with 1 μg/mL PI incubated for 15 minutes at room temperature, and analyzed using the BD FACSVerse cytometer. Cell-cycle distribution was analyzed via 5-bromo-2′-deoxyuridine (BrdU) incorporation. Apoptotic cell death was analyzed by Annexin V/PI staining as described.8
Clonogenic assays in methylcellulose
Colony formation of primary patient AML or green fluorescent protein-positive (GFP+)-murine tumor cells was analyzed in methylcellulose (human M4435 and mouse M3434; Stemcell Technologies) as described.6
Histology, terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL), and May-Grünwald Giemsa staining
Tissues were fixed in 10% neutral buffered formalin, femurs decalcified, and paraffin wax embedded and cut (4 μm sections). Sections were stained with hematoxylin and eosin and TUNEL performed. GFP+-sorted cells were cytospun (2 minutes, 800 rpm), air-dried, and stained with May-Grünwald Giemsa (Grale Scientific). Slides were analyzed using an Olympus BX-61 and images were captured using SPOT Advanced software.
Immunoblotting
Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to immobilon-P (Millipore), blocked with 5% nonfat milk in Tris-buffered saline/0.1% Tween-20 at room temperature, and incubated with primary (supplemental Table 3) then secondary antibodies before detection by enhanced chemiluminescence (GE Healthcare).
Quantitative real-time polymerase chain reaction
RNA was isolated per manufacturer’s instructions (Bioline or Qiagen). Complementary DNA was synthesized using Superscript III (Invitrogen) and random hexamer primers, and quantitative real-time polymerase chain reaction was performed using Fast SYBR Green Master Mix (Applied Biosystems), 100 μmol/L primer on a StepOnePlus Real-time PCR System (Applied Biosystems). Computed tomography values were determined, normalized to housekeeping genes, and fold changes in expression calculated using the 2–ΔΔ computed tomography method. Primer sequences are listed in supplemental Table 4.
RNA sequencing
Sequencing libraries were prepared using the TruSeq RNA Kit (Illumina) and sequenced on an Illumina Genome Analyzer IIx. A total of 50 bp paired-end reads were aligned to the genome using TopHat version 2.0.8b and counted using HTSeq. Differential expression was calculated utilizing the EdgeR package in R (version 3.0.2). Absolute gene expression was defined by determining reads per kilobase per million.11 Pathway analysis was performed using GeneGo.
Statistical tests
Unpaired Student t test and log-rank (Mantel-Cox) test were performed using GraphPad Prism software (version 6.0f).
Results
AML cells are highly sensitive to inhibition of Pol I transcription
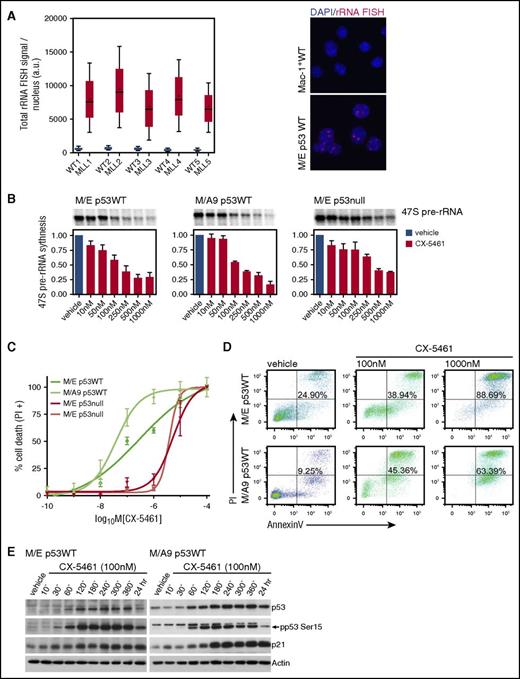
RNA Pol I transcription is upregulated in malignant cells compared with normal myeloid cells, suggesting that these cells might be highly sensitive to drugs that target this process (Figure 1A). We examined the therapeutic potential of the selective Pol I transcription inhibitor CX-54617,8 to treat culture-adapted p53WT and p53null AML cells expressing GFP-tagged MLL fusion proteins MLL/ENL or MLL/AF9 (hereafter termed M/E or M/A9), both together with luciferase-tagged oncogenic NrasG12D.9 Both p53WT and p53null MLL-rearranged leukemia exhibited a dose-dependent reduction in 47S pre-rRNA synthesis after 1 hour of CX-5461 treatment (Figure 1B; supplemental Figure 1A), and cell death was induced (Figure 1C). However, the IC50 for the p53WT cells was in a low nanomolar range (M/A9 37 nM; M/E 280 nM) compared with p53null cells (5328 nM; 3800 nM). p53WT M/E and M/A9 AML apoptosis (Figure 1D) was associated with elevated p53 protein and serine 15 (S15) phosphorylation, and induction of its downstream target p21 (Figure 1E; supplemental Figure 1B). These results are consistent with hyperactivation of Pol I transcription during malignant transformation and activation of the nucleolar stress pathway induced by the disruption of ribosomal DNA (rDNA) transcription.8,12,13
MLL-driven leukemias are sensitive to inhibition of hyperactivated Pol I transcription in vitro. (A) Pol I transcription is upregulated in malignant myeloid (M/E, GFP+-sorted) cells compared with normal myeloid (Mac-1+) cells from the bone marrow (BM) of C57Bl/6 mice. rRNA FISH with DAPI counterstain in malignant myeloid (GFP+) cells compared with normal myeloid (Mac-1+) cells. Images were taken with a ×63 objective. (B) Culture-adapted MLL-driven AML cells (M/E and M/A9) (M/E either p53WT or null) were treated with vehicle or CX-5461. The cells were treated with increasing concentration of CX-5461 for 1 hour, and 47S precursor rRNA synthesis was analyzed by 32P-orthophosphate labeling (average IC50 of 100 to 200 nM in p53WT and 400 to 500 nM in p53null AML). Graph represents mean ± SEM; n = 3. (C) Cell death was determined by PI incorporation after 24 hours of CX-5461 treatment (M/E IC50 = 280 nM; M/A9 IC50 = 37 nM; M/E p53null IC50 = 5328 nM; and M/E p53null IC50 = 3800 nM). Graph represents mean ± SEM; n = 3. (D) Apoptotic cell death was analyzed by Annexin V/PI staining using flow cytometry, after cells were treated with 100 nM and 1000 nM CX-5461 for 24 hours. Representative dot plots from n = 3. (E) Western blot analysis of total p53 and p21 protein after M/E and M/A9 p53WT cells were treated with 100 nM CX-5461 for the times indicated. Representative western blot from n = 3. a.u., arbitrary units; IC50, 50% inhibitory concentration; SEM, standard error of the mean.
MLL-driven leukemias are sensitive to inhibition of hyperactivated Pol I transcription in vitro. (A) Pol I transcription is upregulated in malignant myeloid (M/E, GFP+-sorted) cells compared with normal myeloid (Mac-1+) cells from the bone marrow (BM) of C57Bl/6 mice. rRNA FISH with DAPI counterstain in malignant myeloid (GFP+) cells compared with normal myeloid (Mac-1+) cells. Images were taken with a ×63 objective. (B) Culture-adapted MLL-driven AML cells (M/E and M/A9) (M/E either p53WT or null) were treated with vehicle or CX-5461. The cells were treated with increasing concentration of CX-5461 for 1 hour, and 47S precursor rRNA synthesis was analyzed by 32P-orthophosphate labeling (average IC50 of 100 to 200 nM in p53WT and 400 to 500 nM in p53null AML). Graph represents mean ± SEM; n = 3. (C) Cell death was determined by PI incorporation after 24 hours of CX-5461 treatment (M/E IC50 = 280 nM; M/A9 IC50 = 37 nM; M/E p53null IC50 = 5328 nM; and M/E p53null IC50 = 3800 nM). Graph represents mean ± SEM; n = 3. (D) Apoptotic cell death was analyzed by Annexin V/PI staining using flow cytometry, after cells were treated with 100 nM and 1000 nM CX-5461 for 24 hours. Representative dot plots from n = 3. (E) Western blot analysis of total p53 and p21 protein after M/E and M/A9 p53WT cells were treated with 100 nM CX-5461 for the times indicated. Representative western blot from n = 3. a.u., arbitrary units; IC50, 50% inhibitory concentration; SEM, standard error of the mean.
CX-5461 therapy has significant therapeutic efficacy in aggressive AML in vivo
To determine whether the sensitivity of AML cells to CX-5461 in vitro could be harnessed to treat AML-bearing mice, we transplanted syngeneic recipient mice with p53WT leukemia expressing either M/E or M/A9 and NrasG12D.
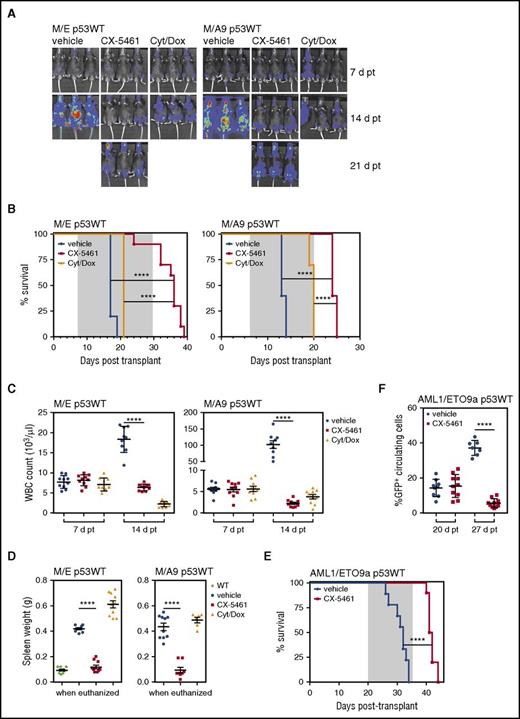
Continuous CX-5461 administration significantly delayed disease progression (Figure 2A) and prolonged survival in p53WT MLL-AML (Figure 2B; supplemental Figure 2). Strikingly, CX-5461 significantly prolonged survival compared with standard treatment of cytarabine/doxorubicin (Figure 2B).9 The therapeutic response to CX-5461 was associated with a non-elevated WBC count at 14 days posttransplant (3 doses) (Figure 2C). However, eventually the mice succumbed to disease with leukemia accumulating both in the head and abdominal region, although interestingly, extramedullary disease was still profoundly reduced, as evident by the absence of splenomegaly (Figure 2A,D). CX-5461 therapy in a model of a different genetic AML subtype, AML1/ETO9a NrasG12D p53WT (AML/ETO),9 also significantly prolonged survival and decreased circulating disease (Figure 2E-F), demonstrating that the therapeutic response was not restricted to MLL-driven AML. The same dosing regimen in nondiseased mice resulted in some myelosuppression that was reversible (supplemental Figure 3A-D), and was not associated with loss of body weight (supplemental Figure 3D). Thus, CX-5461 has robust therapeutic efficacy associated with extended survival in murine models of AML, and is superior to the standard therapy used to treat AML patients.
CX-5461 therapy delays disease progression and significantly extends survival in leukemic mice. (A) Leukemia progression and engraftment of M/E p53WT and M/A9 p53WT AML in recipient C57Bl/6 mice was followed by bioluminescence imaging at day 7 (7 d, baseline), 14 (14 d), and 21 (21 d) posttransplant (pt). Representative images of n = 10 per group. (B) Kaplan-Meier survival curves for CX-5461 (40 mg/kg every 3 days, start of therapy day 7 pt, last dose day 28 pt in M/E p53WT, and last dose day 20 pt in M/A9 p53WT); cytarabine/doxorubicin (5 days of 50 mg/kg cytarabine in combination with the first 3 days of 1.5 mg/kg doxorubicin, start of therapy day 7 pt, and last dose day 11 pt) and vehicle-treated leukemic mice (M/E p53WT median survival, 17 days for vehicle vs 36 days for CX-5461, P < .0001; M/A9 p53WT median survival, 13 days for vehicle vs 24 days for CX-5461, ****P < .0001; n = 8 to 10 per group). (C) WBC prior to therapy initiation and after 3 doses of CX-5461 treatment (****P < .0001; n = 8 to 10 per group). (D) Spleen weights of mice when euthanized (****P < .0001; n = 8 to 10 per group; WT nonmalignant). (E) Overall survival of CX-5461 (40 mg/kg every 3 days, 6 doses total, start of therapy day 20 pt, and last dose day 35 pt) and vehicle-treated AML1/ETO9a Nras p53WT leukemia-bearing mice (****P < .0001; n = 10 per group). (F) GFP+ circulating tumor cells prior to therapy and after 3 doses of treatment in the peripheral blood of leukemic mice (AML1/ETO9a Nras p53WT (****P < .0001; n = 7 to 10 per group). Gray indicates time of CX-5461 and vehicle treatment (B,E). Log-rank test (B,E) and unpaired 2-tailed Student t test were performed (C,D,F). Graphs represent mean ± SEM. Cyt, cytarabine; d, day; Dox, doxorubicin.
CX-5461 therapy delays disease progression and significantly extends survival in leukemic mice. (A) Leukemia progression and engraftment of M/E p53WT and M/A9 p53WT AML in recipient C57Bl/6 mice was followed by bioluminescence imaging at day 7 (7 d, baseline), 14 (14 d), and 21 (21 d) posttransplant (pt). Representative images of n = 10 per group. (B) Kaplan-Meier survival curves for CX-5461 (40 mg/kg every 3 days, start of therapy day 7 pt, last dose day 28 pt in M/E p53WT, and last dose day 20 pt in M/A9 p53WT); cytarabine/doxorubicin (5 days of 50 mg/kg cytarabine in combination with the first 3 days of 1.5 mg/kg doxorubicin, start of therapy day 7 pt, and last dose day 11 pt) and vehicle-treated leukemic mice (M/E p53WT median survival, 17 days for vehicle vs 36 days for CX-5461, P < .0001; M/A9 p53WT median survival, 13 days for vehicle vs 24 days for CX-5461, ****P < .0001; n = 8 to 10 per group). (C) WBC prior to therapy initiation and after 3 doses of CX-5461 treatment (****P < .0001; n = 8 to 10 per group). (D) Spleen weights of mice when euthanized (****P < .0001; n = 8 to 10 per group; WT nonmalignant). (E) Overall survival of CX-5461 (40 mg/kg every 3 days, 6 doses total, start of therapy day 20 pt, and last dose day 35 pt) and vehicle-treated AML1/ETO9a Nras p53WT leukemia-bearing mice (****P < .0001; n = 10 per group). (F) GFP+ circulating tumor cells prior to therapy and after 3 doses of treatment in the peripheral blood of leukemic mice (AML1/ETO9a Nras p53WT (****P < .0001; n = 7 to 10 per group). Gray indicates time of CX-5461 and vehicle treatment (B,E). Log-rank test (B,E) and unpaired 2-tailed Student t test were performed (C,D,F). Graphs represent mean ± SEM. Cyt, cytarabine; d, day; Dox, doxorubicin.
CX-5461 therapy has significant therapeutic efficacy in p53-deficient MLL-driven AML in vivo
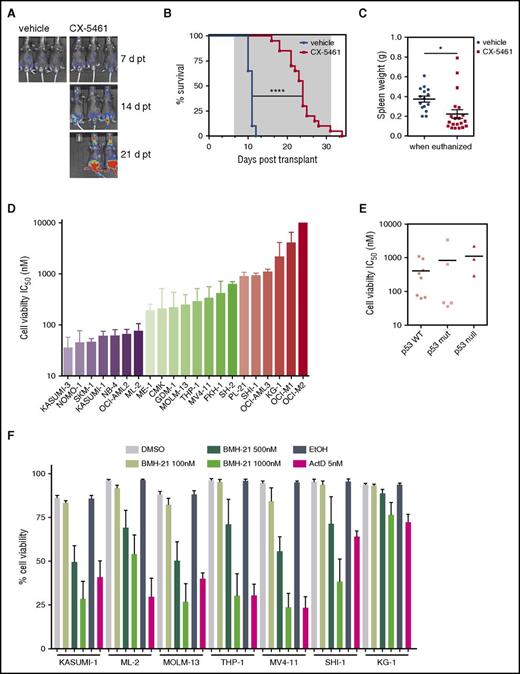
We examined the efficacy of CX-5461 therapy in p53null M/E leukemia. Surprisingly, despite the differences in sensitivity with respect to the induction of cell death in vitro (Figure 1B), inhibition of Pol I transcription in p53null leukemia-bearing mice markedly delayed tumor progression and significantly prolonged survival (Figure 3A-B). Consistent with findings in p53WT AML, CX-5461–treated mice showed reduced splenomegaly (Figure 3C). These results provide compelling evidence that the therapeutic benefit of CX-5461 is mediated through pathways in addition to those downstream of p53.
CX-5461 exhibits therapeutic potential in mouse and human AML independent of their p53 status. (A) Leukemia progression and engraftment of M/E p53null AML in recipient C57Bl/6 mice was followed by bioluminescence imaging at day 7 (7 d) prior to initiation of treatment, 14 (14 d), and 21 (21 d) posttransplant (pt). Representative images from n = 20 per group. (B) Kaplan-Meier survival curves for CX-5461 (35 mg/kg every 3 days, start of therapy day 7 pt, and last dose day 31 pt) and vehicle-treated leukemic mice (median survival, 11 days for vehicle vs 24 days for CX-5461; ****P < .0001; n = 20 per group). Gray indicates time of CX-5461 and vehicle treatment. (C) Spleen weight of mice when euthanized (*P = .0132; n = 20 per group). Log-rank test (B) and unpaired 2-tailed Student t test (C) were performed. Graphs represent mean ± SEM. (D) Human leukemia cell lines differ in their sensitivity to Pol I inhibition 48 hours after drug treatment as assessed by PI exclusion (cell viability). IC50 value, the concentration of CX-5461 that decreases cell viability by 50% compared to the control, was calculated using a three-parameter log vs the inhibition nonlinear regression method in GraphPad Prism software. AML cells are graphed in groups: IC50 <100 nM (blue), IC50 100 nM to 1000 nM (green), and IC50 >1000 nM (red). IC50 values are expressed as the best-fit values for at least n = 3. Graph represents mean ± SEM. (E) The average IC50 for CX-5461 does not correlate with p53 status according to an unpaired 2-tailed Student t test. TP53 mutation status was provided by the Cancer Cell Line Encyclopedia and the Catalogue of Somatic Mutations in Cancer. Only cell lines with known p53 status via these databases are shown in the figure. (F) Cell viability in response to BMH-21 (100 nM, 500 nM, and 1000 nM) and ActD (5 nM) in KASUMI-1, ML-2, MOLM-13, THP-1, MV4-11, SHI-1, and KG-1 was determined by PI exclusion after 48 hours of treatment. DMSO, dimethyl sulfoxide.
CX-5461 exhibits therapeutic potential in mouse and human AML independent of their p53 status. (A) Leukemia progression and engraftment of M/E p53null AML in recipient C57Bl/6 mice was followed by bioluminescence imaging at day 7 (7 d) prior to initiation of treatment, 14 (14 d), and 21 (21 d) posttransplant (pt). Representative images from n = 20 per group. (B) Kaplan-Meier survival curves for CX-5461 (35 mg/kg every 3 days, start of therapy day 7 pt, and last dose day 31 pt) and vehicle-treated leukemic mice (median survival, 11 days for vehicle vs 24 days for CX-5461; ****P < .0001; n = 20 per group). Gray indicates time of CX-5461 and vehicle treatment. (C) Spleen weight of mice when euthanized (*P = .0132; n = 20 per group). Log-rank test (B) and unpaired 2-tailed Student t test (C) were performed. Graphs represent mean ± SEM. (D) Human leukemia cell lines differ in their sensitivity to Pol I inhibition 48 hours after drug treatment as assessed by PI exclusion (cell viability). IC50 value, the concentration of CX-5461 that decreases cell viability by 50% compared to the control, was calculated using a three-parameter log vs the inhibition nonlinear regression method in GraphPad Prism software. AML cells are graphed in groups: IC50 <100 nM (blue), IC50 100 nM to 1000 nM (green), and IC50 >1000 nM (red). IC50 values are expressed as the best-fit values for at least n = 3. Graph represents mean ± SEM. (E) The average IC50 for CX-5461 does not correlate with p53 status according to an unpaired 2-tailed Student t test. TP53 mutation status was provided by the Cancer Cell Line Encyclopedia and the Catalogue of Somatic Mutations in Cancer. Only cell lines with known p53 status via these databases are shown in the figure. (F) Cell viability in response to BMH-21 (100 nM, 500 nM, and 1000 nM) and ActD (5 nM) in KASUMI-1, ML-2, MOLM-13, THP-1, MV4-11, SHI-1, and KG-1 was determined by PI exclusion after 48 hours of treatment. DMSO, dimethyl sulfoxide.
To investigate whether these findings translate to human AML, we tested a panel of human AML cell lines for CX-5461 sensitivity in vitro. Although the average IC50 assessed by cell viability at 48 hours posttreatment differs greatly among the human cell lines (IC50, 45 nM to 2182 nM; average IC50, 506 nM), there was no correlation with functional p53 (Figure 3D-E; supplemental Table 1). Thus, like p53null M/E, CX-5461 had robust activity in human AML with loss of p53 function. Moreover, the sensitivity was not restricted to MLL-driven AML. To support our findings that the antileukemic effect of CX-5461 is due to on-target inhibition of Pol I transcription, 7 AML cell lines were tested for their sensitivity to 2 additional Pol I inhibitors, BMH-2114 and low-dose ActD,15 and an inactive compound (CX-5447), which is structurally similar to CX-5461.7,16 Both BMH-21 and ActD decreased cell viability after 48 hours in 7 AML cell lines with the level of sensitivity broadly correlating with that of CX-5461 (Figure 3F). As expected, CX-5447 at equivalent concentrations to efficacious doses of CX-5461 did not decrease cell viability in any of the 7 AML cell lines (supplemental Figure 4A), nor altered Pol I transcription (supplemental Figure 4B; representative cell lines SHI-1 and KG-1 shown). Together, these findings demonstrate that specific inhibition of Pol I transcription is a promising therapeutic target for the treatment of AML.
Acute inhibition of rDNA transcription reduces leukemic burden in M/E AML in vivo
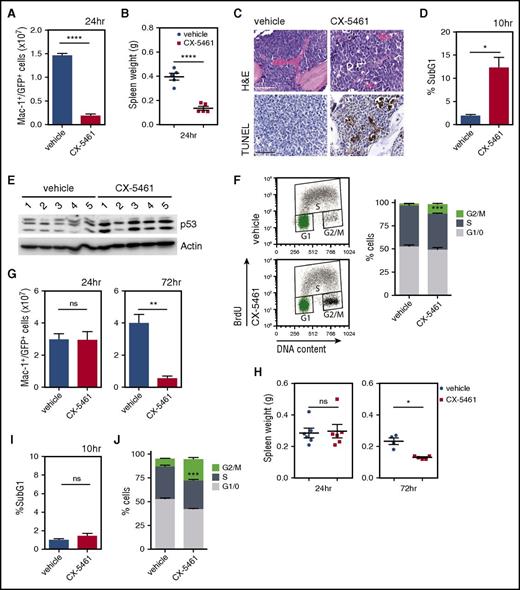
To understand mechanistically how CX-5461 treats AML in vivo, we characterized the immediate (1 to 72 hour) cellular response to CX-5461 in leukemic mice. A significant reduction in the rate of rDNA transcription was confirmed at 1 hour posttreatment in the peripheral blood (supplemental Figure 5A). A single dose of CX-5461 significantly reduced the total number of Mac-1+/GFP+ double-positive tumor cells within the BM of p53WT M/E leukemic mice after 24 hours (Figure 4A). Moreover, acute inhibition of Pol I transcription was associated with a rapid decrease in spleen size to within a normal range (Figure 4B). This rapid reduction in medullary and extramedullary disease was associated with cell death 10 to 12 hours posttreatment, and upregulation of total p53 protein in CX-5461–treated mice (Figure 4C-E). Importantly, CX-5461 administration did not induce a p53 response in the Mac-1+/Gr-1+ cells isolated from the BM of nondisease-bearing mice (supplemental Figure 3E), attesting to the tumor specificity of the therapeutic response to Pol I inhibition. The drug-induced reduction in leukemic burden was also associated with a population of nonapoptotic tumor cells with a significant population accumulating in the G2/M phase of the cell cycle (Figure 4F). Comparable levels of tumor cell death, p53 total protein induction, and G2/M cell-cycle delay was observed in CX-5461–treated p53WT M/A9 leukemic mice (supplemental Figure 6A-F). Thus, CX-5461 treatment of p53WT AML-bearing mice induced a rapid tumor cell response characterized by both cell death and aberrant cell-cycle progression.
A single CX-5461 administration reduces the tumor burden in M/E leukemic mice. M/E p53WT AML was transplanted into recipient C57Bl/6 mice (A-E). (A) Mac-1+/GFP+ double-positive tumor cells within the BM (1.45 × 107 ± 0.05 cells for vehicle vs 0.18 × 107 ± 0.04 cells for CX-5461; ****P < .0001; n = 5) and (B) spleen weights were determined after 24 hours of CX-5461 treatment (40 mg/kg) (****P < .0001; n = 5). (C) Analysis of apoptotic cell death via TUNEL staining of femoral BM sections from mice treated for 12 hours and stained with hematoxylin and eosin (H&E). Sections shown are representative of n = 3. Scale bar represents 50 μm (D) Cell death was determined by examining the SubG1 DNA content in the BM 10 hours post-drug administration (12.2% ± 2.2 for CX-5461 vs 1.9% ± 0.3 for vehicle; *P = .0102; n = 5). (E) Total p53 protein induction in response to CX-5461 single-dose treatment in the BM of M/E leukemic mice (n = 5 per group). (F) Quantitation of cell-cycle distribution in the BM cells by BrdU incorporation 24 hours post–CX-5461 treatment (***P = .0005; n = 5). Graph represents mean ± SEM. M/E p53null AML was injected into recipient C57Bl/6 mice (G-J). (G) The total number of Mac1+/GFP+ double-positive tumor cells within the BM (3.2 × 107 ± 0.2 cells for vehicle vs 3.1 × 107 ± 0.3 cells for CX-5461 at 24 hours, and 3.9 × 107 ± 0.5 cells for vehicle and 0.5 × 107 ± 0.2 cells for CX-5461 at 72 hours; **P = .0053), and (H) spleen weights (*P = .0134; n = 4 to 6) were determined after CX-5461 administration (35 mg/kg). Graphs represent mean ± SEM. Cell death (I) was analyzed by determining SubG1 DNA content in the BM 10 hours post–CX-5461 treatment (n = 6). Quantitation of cell-cycle distribution (J) in the BM cell was analyzed by BrdU incorporation 24 hours post–CX-5461 administration (***P = .0007). Graphs represent mean ± SEM (n = 5). In all cases, an unpaired 2-tailed Student t test was used. n.s., not significant.
A single CX-5461 administration reduces the tumor burden in M/E leukemic mice. M/E p53WT AML was transplanted into recipient C57Bl/6 mice (A-E). (A) Mac-1+/GFP+ double-positive tumor cells within the BM (1.45 × 107 ± 0.05 cells for vehicle vs 0.18 × 107 ± 0.04 cells for CX-5461; ****P < .0001; n = 5) and (B) spleen weights were determined after 24 hours of CX-5461 treatment (40 mg/kg) (****P < .0001; n = 5). (C) Analysis of apoptotic cell death via TUNEL staining of femoral BM sections from mice treated for 12 hours and stained with hematoxylin and eosin (H&E). Sections shown are representative of n = 3. Scale bar represents 50 μm (D) Cell death was determined by examining the SubG1 DNA content in the BM 10 hours post-drug administration (12.2% ± 2.2 for CX-5461 vs 1.9% ± 0.3 for vehicle; *P = .0102; n = 5). (E) Total p53 protein induction in response to CX-5461 single-dose treatment in the BM of M/E leukemic mice (n = 5 per group). (F) Quantitation of cell-cycle distribution in the BM cells by BrdU incorporation 24 hours post–CX-5461 treatment (***P = .0005; n = 5). Graph represents mean ± SEM. M/E p53null AML was injected into recipient C57Bl/6 mice (G-J). (G) The total number of Mac1+/GFP+ double-positive tumor cells within the BM (3.2 × 107 ± 0.2 cells for vehicle vs 3.1 × 107 ± 0.3 cells for CX-5461 at 24 hours, and 3.9 × 107 ± 0.5 cells for vehicle and 0.5 × 107 ± 0.2 cells for CX-5461 at 72 hours; **P = .0053), and (H) spleen weights (*P = .0134; n = 4 to 6) were determined after CX-5461 administration (35 mg/kg). Graphs represent mean ± SEM. Cell death (I) was analyzed by determining SubG1 DNA content in the BM 10 hours post–CX-5461 treatment (n = 6). Quantitation of cell-cycle distribution (J) in the BM cell was analyzed by BrdU incorporation 24 hours post–CX-5461 administration (***P = .0007). Graphs represent mean ± SEM (n = 5). In all cases, an unpaired 2-tailed Student t test was used. n.s., not significant.
In contrast to p53WT AML, CX-5461 treatment of p53null leukemia did not immediately reduce medullary and extramedullary disease as assessed by the total number of tumor cells within the BM and spleen weight 24 hours posttreatment (Figure 4G-H). However by 72 hours, all mice displayed a significant decrease in their leukemic load in the absence of cell death, as assessed by SubG1 DNA content analysis (Figure 4G-I; supplemental Figure 7A), consistent with an unchanged BM cellularity at 10 and 24 hours posttreatment (supplemental Figure 7B). Significantly, p53null leukemia displayed a robust G2/M cell-cycle delay following CX-5461 treatment, similar to that observed in p53WT AML (Figure 4J). This suggests that aberrant cell-cycle progression in the tumor cell population contributed to the prolonged survival of mice bearing p53null M/E following CX-5461 therapy (Figure 3B).
CX-5461 activates p53-independent cell-cycle checkpoint signaling
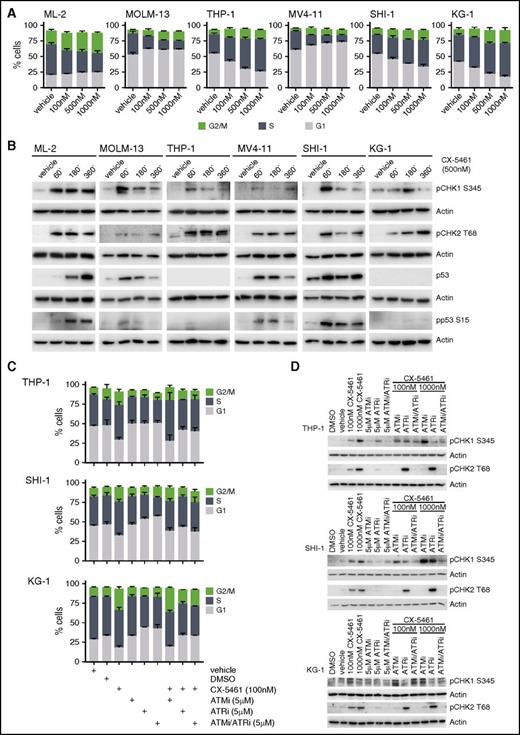
In order to further validate signaling pathways activated in response to Pol I inhibition, in addition to p53, we analyzed cell-cycle progression in human AML cell lines with a different p53 status. THP-1 (p53null), SHI-1 (p53WT), and KG-1 (p53null) exhibited aberrant cell-cycle progression in response to CX-5461 with cells accumulating in late S and G2/M phase (Figure 5A). In line with our recent study demonstrating CX-5461–mediated activation of ataxia telangiectasia-mutated and Rad31-related kinase (ATM/ATR) signaling in the absence of global DNA damage in fibroblasts,17 and consistent with aberrant G2/M transition, Pol I inhibition induced phosphorylation of the CHK1 on S345 and CHK2 threonine 68, which are ATM-/ATR-dependent sites, respectively (Figure 5B). Moreover, pharmacologic inhibition of ATM/ATR signaling partially rescued the delay in late S and the G2/M phase arrest in THP-1, SHI-1, and KG-1 cell lines (Figure 5C-D). Taken together, these data strongly suggest that inhibition of Pol I transcription exhibits a robust therapeutic efficacy in the absence of functional p53 mediated, at least in part, through activation of ATM/ATR-CHK1/2 signaling and a subsequent S- and G2/M progression defect.
Pol I inhibition activates ATM/ATR-dependent signaling and alters cell-cycle progression in human AML cell lines. (A) Cell-cycle analysis by BrdU incorporation after 24 hours of Pol I inhibition (n = 4). (B) Western blot analysis of CHK1 S345, CHK2 T68, and pp53 S15 phosphorylation, and total p53 abundance in 6 human AML cell lines with varied p53 status after treatment with 500 nM CX-5461 for the times indicated (n = 2). (C) Quantitation of cell-cycle distribution by BrdU incorporation in SHI-1, KG-1, and THP-1 after 30 minutes of pre-treatment with either ATMi (5 μM KU-5593), ATRi (5 μM VE-821), or both ATMi/ATRi (5 μM) in combination with CX-5461 (100 nM) treatment of 24 hours (n = 3). (D) Analysis of CHK1 S345, and CHK2 T68 phosphorylation in THP-1 and SHI-1 after 30 minutes pre-treatment with either ATMi (5 μM KU-5593), ATRi (5 μM VE-821), or both ATMi/ATRi (5 μM) in combination with 100 nM or 1000 nM CX-5461 for 1 hour by western blotting (n = 2). Graphs represent mean ± SEM.
Pol I inhibition activates ATM/ATR-dependent signaling and alters cell-cycle progression in human AML cell lines. (A) Cell-cycle analysis by BrdU incorporation after 24 hours of Pol I inhibition (n = 4). (B) Western blot analysis of CHK1 S345, CHK2 T68, and pp53 S15 phosphorylation, and total p53 abundance in 6 human AML cell lines with varied p53 status after treatment with 500 nM CX-5461 for the times indicated (n = 2). (C) Quantitation of cell-cycle distribution by BrdU incorporation in SHI-1, KG-1, and THP-1 after 30 minutes of pre-treatment with either ATMi (5 μM KU-5593), ATRi (5 μM VE-821), or both ATMi/ATRi (5 μM) in combination with CX-5461 (100 nM) treatment of 24 hours (n = 3). (D) Analysis of CHK1 S345, and CHK2 T68 phosphorylation in THP-1 and SHI-1 after 30 minutes pre-treatment with either ATMi (5 μM KU-5593), ATRi (5 μM VE-821), or both ATMi/ATRi (5 μM) in combination with 100 nM or 1000 nM CX-5461 for 1 hour by western blotting (n = 2). Graphs represent mean ± SEM.
CX-5461 induces myeloid differentiation
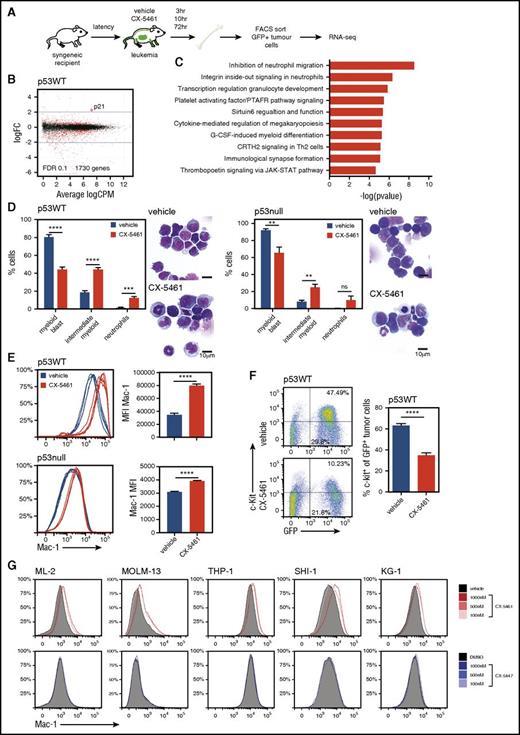
To further explore the molecular response induced by CX-5461, we used RNA deep sequencing to analyze the global messenger RNA transcriptome signature. GFP+ tumor cells were isolated from the BM of vehicle and CX-5461–treated p53WT or p53null M/E leukemic mice (Figure 6A). Using a false discovery rate of 0.1, we observed no significant changes in messenger RNA abundance at 3 hours post-drug treatment (M/E p53WT, data not shown), attesting to the selectivity of CX-5461 to inhibit Pol I and not Pol II transcription. After 10 hours of treatment, we identified 1730 differentially expressed Pol II-regulated genes in p53WT AML, presumably as an indirect response to inhibition of Pol I transcription (Figure 6B). Functional ontology enrichment analysis (GeneGo) revealed in p53WT M/E gene signatures involved in neutrophil migration, transcriptional regulation of granulocyte development and myeloid differentiation, which were not differentially expressed in p53null M/E at 10 hours (Figure 6C; supplemental Figure 8A,C-E). Genes upregulated in response to CX-5461 in p53WT M/E also included p53 target genes associated with apoptosis and cell-cycle regulation (p21, MDM2, Puma, and Bax), which were validated by quantitative real-time polymerase chain reaction in vitro (supplemental Figure 8B).
Transcriptome analysis of M/E p53WT AML treated with CX-5461 by RNA sequencing revealed induction of myeloid maturation. (A) Schematic diagram of the experimental design (n = 3 mice per group). M/E p53WT AML were treated with a single dose of CX-5461 (40 mg/kg). (B) Scatter-plots for p53WT M/E illustrating logFC over CPM at 10 hours, and (C) GeneGo pathway analysis. (D) M/E p53WT or p53null AML were engrafted into recipient C57Bl/6 mice and treated with CX-5461 for 48 hours. Differential cell classification was performed on May-Grünwald Giemsa-stained cytospins prepared from GFP+-sorted BM (****P < .0001, ***P = .0008, **P = .0066; n.s. P = .0036; n = 5). Graphs represent mean ± SEM. Insert is a representative image from 1 mouse per group. Scale bar represents 10 μm. (E) Expression of Mac-1, as determined by flow cytometry, in the BM-derived tumor cells (M/E p53WT and p53null, MFI, ****P < .0001; n = 5). (F) Representative flow cytometry dot plots from 1 mouse per group and quantitation of c-Kit expression in GFP+ tumor cells from the BM of M/E p53WT leukemic mice (****P < .0001; n = 5). (G) Mac-1 cell surface expression in KG-1, THP-1, ML-2, and MOLM-13 cells in response to CX-5461 or CX-5447 treatment after 24 hours (n = 3). Graph represents mean ± SEM. Unpaired 2-tailed Student t test (A,D) was used. CPM, counts per million; CRTH2, chemoattractant receptor-homologous molecule expressed on T helper type 2 cells; FACS, fluorescence-activated cell sorting; FDR, false discovery rate; G-CSF, granulocyte colony-stimulating factor; logFC, log fold-change; MFI, mean fluorescence intensity; n.s., not significant; PTAFR, platelet-activating factor receptor; RNA-seq, RNA sequencing; Th2, T helper 2.
Transcriptome analysis of M/E p53WT AML treated with CX-5461 by RNA sequencing revealed induction of myeloid maturation. (A) Schematic diagram of the experimental design (n = 3 mice per group). M/E p53WT AML were treated with a single dose of CX-5461 (40 mg/kg). (B) Scatter-plots for p53WT M/E illustrating logFC over CPM at 10 hours, and (C) GeneGo pathway analysis. (D) M/E p53WT or p53null AML were engrafted into recipient C57Bl/6 mice and treated with CX-5461 for 48 hours. Differential cell classification was performed on May-Grünwald Giemsa-stained cytospins prepared from GFP+-sorted BM (****P < .0001, ***P = .0008, **P = .0066; n.s. P = .0036; n = 5). Graphs represent mean ± SEM. Insert is a representative image from 1 mouse per group. Scale bar represents 10 μm. (E) Expression of Mac-1, as determined by flow cytometry, in the BM-derived tumor cells (M/E p53WT and p53null, MFI, ****P < .0001; n = 5). (F) Representative flow cytometry dot plots from 1 mouse per group and quantitation of c-Kit expression in GFP+ tumor cells from the BM of M/E p53WT leukemic mice (****P < .0001; n = 5). (G) Mac-1 cell surface expression in KG-1, THP-1, ML-2, and MOLM-13 cells in response to CX-5461 or CX-5447 treatment after 24 hours (n = 3). Graph represents mean ± SEM. Unpaired 2-tailed Student t test (A,D) was used. CPM, counts per million; CRTH2, chemoattractant receptor-homologous molecule expressed on T helper type 2 cells; FACS, fluorescence-activated cell sorting; FDR, false discovery rate; G-CSF, granulocyte colony-stimulating factor; logFC, log fold-change; MFI, mean fluorescence intensity; n.s., not significant; PTAFR, platelet-activating factor receptor; RNA-seq, RNA sequencing; Th2, T helper 2.
Because the inhibition of Pol I transcription altered the expression of genes involved in granulocyte development and myeloid differentiation (Figure 6C; supplemental Figure 8A), we examined the effect of CX-5461 on AML tumor cell differentiation. Strikingly, within 48 hours of treatment, CX-5461 induced morphologic and immunophenotypic changes independent of p53, characterized by a robust decrease in the number of leukemic myeloid blasts, a significant increase in an intermediate myeloid population (promyelocytes and myelocytes), and elevated neutrophils and increased expression of Mac-1, a myeloid lineage marker (Figure 6D-E). Moreover, CX-5461–treated p53WT AML exhibited a significantly lower c-Kit expression, a member of the tyrosine kinase receptor family expressed by hematopoietic progenitor cells (Figure 6F), further suggesting that inhibition of Pol I transcription promotes myeloid maturation in leukemic blasts. In p53null AML, c-Kit is expressed only on a small subpopulation, which did not change with treatment (data not shown). This myeloid differentiation phenotype is consistent with a gene signature observed in p53WT AML after 72 hours of CX-5461 treatment involved in granulocyte development (C/EBPalpha, ε, c-Myc, and Mxd1) (supplemental Figure 9A-C).
To confirm that Pol I inhibition induces myeloid differentiation rather than selectively killing malignant myeloid blasts, human AML cell lines were treated with CX-5461 or CX-5447 for 24 hours. Consistent with our in vivo findings, the inhibition of Pol I transcription increased Mac-1 expression in all AML cell lines tested, independent of the p53 status (Figure 6G) and in the absence of cell death. In contrast, the inactive compound CX-5447 did not alter Mac-1 expression (Figure 6G). Together, these data provide strong evidence that AML are reliant on elevated rates of Pol I transcription to maintain an immature phenotype, and its inhibition induces p53-independent differentiation in leukemic blasts.
CX-5461 reduces LICs and their clonogenic potential in murine and primary human AML
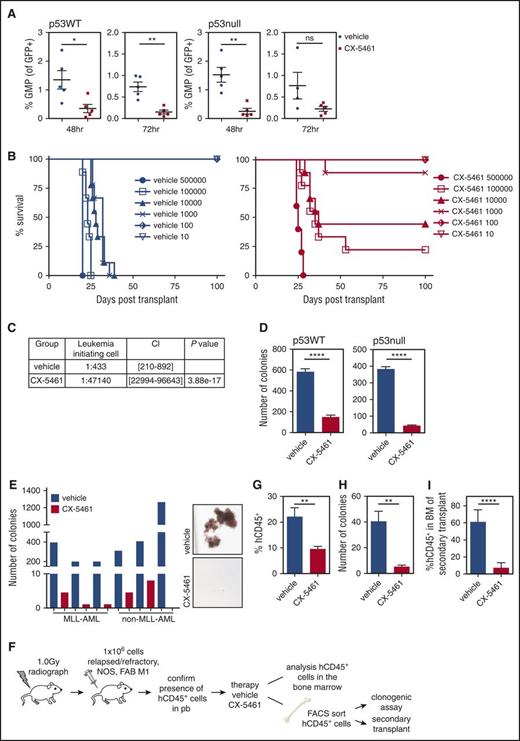
AML are thought to possess a population of cancer-initiating cells. In M/A9 mouse transplantation models, these LICs are a committed progenitor population characterized by aberrant self-renewal capacity and the ability to initiate disease.18 Although the relative frequency of LICs is still a matter of scientific discussion, it has been reported to account for ∼25% to 30% of the tumor at late-stage disease.19 Eradication of LICs is likely to be a critical element of successful AML therapy. The L-GMP is reported to contain a population of LICs.18 We examined the effect of Pol I transcription inhibition on the L-GMP population in p53WT and null M/E leukemia using a defined set of immunophenotypic markers (tumor cell-specific GFP+, lineage−, c-kit+, Sca-1−, FcγRII/III+, and CD34+; supplemental Figure 10A).6,18 In AML, a single dose of CX-5461 was sufficient to reduce the L-GMP population, suggesting that the extension in survival observed was, at least in part, caused by this reduction (Figure 7A). To assess whether CX-5461 targets the LICs and suppresses their clonogenic capacity, we performed a limiting dilution transplantation analysis where recipient mice were re-injected with a defined number of viable GFP+-sorted p53WT M/E tumor cells (range, 10,000 to 500 000 cells per mouse), isolated from disease-bearing mice treated with a single dose of CX-5461 or vehicle. Importantly, recipient mice transplanted with leukemic cells obtained from a CX-5461–treated mouse displayed delayed disease onset with a reduced frequency of tumor formation (Figure 7B).
CX-5461 reduces the L-GMP and LIC population in MLL-driven AML. M/E p53WT or p53null AML cells were engrafted into recipient C57Bl/6 mice and treated with CX-5461 for 48 or 72 hours (p53WT 40 mg/kg, p53null 35 mg/kg). (A) The L-GMP population (% of the GFP+ tumor cells that are GMP+) was determined by flow cytometry at 48 and 72 hours (p53WT: *P = .0224, **P = .0011, n = 5; p53null: **P = .0017; n = 4). (B) Serial dilution transplant experiment. M/E tumor cells from the BM of either a CX-5461 or vehicle-treated mouse were re-injected into recipient C57Bl/6 mice (500 000 cells: n = 5 per group; 10 to 100 000 cells: n = 9 per group) (500 000 cells vehicle vs CX-5461, P = .0027; 100 000 cells vehicle vs CX-5461, P < .0001; 10 000 cells vehicle vs CX-5461, P = .0078; and 1000 cells vehicle vs CX-5461, P < .0001). (C) Single-dose CX-5461 treatment reduces LIC frequency. (D) Drug treatment reduces the clonogenic capacity in methylcellulose (****P < .0001; n = 5). (E) CX-5461 (500 nM) reduces colony forming potential in primary human MLL and non-MLL rearranged AML. Representative image from 1 patient sample. (F) Schematic overview of the patient-derived AML (relapsed/refractory, NOS, FAB M1, and non-MLL) xenograft experiments. (G) CX-5461 (40 mg/kg) therapy (total of 6 doses) reduced the percentage of hCD45+ tumor cells in the BM of mice with established human disease (**P = .0075; n = 6). (H) Colony formation of sorted hCD45+ tumor cells is significantly reduced post–CX-5461 therapy ex vivo (**P = .0048; n = 4). (I) Delayed disease latency in secondary recipient transplanted with sorted CX-5461–treated hCD45+ tumor cells assessed by analysis of disease burden in the BM of secondary recipient (****P < .0001; n = 6). Unpaired 2-tailed Student t test. Graphs represent mean ± SEM. CI, confidence interval; FAB, French-American-British; FACS, fluorescence-activated cell sorting; NOS, not otherwise specified; n.s., not significant; pb, peripheral blood.
CX-5461 reduces the L-GMP and LIC population in MLL-driven AML. M/E p53WT or p53null AML cells were engrafted into recipient C57Bl/6 mice and treated with CX-5461 for 48 or 72 hours (p53WT 40 mg/kg, p53null 35 mg/kg). (A) The L-GMP population (% of the GFP+ tumor cells that are GMP+) was determined by flow cytometry at 48 and 72 hours (p53WT: *P = .0224, **P = .0011, n = 5; p53null: **P = .0017; n = 4). (B) Serial dilution transplant experiment. M/E tumor cells from the BM of either a CX-5461 or vehicle-treated mouse were re-injected into recipient C57Bl/6 mice (500 000 cells: n = 5 per group; 10 to 100 000 cells: n = 9 per group) (500 000 cells vehicle vs CX-5461, P = .0027; 100 000 cells vehicle vs CX-5461, P < .0001; 10 000 cells vehicle vs CX-5461, P = .0078; and 1000 cells vehicle vs CX-5461, P < .0001). (C) Single-dose CX-5461 treatment reduces LIC frequency. (D) Drug treatment reduces the clonogenic capacity in methylcellulose (****P < .0001; n = 5). (E) CX-5461 (500 nM) reduces colony forming potential in primary human MLL and non-MLL rearranged AML. Representative image from 1 patient sample. (F) Schematic overview of the patient-derived AML (relapsed/refractory, NOS, FAB M1, and non-MLL) xenograft experiments. (G) CX-5461 (40 mg/kg) therapy (total of 6 doses) reduced the percentage of hCD45+ tumor cells in the BM of mice with established human disease (**P = .0075; n = 6). (H) Colony formation of sorted hCD45+ tumor cells is significantly reduced post–CX-5461 therapy ex vivo (**P = .0048; n = 4). (I) Delayed disease latency in secondary recipient transplanted with sorted CX-5461–treated hCD45+ tumor cells assessed by analysis of disease burden in the BM of secondary recipient (****P < .0001; n = 6). Unpaired 2-tailed Student t test. Graphs represent mean ± SEM. CI, confidence interval; FAB, French-American-British; FACS, fluorescence-activated cell sorting; NOS, not otherwise specified; n.s., not significant; pb, peripheral blood.
This correlated with a reduced LIC frequency, ∼1:47 140 compared with 1:433 in drug-naïve M/E (Figure 7C).20 Secondary recipient mice injected with BM cells from CX-5461–treated mice demonstrated significantly increased survival when compared with secondary recipients injected with cells from vehicle-treated mice. Strikingly, the transplantation of 1000 drug-naïve cells led to 100% lethality in its recipients, whereas only 1 in 9 mice injected with drug-exposed cells developed AML (Figure 7B; supplemental Table 2). Together, these findings indicate that CX-5461 significantly reduces the frequency of LICs in CX-5461–treated mice compared with drug-naïve M/E (Figure 7C) mice.20 Consistent with these findings, ex vivo colony formation in M/E tumor cells was significantly reduced post–single-drug administration, regardless of p53 status (Figure 7D).
Testing the therapeutic potential in primary human MLL- and non-MLL AML revealed that CX-5461 (500 nM) almost completely abrogated colony formation independent of p53 activation (Figure 7E; supplemental Figure 10C). Most importantly, CX-5461 single-agent therapy in PDX AML significantly reduced the leukemic load (hCD45+) in the BM of engrafted mice (Figure 7F-G; supplemental Figure 11A). Moreover, hCD45+ BM cells isolated from CX-5461–treated mice had reduced clonogenic capacity (Figure 7H). Most remarkable however, CX-5461 depleted the LICs population in human AML efficiently, because recipient mice re-injected with sorted hCD45+ tumor cells from CX-5461–treated mice displayed a significant lower tumor burden (hCD45+) in their BM compared with those re-injected with vehicle-treated AML (Figure 7I). Together, these data significantly extend the earlier study of CX-5461 in a xenografted human cell line, MV4-11 AML,16 and provide compelling evidence of the therapeutic potential of pharmacologic inhibition of Pol I transcription in human AML, mediated in part by reducing the LIC population and suppression of the clonogenic potential.
Discussion
This study provides strong preclinical evidence that inhibition of Pol I transcription possesses significant therapeutic potential to treat aggressive AML, including MLL-rearranged AML. Most strikingly, using different mouse models of AML, CX-5461 significantly improved survival compared with standard cytotoxic therapy. Mechanistically, we show that pharmacologic inhibition of Pol I transcription robustly reduced clonogenic capacity of AML in vitro and LICs in vivo. These findings are of significant clinical interest because the failure of current therapies are attributed to inadequate elimination of LICs, which are suggested as the basis of acquired resistance and posttreatment relapse.19
One of the main processes thought to be responsible for the refractory nature of MLL-driven AML to standard chemotherapeutic regimens is the ability of MLL fusion-oncoproteins to attenuate the activation of p53 pathways.9,21 In contrast, we demonstrate that CX-5461 induced p53-mediated apoptosis in MLL-AML. Thus, the superior efficacy of CX-5461 in MLL-AML, compared with cytotoxic therapies is, at least in part, due to p53 activation in these tumors.
However, mice bearing p53null MLL-AML also exhibited increased survival with CX-5461, suggesting that Pol I inhibition reduced tumor cell survival through pathways other than p53-induced apoptosis. These observations are significant because although p53 mutations are rare at diagnosis (3% to 8% AML; 10% to 20% chronic lymphocytic leukemia), their frequency increases in relapsed AML, therapy-related AML, and therapy-related myelodysplastic syndrome (>35%).22,23 Moreover, AML with de novo p53 genetic alterations are associated with more aggressive disease and therapy resistance.24,25 Thus, therapies targeting Pol I transcription may represent a valuable new treatment option in the context of malignancies carrying de novo and acquired p53 mutations.
The increase in survival of p53null AML in response to CX-5461 was associated with activation of CHK1/2 signaling and accumulation of cells in the late S and G2/M phase. These data, together with our previous studies,8 demonstrate that acute inhibition of Pol I transcription leads to activation of multiple cell-cycle checkpoints17 ; and specifically, a G1 checkpoint characterized by activation of p53 and a G2/M checkpoint associated with CHK1/CHK2 signaling, downstream of ATM/ATR activation. Thus, mammalian cells appear to have evolved a number of mechanisms to monitor rRNA synthesis and inactivate cell-cycle progression if rRNA output does not match cellular demand. Tumor cells appear to be particularly sensitive to activation of these checkpoints. Interestingly, recent studies reported a p53-independent nucleolar stress response activated by Pol I inhibition involving the Wnt-target Peter Pan, nucleophosmin, Bax, and ATM/ATR signaling in the absence of global DNA damage.26-28 Here, we demonstrate that ATM/ATR signaling contributes directly to the therapeutic efficacy of Pol I inhibition in the absence of p53.
Furthermore, we observed a robust induction of myeloid differentiation in leukemic blasts following inhibition of Pol I transcription in vivo and in vitro. Therefore, reducing rDNA transcription rescues, at least partially, the myeloid differentiation block in AML. Consistent with this concept, recent publications suggested that altered rRNA synthesis is sufficient to trigger differentiation in human AML-derived cancer cell lines, and determine cell fate within the stem cell lineage in Drosophila.29-31 Differentiation therapy is a favorable nongenotoxic treatment option and well established for PML-RARα–driven acute promyelocytic leukemia, although all-trans retinoic acid-induced myeloid differentiation in MLL/AF9 but not MLL/AF4 AML.32 The complex cellular response to CX-5461 involving cell death, cell-cycle delay, and blast differentiation is likely to explain the superior therapeutic efficacy of CX-5461 over cytotoxic therapy. What is less clear is why one AML subpopulation undergoes cell death, whereas another undergoes aberrant cell-cycle progression and/or differentiate in response to CX-5461. Whether this is due to intrinsic tumor heterogeneity or external signals from the microenvironment remains to be established.
In conclusion, our work demonstrates that CX-5461 treats aggressive hematologic malignancies. Most strikingly, we show that targeting rDNA transcription effectively prolongs survival by reducing the leukemia-initiating potential, and suppresses the clonogenic capacity of the LICs population in primary human and murine models of AML. The therapeutic efficacy is mediated through p53-dependent cell death, and p53-independent cell-cycle defects and myeloid differentiation. These data strongly suggest that inhibition of rDNA transcription may be broadly beneficial for the treatment of a variety of hematologic cancer types characterized by TP53 alterations. These findings and favorable toxicology studies had led us to initiate a phase 1 study of CX-5461 (Australian New Zealand Clinical Trials Registry, #12613001061729) in patients with advanced hematologic malignancies, including AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Scott W. Lowe, Megan Bywater, Jessica Salmon, and Kerry Ardley for providing essential reagents, scientific discussion, and technical assistance.
This work was supported by project grants from the National Health and Medical Research Council (NHMRC) of Australia (#1043884, 251608, 566702, 166908, 251688, 509087, 400116, 400120, and 566876) and an NHMRC program grant (#1053792), and the Cancer Council of Victoria grant-in-aid (#1084545). The researchers were funded by the following: NHMRC fellowships (R.D.H., R.B.P., R.W.J., and G.A.M.), Cancer Council of Victoria, Cancer Council of Australian Capital Territory, Sir Edward Weary Dunlop fellowship (G.A.M.), and the Leukemia Foundation of Australia (grant-in-aid to M.W., and Ph.D. scholarships to D.P.C. and J.D.).
Authorship
Contribution: N.H. and R.D.H. were responsible for the concept and design of experiments; J.R.D., D.D., M.A.D., J.Z., I.V., A.B., S.J.H., A.W., E.S., J.Q., S.M.P., and R.W.J. provided essential materials; N.H., D.P.C., J.S., N-Y.N.N., C.Y.F., M.P., J.A.P., and K.M.H. were responsible for the collection and data assembly; N.H., D.P.C., J.S., J.A.P., C.Y.F., N-Y.N.N., M.W., M.A.D., C.C., J.D., G.P., A.J.G., M.A.G., G.A.M., R.B.P., J.D.R., and R.D.H. were involved in data analysis and interpretation; N.H. and R.D.H. wrote the manuscript; and all authors approved the final form of the manuscript. D.D. was affiliated with Cylene Pharmaceuticals. However, this did not influence the conduct of the research in this manuscript, and had no bearing on the decision to submit this paper for publication.
Conflict-of-interest disclosure: R.D.H. is Chief Scientific Advisor and D.D. is VP (R&D) of Pimera Inc, San Diego, CA; R.W.J. has a commercial research grant and has received honoraria for service on the speakers’ bureau for Novartis; and G.A.M. has received commercial research grants from Celgene and Pfizer. The remaining authors declare no competing financial interests.
Correspondence: Nadine Hein, Australian National University, Garran Rd, Canberra, ACT 2601, Australia; e-mail: nadine.hein@anu.edu.au; and Ross D. Hannan, Australian Cancer Research Foundation Department of Cancer Biology and Therapeutics, The John Curtin School of Medical Research, Australian National University, Building 131, Garran Rd, Canberra, ACT 2601, Australia; e-mail: ross.hannan@anu.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal