Key Points
PI3K/mTOR inhibition potently inhibited leukemia proliferation and signal transduction in vivo in human Ph-like ALL xenograft models.
Combined PI3K/mTOR and JAK or ABL inhibition was superior to monotherapy in CRLF2/JAK-mutant and ABL/PDGFR-mutant Ph-like ALL models.
Abstract
Philadelphia chromosome (Ph)–like B-cell acute lymphoblastic leukemia (Ph-like ALL) is associated with activated JAK/STAT, Abelson kinase (ABL), and/or phosphatidylinositol 3-kinase (PI3K) signaling and poor clinical outcomes. PI3K pathway signaling inhibitors have been minimally investigated in Ph-like ALL. We hypothesized that targeted inhibition of PI3Kα, PI3Kδ, PI3K/mTOR, or target of rapamycin complex 1/2 (TORC1/TORC2) would decrease leukemia proliferation and abrogate aberrant kinase signaling and that combined PI3K pathway and JAK inhibition or PI3K pathway and SRC/ABL inhibition would have superior efficacy compared to inhibitor monotherapy. We treated 10 childhood ALL patient-derived xenograft models harboring various Ph-like genomic alterations with 4 discrete PI3K pathway protein inhibitors and observed marked leukemia reduction and in vivo signaling inhibition in all models. Treatment with dual PI3K/mTOR inhibitor gedatolisib resulted in near eradication of ALL in cytokine receptor-like factor 2 (CRLF2)/JAK-mutant models with mean 92.2% (range, 86.0%-99.4%) reduction vs vehicle controls (P < .0001) and in prolonged animal survival. Gedatolisib also inhibited ALL proliferation in ABL/platelet-derived growth factor receptor (PDGFR)-mutant models with mean 66.9% (range, 42.0%-87.6%) reduction vs vehicle (P < .0001). Combined gedatolisib and ruxolitinib treatment of CRLF2/JAK-mutant models more effectively inhibited ALL proliferation than either inhibitor alone (P < .001) and further enhanced survival. Similarly, superior efficacy of combined gedatolisib and dasatinib was observed in ABL/PDGFR-mutant models (P < .001). Overall, PI3K/mTOR inhibition potently decreased ALL burden in vivo; antileukemia activity was further enhanced with combination inhibitor therapy. Clinical trials testing combinations of kinase inhibitors in Ph-like ALL patients are indicated.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL), the most common childhood cancer, is caused by somatic genetic mutations that result in aberrant arrest of normal lymphoid maturation, dysregulated cellular proliferation, and evasion of programmed cell death.1-3 Increased understanding of the biologic heterogeneity of childhood acute lymphoblastic leukemia (ALL) has led to modern risk stratification, which incorporates the critical contributions of genetic subgroups and induction chemotherapy responses to deliver appropriately intensive therapy to achieve cure.4-6 Unfortunately, ∼15% of children with ALL have recurrent disease, and relapsed ALL remains a leading cause of pediatric cancer mortality.7 Adults with ALL fare even more poorly with >50% relapse rates and 20% to 40% overall survival.8,9
Genomic profiling of high-risk (HR) ALL cases has identified the Philadelphia chromosome (Ph)-like subtype of B-ALL (Ph-like ALL), which comprises 10% to 20% of HR B-ALL in children and adolescents and nearly 30% in young adults.10-15 Ph-like ALL is defined by lack of BCR–Abelson kinase 1 (ABL1) fusion (as in Ph+ ALL), but has a kinase-activated gene expression signature similar to that of Ph+ ALL, frequent deletion of IKZF1, and mutations in other kinase and cytokine receptor signaling pathway genes.12-14,16-18 Rearrangements of cytokine receptor-like factor 2 (CRLF2) occur in ∼50% of Ph-like ALL cases and result in CRLF2 overexpression.19-21 Half of CRLF2-rearranged ALLs have concomitant alterations in JAK pathway-associated genes, including JAK1 and JAK2, and a small number of these cases also harbor interleukin-7 receptor α (IL7RA) mutations.22-24 JAK2 point mutations are the most frequent coexisting genetic abnormality in CRLF2-rearranged ALL and also occur in non-Ph-like ALL, including trisomy 21–associated B-ALL and T-cell ALL.25,26 Genomic rearrangements of ABL1 and ABL2, colony-stimulating factor-1 receptor (CSF1R), platelet-derived growth factor receptor-β (PDGFRB), JAK2, erythropoietin receptor (EPOR), and other genes account for an additional 30% to 40% of Ph-like ALL and also drive constitutive kinase signaling.12-14,26,27 Patients with Ph-like ALL often have high rates of minimal residual disease at the end of induction chemotherapy with some patients experiencing overt induction failure.14,21,28 Retrospective analyses of HR children and adolescents and young adults with Ph-like ALL demonstrate greatly increased risk of treatment failure and poor overall survival regardless of underlying Ph-like genomic alterations.10,13,14 New therapies are indicated for these patients.
All Ph-like ALL-associated mutations identified to date activate kinase signaling, particularly of CRLF2-, JAK-, ABL-, and PDGFRB-associated pathways. Biochemical sequelae of these mutations have been primarily studied to date in the most common CRLF2-rearranged subset of Ph-like ALL with the goal of identifying potential signal transduction inhibitor therapies, but perturbed intracellular signaling mechanisms of these genomic alterations remain incompletely characterized. We and others previously reported constitutive and/or cytokine-inducible JAK/STAT and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR) signaling in CRLF2-rearranged ALL.29,30 We further observed robust inhibition of constitutively activated signal transduction with JAK or mTOR inhibition in vitro and in vivo.24,31-33 More recent studies focused upon non-CRLF2-rearranged Ph-like ALL cases have demonstrated in vitro and in vivo efficacy of other kinase inhibitors, including anecdotal clinical responses in children and adults with Ph-like ALL harboring ABL1, ABL2, or PDGFRB rearrangements and fusion proteins (“ABL class” rearrangements) treated with imatinib or dasatinib.14,34,35
Although preclinical32,36 and early clinical studies of JAK inhibition in CRLF2/JAK-mutant and SRC/ABL inhibition in ABL/PDGFR-mutant Ph-like ALL are ongoing (NCT02723994, NCT02883049), therapeutic disruption of aberrant PI3K pathway signaling has been minimally investigated. Clinical efficacy of the mTOR inhibitor (mTORi) rapamycin and its analogs has proven suboptimal in various cancers, at least in part due to upregulation of Akt signaling, a known sequela of mTORi monotherapy and a common resistance mechanism.37 Newer-generation kinase inhibitors that target multiple PI3K pathway signaling proteins or that selectively inhibit PI3K isoforms may have superior antileukemia cytotoxicity and may avoid compensatory upregulation of salvage signaling pathways.38,39 Such “next-generation” PI3K pathway inhibitors (PI3Ki’s) have been minimally evaluated in ALL to date.39 Furthermore, the efficacy of simultaneously targeting multiple oncogenic signaling networks in Ph-like ALL, such as combination therapy with PI3Ki’s and JAK inhibitors (JAKi’s), has not been investigated.
Using patient-derived xenograft (PDX) models of childhood Ph-like ALL, we demonstrate the in vivo therapeutic efficacy of, and pharmacodynamic signaling inhibition by, 4 clinically promising PI3Ki’s with particularly potent efficacy of the dual PI3K/mTORi gedatolisib. We further demonstrate augmented leukemia cytotoxicity in vivo with combined gedatolisib and ruxolitinib (JAK1/2i) treatment of CRLF2/JAK-mutant Ph-like ALL and with gedatolisib and dasatinib (SRC/ABL inhibitor [SRC/ABLi]) treatment of ABL/PDGFR-mutant Ph-like ALL. These data provide compelling rationale for testing combinations of kinase inhibitors without or with multiagent cytotoxic chemotherapy in children and adults with Ph-like ALL.
Methods
Ph-like ALL specimens
Viably cryopreserved leukemia cells from children and adolescents and young adults with de novo Ph-like ALL (n = 8) were obtained from the Children’s Oncology Group (COG) for xenotransplantation studies as described.12,14,32 Additional specimens from patients with multiply relapsed Ph-like ALL (n = 2) were obtained from the Children’s Hospital of Philadelphia (CHOP) and University of California San Francisco leukemia biorepositories under approved institutional research protocols after obtainment of written informed consent in accordance with the Declaration of Helsinki (Table 1). Ph-like genomic alterations were identified by polymerase chain reaction (PCR) and Sanger sequencing and/or fluorescence in situ hybridization assays as described.21,40,41 RNA from primary and corresponding xenografted leukemia specimens were also assessed for an activated kinase Ph-like ALL gene expression signature using a 15-gene low-density microarray classifier as described.40
Genomic characteristics of ALL specimens used for xenograft studies
| PDX . | USI . | Disease status . | Ph-like ALL genomic lesions . | ||
|---|---|---|---|---|---|
| CRLF2 . | JAK . | ABL class . | |||
| NH362 | PALTWS | D | IGH@-CRLF2* | ||
| JH331 | PAMDKS | D | IGH@-CRLF2 | JAK2R683G | |
| JH612 | PAMDRM | D | IGH@-CRLF2 | JAK2GPinsR683 | |
| ALL121 | n/a | R | IGH@-CRLF2 | JAK2R683G | |
| ALL4364 | PAWAKV | R | P2RY8-CRLF2 | JAK2R683G | |
| JL491 | PAKMVD | D | JAK1S646F | ||
| NL482A | PAKYEP | D | BCR-JAK2 | ||
| NL432 | PAKKCA | D | EBF1-PDGFRB | ||
| NH011 | PAKVKK | D | NUP214-ABL1 | ||
| PHL3 | PANSFD | D | ETV6-ABL1 | ||
| PDX . | USI . | Disease status . | Ph-like ALL genomic lesions . | ||
|---|---|---|---|---|---|
| CRLF2 . | JAK . | ABL class . | |||
| NH362 | PALTWS | D | IGH@-CRLF2* | ||
| JH331 | PAMDKS | D | IGH@-CRLF2 | JAK2R683G | |
| JH612 | PAMDRM | D | IGH@-CRLF2 | JAK2GPinsR683 | |
| ALL121 | n/a | R | IGH@-CRLF2 | JAK2R683G | |
| ALL4364 | PAWAKV | R | P2RY8-CRLF2 | JAK2R683G | |
| JL491 | PAKMVD | D | JAK1S646F | ||
| NL482A | PAKYEP | D | BCR-JAK2 | ||
| NL432 | PAKKCA | D | EBF1-PDGFRB | ||
| NH011 | PAKVKK | D | NUP214-ABL1 | ||
| PHL3 | PANSFD | D | ETV6-ABL1 | ||
D, de novo; n/a, not available; R, relapse; USI, unique specimen identifier for primary patient sample.
Non–Ph-like by prediction analysis of microarrays.
PDX models
Studies to date have largely focused upon studying the most common Ph-like ALL subset harboring CRLF2 rearrangements. However, Ph-like ALL is now known to be highly genetically diverse with a variety of mutations that induce kinase hyperactivation.14 In these studies, we thus assessed the efficacy of PI3K pathway inhibition in PDX models of CRLF2-rearranged, JAK1-mutant, JAK2-rearranged, ABL1-rearranged, and PDGFRB-rearranged Ph-like ALL (Table 1).
Individual PDX models were established in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice as described.32,42 To assess initial ALL engraftment and during therapeutic efficacy analyses, peripheral venous blood or splenocyte suspensions from vehicle or PI3Ki-treated mice were erythrocyte-lysed, Fc-blocked, and stained with mouse anti-human CD10-phycoerythrin (PE)-cyanine-7, CD19- and CD45-allophycocyanin antibodies (eBioscience). Specimens were analyzed on an Accuri C6 flow cytometer (BD Biosciences) with CountBright absolute counting beads (Thermo Fisher) for quantification of human leukemia cells as described.32
Experimental randomization to treatment with vehicle or PI3Ki commenced when all mice demonstrated ≥5% ALL in peripheral blood. Animals were sacrificed humanely at planned study end point or if ill-appearing in accordance with the Panel on Euthanasia of the American Veterinary Medical Association’s guidelines. Animal studies were conducted under a CHOP Institutional Animal Use and Care Committee–approved protocol.
In vivo therapeutic efficacy studies
For each of the PDX models, cohorts of 5 engrafted mice were randomized to daily treatment with vehicle (0.5% hydroxypropyl methylcellulose/0.2% Tween 80 in water via oral gavage), vehicle (0.3% lactic acid in 5% dextrose in water intraperitoneally), PI3Kαi BYL719 (50 mg/kg per dose via oral gavage), PI3Kδi idelalisib (formerly CAL-101; 50 mg/kg per dose via oral gavage), PI3K/mTORi gedatolisib (formerly PKI-587 or PF-05212384; 10 mg/kg intraperitoneally), or target of rapamycin complex 1/2 inhibitor (TORC1/2i) AZD2014 (10 mg/kg intraperitoneally).
For combination studies in CRLF2/JAK-mutant ALL models, animals were treated as described in the previous paragraph with vehicle, gedatolisib, ruxolitinib (2 g/kg ruxolitinib-infused rodent chow continuously provided), or simultaneous gedatolisib and ruxolitinib. For combination studies in ABL/PDGFR-mutant ALL models, animals were treated with vehicle, gedatolisib, dasatinib (10 mg/kg per dose twice daily via oral gavage), or simultaneous gedatolisib and dasatinib. Studies were terminated and all mice sacrificed after 28 days of treatment or sooner if ill-appearing. Spleens and bone marrow were harvested as described for quantification of human ALL and for cryopreservation for future studies.32
Survival analyses were conducted in some PDX models with vehicle, gedatolisib, ruxolitinib, dasatinib, combined gedatolisib and ruxolitinib, or combined gedatolisib and dasatinib treatment (n = 5 mice per cohort) as described in this section’s first paragraph for up to 120 days to assess potential longer-term therapeutic efficacy of inhibitor therapies.
Vehicle reagents were obtained from Sigma-Aldrich. BYL719, idelalisib, and AZD2014 were purchased from Active Biochem. Gedatolisib and ruxolitinib were provided by Pfizer and Incyte, respectively. Dasatinib was purchased from LC Laboratories.
Pharmacodynamic assessment of signal transduction inhibition
For measurements of in vivo signaling inhibition, additional PDX mice (n = 3-4 animals per treatment) were administered vehicle or inhibitors as described in “In vivo therapeutic efficacy studies” for 72 hours, then sacrificed at 1 hour following final inhibitor dose for leukemia cell-specific phosphoflow cytometry analysis as described.32 Fixed and permeabilized splenocytes from vehicle- and PI3Ki-treated animals were Fc-blocked and stained with mouse anti-human CD10-PE-Cy7, CD19-allophycocyanin-Cy7, and/or thymic stromal lymphopoietin protein receptor (TSLPR)-PE antibodies (BD Biosciences or eBioscience). Cells were also stained with rabbit anti-human antibodies against phosphorylated 4EBP1T37/46 (p4EBP1T37/46) or pAktS473 (Cell Signaling Technology) with secondary Pacific Blue–conjugated immunoglobulin G H+L (Invitrogen/Life Technologies) and with pS6S240/244-Ax488 (Cell Signaling Technology), phosphorylated extracellular signal-regulated kinase T202/Y204 (pERKT202/Y204)-Ax647 (BD Biosciences), or pSTAT5Y694-Ax647 (BD Biosciences). Data were compensated, acquired, and analyzed on an LSRII flow cytometer (BD Biosciences) as described.29,32 Phosphosignaling data were gated and measured using fluorescence-minus-one (FMO) controls for each phosphoprotein fluorophore.43 Immunoblotting of splenic lysates from end-study animals (after 3 or 4 weeks of treatment) for phosphorylated pSTAT5Y694, pS6S240/244, 4EBP1T37/46, corresponding total proteins, and β-actin (all antibodies from Cell Signaling Technology) was performed as described for the combination inhibitor experiments.32 For experiments with dasatinib treatment, immunoblotting was also performed for total and pCrkLY207 (Cell Signaling Technology), a downstream effector of ABL-mediated signaling.44,45
Statistical analyses
For therapeutic efficacy studies, mean human ALL cell numbers in peripheral blood of inhibitor- and vehicle-treated animals for each xenograft model were calculated at each measured time point. Kaplan-Meier curves for survival analyses were compared using the log-rank test. Human ALL cell number in murine spleens (efficacy studies) and phosphoflow cytometry data for each phosphoprotein (pharmacodynamic studies) were compared via analysis of variance (ANOVA) with Dunnett or Tukey posttest for multiple comparisons. All statistical analyses were conducted using Prism version 6.0h for Mac (GraphPad).
Results
Genetic characterization of Ph-like ALL PDX models
Low-density microarray data confirmed the Ph-like signature in relevant PDX models (n = 9 Ph-like, n = 1 non–Ph-like), which were concordant with gene expression signature patterns of primary ALL specimens from which PDX models were derived (supplemental Figure 1, available on the Blood Web site). Known Ph-like driver lesions (eg, CRLF2 rearrangements, JAK1 and JAK2 point mutations, ABL1 or PDGFRB fusions) were also identified in all xenografted leukemias by fluorescence in situ hybridization and reverse transcription PCR or PCR with Sanger sequencing, confirming genetic fidelity of studied models (not shown).
PI3K pathway inhibitors robustly inhibit leukemia proliferation in Ph-like ALL
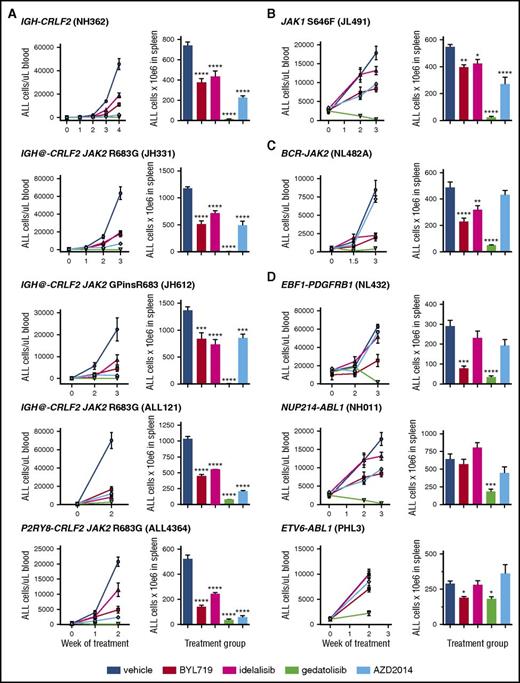
Based upon our prior studies demonstrating aberrant PI3K pathway signaling in CRLF2-rearranged ALL and in vitro signaling inhibition with kinase inhibitors,29,32 we hypothesized that “next-generation” PI3Ki could robustly inhibit in vivo leukemia proliferation and oncogenic signal transduction. We tested 4 clinically available, selective inhibitors of PI3K pathway proteins to identify the most potent therapeutic approach(es) of targeting activated PI3K signaling in Ph-like ALL. In therapeutic efficacy analyses, engrafted mice were treated daily with vehicle or PI3Ki for 2 to 4 weeks depending upon the rapidity of leukemia progression in control animals. We assessed tumor burden in peripheral blood during treatment (line graphs) and in spleens at the conclusion of treatment (Figure 1 column graphs). In total, 10 PDX models representing genetically diverse subsets of Ph-like ALL were studied. Importantly, all PI3Ki-treated models demonstrated inhibition of ALL proliferation in blood and spleens in comparison with vehicle-treated control animals (Figure 1 and representative flow cytometry data in supplemental Figure 2). Inhibitor treatment appeared well tolerated with normal physical appearance and stability of weight in animals during studies (supplemental Figure 3). Therapeutic responses varied modestly among PDX models for each PI3Ki tested depending upon the underlying genomic alteration(s) and the disease status of the primary ALL specimen engrafted (de novo vs relapse).
In vivo efficacy of PI3K pathway inhibition in Ph-like ALL. PDX models were treated with vehicle, PI3Kα inhibitor BYL719, PI3Kδ inhibitor idelalisib, PI3K/mTOR inhibitor gedatolisib, or TORC1/TORC2 inhibitor AZD2014 (n = 5 mice/treatment arm) daily for 2 to 4 weeks depending on rapidity of leukemia progression in vehicle-treated control animals. PI3K isoform or TORC1/TORC2 inhibition resulted in significant suppression of leukemia proliferation in peripheral blood and end-of-study spleens of most PDX models of (A) CRLF2-rearranged, (B) JAK1-mutant, (C) JAK2 fusion, and (D) ABL/PDGFR-mutant ALL vs vehicle controls. Gedatolisib most effectively inhibited ALL proliferation with near eradication of leukemia in CRLF2/JAK-mutant models and marked suppression in ABL/PDGFR-mutant models. Treatment groups were compared with the vehicle control for each PDX model via 1-way ANOVA with the Dunnett posttest for multiple comparisons with α = 0.05. *P < .05, **P < .01, ***P < .001, ****P < .0001.
In vivo efficacy of PI3K pathway inhibition in Ph-like ALL. PDX models were treated with vehicle, PI3Kα inhibitor BYL719, PI3Kδ inhibitor idelalisib, PI3K/mTOR inhibitor gedatolisib, or TORC1/TORC2 inhibitor AZD2014 (n = 5 mice/treatment arm) daily for 2 to 4 weeks depending on rapidity of leukemia progression in vehicle-treated control animals. PI3K isoform or TORC1/TORC2 inhibition resulted in significant suppression of leukemia proliferation in peripheral blood and end-of-study spleens of most PDX models of (A) CRLF2-rearranged, (B) JAK1-mutant, (C) JAK2 fusion, and (D) ABL/PDGFR-mutant ALL vs vehicle controls. Gedatolisib most effectively inhibited ALL proliferation with near eradication of leukemia in CRLF2/JAK-mutant models and marked suppression in ABL/PDGFR-mutant models. Treatment groups were compared with the vehicle control for each PDX model via 1-way ANOVA with the Dunnett posttest for multiple comparisons with α = 0.05. *P < .05, **P < .01, ***P < .001, ****P < .0001.
Treatment with the PI3K/mTORi gedatolisib was remarkably efficacious in all tested models with mean reduction in splenic ALL cell counts of 91.8% (range, 82.5%-99.4%) in comparison with vehicle-treated mice (P < .0001 for all models). Treatment with the PI3Kαi BYL719, PI3Kδi idelalisib, or TORC1/TORC2i AZD2014 inhibited ALL proliferation with mean leukemia reductions of 55.0% (range, 27.5%-72.6%), 38.1% (range, 13.9%-53.1%), and 52.8% (range, 20.1%-88.7%), respectively, vs vehicle controls. Leukemia inhibition by the 2 isoform-selective PI3K inhibitors, BYL719 and idelalisib, were generally similar with greater responses to BYL719 in some models (Figure 1). Therapeutic responses to AZD2014 were greater in the 2 tested relapsed ALL models (ALL121, ALL4364) in which high basal p4EBP1 activation was observed in the pharmacodynamic studies reported in the next section. Overall, these data suggest that Ph-like ALL requires PI3K pathway signaling activation for survival in vivo and demonstrate potent efficacy of PI3K/mTORi gedatolisib in suppressing ALL cell growth.
Pharmacodynamic inhibition of signaling proteins
We next addressed the question of whether PI3Ki could abrogate constitutive PI3K pathway signaling in vivo and whether compensatory upregulation of alternative pathways occurs after drug administration (Figure 2A). Long-term treatment of human cancer cells with kinase inhibitors can lead to selection for alternative signaling activation.39,46 Engrafted PDX mice were treated with vehicle or PI3Ki for 72 hours, then sacrificed for immediate quantification of target protein phosphorylation. We first defined basal activation of PI3K pathway phosphoproteins, pERK, and pSTAT5 by leukemia cell-specific phosphoflow cytometry measurements and displayed data as the percentage of cells in each positive FMO phosphoprotein gate (Figure 2B). Overall, constitutive activation of pSTAT5, pS6, and pERK was observed in most models regardless of underlying Ph-like genetic alteration(s). Basal activation of pAktS473 and p4EBP1 was more heterogeneous across models (Figure 2C).
Constitutive activation of signaling phosphoproteins in Ph-like ALL PDX models. (A) Schema of signaling nodes assessed and targets of kinase inhibitors. (B) Gating strategy for phosphoflow cytometry analyses: live singlet CD10+CD19+ (and TSLPR+ if CRLF2-rearranged) human ALL cells in murine spleens were gated. FMO controls were used for each fluorophore-conjugated phosphoprotein antibody to set negative (FMO−) and positive (FMO+) gates. The percentage of cells in the FMO+ gate was calculated for all phosphoproteins for each STI treatment and displayed graphically. All studied ALL samples were uniformly CD10+. (C) Basal levels of each measured phosphoprotein in vehicle-treated mice from each PDX model are displayed as percentages of human ALL cells in FMO+ gates. Data are depicted for each PDX model (colored columns) as mean percentage of cells in FMO+ gate on the y-axis with standard error of the mean (black bars). FSC-A, forward scatter area; SSC-A, side scatter area; SSC-W, side scatter width.
Constitutive activation of signaling phosphoproteins in Ph-like ALL PDX models. (A) Schema of signaling nodes assessed and targets of kinase inhibitors. (B) Gating strategy for phosphoflow cytometry analyses: live singlet CD10+CD19+ (and TSLPR+ if CRLF2-rearranged) human ALL cells in murine spleens were gated. FMO controls were used for each fluorophore-conjugated phosphoprotein antibody to set negative (FMO−) and positive (FMO+) gates. The percentage of cells in the FMO+ gate was calculated for all phosphoproteins for each STI treatment and displayed graphically. All studied ALL samples were uniformly CD10+. (C) Basal levels of each measured phosphoprotein in vehicle-treated mice from each PDX model are displayed as percentages of human ALL cells in FMO+ gates. Data are depicted for each PDX model (colored columns) as mean percentage of cells in FMO+ gate on the y-axis with standard error of the mean (black bars). FSC-A, forward scatter area; SSC-A, side scatter area; SSC-W, side scatter width.
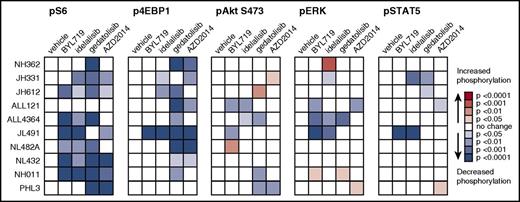
We then compared leukemia signaling in PI3Ki-treated animals to vehicle-treated controls to assess potential in vivo phosphoprotein inhibition (Figure 3; supplemental Figure 5). Treatment with the PI3Kα and PI3Kδ isoform-selective inhibitors BYL719 and idelalisib resulted in significant inhibition of pS6 in 6 of 10 and 7 of 10 PDX models, respectively (P < .05 for each). Both PI3Ki also decreased p4EBP1 (1 of 10 models for BYL719, 3 of 10 for idelalisib), pAkt (2 of 10 models for each inhibitor), and pERK (3 of 10 models for each inhibitor), suggesting potent inhibition of downstream mTORC1 signaling and potential crosstalk with or direct inhibition of the Ras/MAPK/ERK pathway (Figure 3). The PI3K/mTORi gedatolisib also potently inhibited pS6 and p4EBP1 in vivo in 8 of 10 models vs vehicle controls, as well as decreased pAktS473 (3 of 10 models) and pERK (1 model) (P < .05 for each; Figure 3). Importantly, minimal upregulation of pAktS473 was observed in BYL719-, idelalisib-, or gedatolisib-treated animals (Figure 3), suggesting generally effective blockade of potential TORC2-mediated feedback signaling loops that may reactivate proximal PI3K pathway signaling. Similarly, treatment with the TORC1/TORC2i AZD2014 significantly inhibited pS6 and p4EBP1 in 6 of 10 models (P < .05). Surprisingly, minimal inhibition of the TORC2 phosphoprotein pAktS473 was observed in AZD2014-treated animals (1 of 10 models), although many PDX models did not have basal pAktS473 activation (Figure 2C).
PI3K pathway inhibitors inhibit signaling phosphoproteins and induce minimal compensatory signaling upregulation. Ph-like ALL PDX models were treated with vehicle, BYL719, idelalisib, gedatolisib, or AZD2014 (n = 3-4 mice per treatment) for 72 hours, then sacrificed at 1 hour after final dose for pharmacodynamic measurement of in vivo target inhibition via phosphoflow cytometry. Heatmap data depict significant changes in phosphoprotein levels in inhibitor- vs vehicle-treated controls using the percentage of FMO+ cells (described in Figure 2) for each PDX model. Colorimetric scale depicts normalization of each model to vehicle controls (white squares; leftmost columns) with statistically significant decreased phosphorylation (blue colors) or increased phosphorylation (green colors) with inhibitor treatment as calculated by ANOVA with the Dunnett posttest for multiple comparisons. FMO+ data for all individual mice are displayed in greater detail in supplemental Figure 5.
PI3K pathway inhibitors inhibit signaling phosphoproteins and induce minimal compensatory signaling upregulation. Ph-like ALL PDX models were treated with vehicle, BYL719, idelalisib, gedatolisib, or AZD2014 (n = 3-4 mice per treatment) for 72 hours, then sacrificed at 1 hour after final dose for pharmacodynamic measurement of in vivo target inhibition via phosphoflow cytometry. Heatmap data depict significant changes in phosphoprotein levels in inhibitor- vs vehicle-treated controls using the percentage of FMO+ cells (described in Figure 2) for each PDX model. Colorimetric scale depicts normalization of each model to vehicle controls (white squares; leftmost columns) with statistically significant decreased phosphorylation (blue colors) or increased phosphorylation (green colors) with inhibitor treatment as calculated by ANOVA with the Dunnett posttest for multiple comparisons. FMO+ data for all individual mice are displayed in greater detail in supplemental Figure 5.
Consistent with our earlier studies,32 we observed marked constitutive pSTAT5 activation in all tested CRLF2/JAK-mutant or ABL/PDGFR-mutant Ph-like ALL PDX models (Figure 2C). Perhaps surprisingly, in vivo inhibition of pSTAT5 occurred with BYL719 (1 model), idelalisib (2 models), gedatolisib (2 models), and AZD2014 (1 model) treatment (Figure 3). These data suggest potential off-target effects of PI3K pathway inhibitors in these models and/or possible interconnection of PI3K/Akt/mTOR and JAK/STAT signaling networks previously described in Ph-like ALL.29,30,32
Three conclusions can be drawn from these experiments. First, the remarkable efficacy of gedatolisib treatment correlates with inhibition of pS6 and p4EBP1 in Ph-like ALL models. Second, minimal upregulation of AktS473 occurs after PI3K pathway inhibition with tested agents. Third, a complex interplay of signaling pathway regulation apparently occurs after in vivo administration of PI3K pathway inhibitors, which may be difficult to assess fully at a single optimized pharmacodynamic time point in these preclinical models.
Superior antileukemia efficacy of combination signal transduction inhibitor treatment
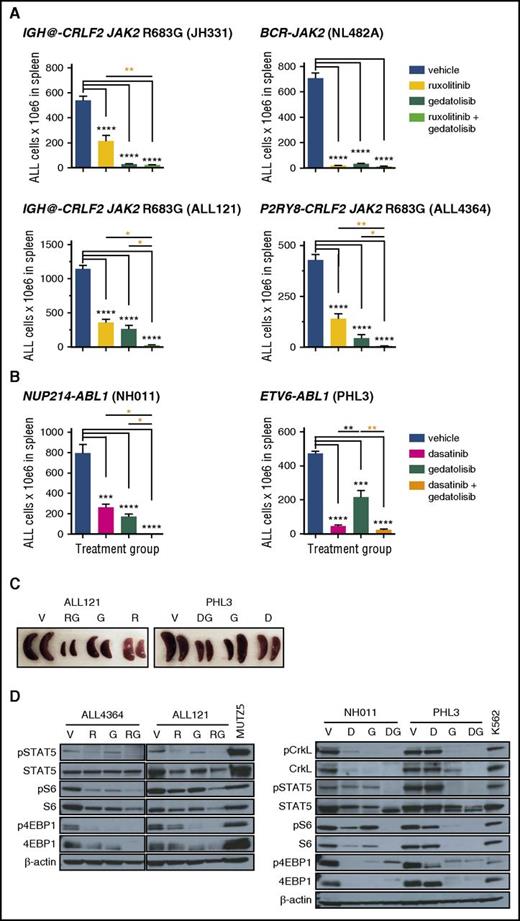
Given the potent, but incomplete, therapeutic efficacy and pharmacodynamic effects of PI3K/mTORi monotherapy in Ph-like ALL models, we further assessed the potential for enhanced efficacy of gedatolisib when coadministered with ruxolitinib (CRLF2/JAK-mutant models; Figure 4A) or dasatinib (ABL1/PDGFRB-mutant models; Figure 4B). Indeed, combined inhibitor therapy resulted in superior inhibition of ALL proliferation in murine spleens than was observed with inhibitors alone with near-complete eradication of leukemia in most models (Figure 4A-B [orange asterisks] and C). Furthermore, greater in vivo inhibition of JAK/STAT, PI3K, and/or ABL pathway signaling phosphoproteins was also observed with combination inhibitor treatment, as shown by immunoblotting of splenic lysates from vehicle- and inhibitor-treated animals after 3 to 4 weeks of treatment (Figure 4D). Combination inhibitor therapy also appeared well tolerated with stability of animal weights in most PDX models during treatment (supplemental Figure 4). Taken together, these data demonstrate enhanced antileukemia efficacy of simultaneous inhibition of multiple signaling nodes in Ph-like ALL and provide strong preclinical rationale to advance such treatment approaches to clinical testing.
Superior efficacy of combination STI treatment. Mice engrafted with (A) CRLF2/JAK-mutant Ph-like ALL were treated with vehicle, ruxolitinib, gedatolisib, or both ruxolitinib and gedatolisib for 3 or 4 weeks. Human ALL in murine spleens after treatment completion was measured by quantitative flow cytometry as in Figure 1. Significantly greater inhibition of ALL proliferation was observed with combined ruxolitinib and gedatolisib treatment (orange asterisks), as measured by 1-way ANOVA with the Tukey posttest for multiple comparisons with α = 0.05. (B) ABL1-rearranged Ph-like ALL PDX models were treated with vehicle, dasatinib, gedatolisib, or both dasatinib and gedatolisib for 3 weeks. Enhanced antileukemia efficacy was also observed in these models with combined dasatinib and gedatolisib treatment (orange asterisks) vs dasatinib alone or gedatolisib alone. (C) Combination inhibitor treatment markedly reduced splenomegaly in ALL-engrafted PDX mice treated as in panels A or B. (D) Immunoblotting of total and phosphorylated signal transduction proteins from murine splenic lysates (obtained after 3 or 4 weeks of treatment) demonstrate greatest inhibition of target phosphoproteins with combination inhibitor treatment of PDX models. Total protein loss is observed with inhibitor treatment in some models. MUTZ5 (a CRLF2/JAK2-mutant ALL cell line) and K562 (a BCR-ABL1–rearranged chronic myeloid leukemia cell line) lysates were used as positive signaling controls, and β-actin immunoblotting was used as a protein loading control. *P < .05, **P < .01, ***P < .001, ****P < .0001. D, dasatinib; DG, dasatinib + gedatolisib combination; G, gedatolisib; R, ruxolitinib; RG, ruxolitinib + gedatolisib combination; V, vehicle.
Superior efficacy of combination STI treatment. Mice engrafted with (A) CRLF2/JAK-mutant Ph-like ALL were treated with vehicle, ruxolitinib, gedatolisib, or both ruxolitinib and gedatolisib for 3 or 4 weeks. Human ALL in murine spleens after treatment completion was measured by quantitative flow cytometry as in Figure 1. Significantly greater inhibition of ALL proliferation was observed with combined ruxolitinib and gedatolisib treatment (orange asterisks), as measured by 1-way ANOVA with the Tukey posttest for multiple comparisons with α = 0.05. (B) ABL1-rearranged Ph-like ALL PDX models were treated with vehicle, dasatinib, gedatolisib, or both dasatinib and gedatolisib for 3 weeks. Enhanced antileukemia efficacy was also observed in these models with combined dasatinib and gedatolisib treatment (orange asterisks) vs dasatinib alone or gedatolisib alone. (C) Combination inhibitor treatment markedly reduced splenomegaly in ALL-engrafted PDX mice treated as in panels A or B. (D) Immunoblotting of total and phosphorylated signal transduction proteins from murine splenic lysates (obtained after 3 or 4 weeks of treatment) demonstrate greatest inhibition of target phosphoproteins with combination inhibitor treatment of PDX models. Total protein loss is observed with inhibitor treatment in some models. MUTZ5 (a CRLF2/JAK2-mutant ALL cell line) and K562 (a BCR-ABL1–rearranged chronic myeloid leukemia cell line) lysates were used as positive signaling controls, and β-actin immunoblotting was used as a protein loading control. *P < .05, **P < .01, ***P < .001, ****P < .0001. D, dasatinib; DG, dasatinib + gedatolisib combination; G, gedatolisib; R, ruxolitinib; RG, ruxolitinib + gedatolisib combination; V, vehicle.
Survival benefit of inhibitor treatment
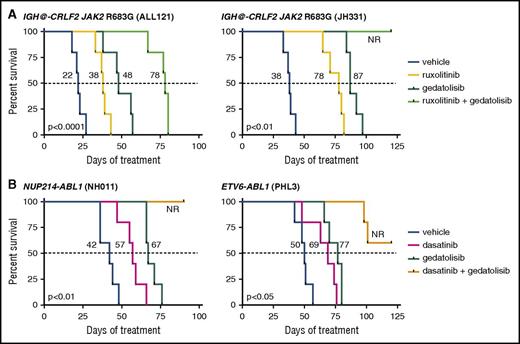
Given the robust potency of gedatolisib monotherapy and enhanced signaling inhibition when combined with ruxolitinib or dasatinib, we next queried whether inhibitor treatment of established leukemia could confer a survival benefit. To address this question, we conducted longer-term survival analyses of vehicle, gedatolisib monotherapy, and combination inhibitor treatment in 4 PDX ALL models (2 CRLF2/JAK, 2 ABL/PDGFR). Mice were first injected with human ALL cells and monitored for engraftment. Once ≥5% leukemia was detected in peripheral blood, daily treatment with vehicle or inhibitor(s) was commenced, and animals were followed by weekly peripheral blood analysis as in the PI3Ki monotherapy studies.
As anticipated, gedatolisib monotherapy significantly prolonged survival vs vehicle controls in all tested models (Figure 5; P < .05 for each). Similar survival benefit was also observed with ruxolitinib monotherapy treatment (Figure 5A; ALL121 and JH331 models) and with dasatinib monotherapy treatment (Figure 5B; NH011 and PHL3 models). Enhanced survival was further observed with combined gedatolisib and ruxolitinib or gedatolisib and dasatinib treatment of up to 120 days. When possible, spleens from expiring animals were analyzed by flow cytometry, which confirmed presence of residual or recurrent human leukemia. These studies thus further demonstrate superior efficacy of combination kinase inhibitor approaches in Ph-like ALL.
Survival advantage with inhibitor monotherapy and combination inhibitor treatment. Animals engrafted with (A) relapsed (ALL121) or de novo (JH331) CRLF2/JAK-mutant Ph-like ALL were treated with vehicle, ruxolitinib chow, gedatolisib 10 mg/kg intraperitoneally daily, or both ruxolitinib and gedatolisib until moribund (n = 5 mice per treatment cohort) for up to 120 days. Similarly, animals engrafted with (B) de novo ABL1-mutant ALL (NH011, PHL3) were treated with vehicle, dasatinib 10 mg/kg twice daily via oral gavage, gedatolisib, or both dasatinib and gedatolisib for up to 120 days. X-axes depict duration of inhibitor treatment (week 0 = treatment initiation) in mice after documentation of human leukemia engraftment (≥5% ALL in peripheral blood). Kaplan-Meier survival curves for each xenograft model were compared statistically using the log-rank test (P values indicated on graphs for each model). Dotted lines delineate median survival in each study with corresponding values for listed each treatment (black numbers). NR, 50% survival not reached.
Survival advantage with inhibitor monotherapy and combination inhibitor treatment. Animals engrafted with (A) relapsed (ALL121) or de novo (JH331) CRLF2/JAK-mutant Ph-like ALL were treated with vehicle, ruxolitinib chow, gedatolisib 10 mg/kg intraperitoneally daily, or both ruxolitinib and gedatolisib until moribund (n = 5 mice per treatment cohort) for up to 120 days. Similarly, animals engrafted with (B) de novo ABL1-mutant ALL (NH011, PHL3) were treated with vehicle, dasatinib 10 mg/kg twice daily via oral gavage, gedatolisib, or both dasatinib and gedatolisib for up to 120 days. X-axes depict duration of inhibitor treatment (week 0 = treatment initiation) in mice after documentation of human leukemia engraftment (≥5% ALL in peripheral blood). Kaplan-Meier survival curves for each xenograft model were compared statistically using the log-rank test (P values indicated on graphs for each model). Dotted lines delineate median survival in each study with corresponding values for listed each treatment (black numbers). NR, 50% survival not reached.
Discussion
PI3K/Akt/mTOR signal transduction in human leukemias
PI3K pathway proteins play a central, highly complex role in the metabolic regulation and survival of cancer cells. Constitutive activation of PI3K/Akt/mTOR signaling occurs frequently in a variety of human cancers and likely contributes to avoidance of normal apoptotic death mechanisms following cellular injury.47,48 Dysregulated PI3K/Akt/mTOR signaling has also been associated with resistance to cytotoxic chemotherapy.49,50 However, the specific role and necessity of PI3K pathway signaling in B-precursor ALL remains inadequately characterized.39
We previously observed constitutive PI3K/Akt/mTOR activation in childhood Ph-like ALL and signaling inhibition with in vitro treatment of primary leukemia cells with PI3K pathway inhibitors.29 We also previously demonstrated in vivo efficacy of rapamycin treatment in some Ph-like ALL xenograft models also used in the current study (JH331, JH612, NH362, NL482A, JL491), although therapeutic responses to rapamycin were incomplete and did not appear to correlate with pharmacodynamic inhibition of TORC1 phosphoproteins.32 In the current studies, we demonstrate that treatment with the PI3K/mTORi gedatolisib profoundly inhibited in vivo Ph-like ALL proliferation. Furthermore, extended gedatolisib treatment was well tolerated in PDX models and significantly prolonged animal survival. Treatment with BYL719 and idelalisib reduced ALL burden by approximately half of that in vehicle-treated animals in most models. The comparable efficacy of both PI3Kαi and PI3Kδi is perhaps surprising, as B-cell lymphoid malignancies are not generally PI3KCA mutation-driven and have higher expression of PI3Kδ.38,51 However, PI3Kα dependence of Ras-driven myeloid leukemias and therapeutic activity of BYL719 in AML cell line xenograft models was recently reported, providing additional rationale for further study of PI3Kαi in human leukemias.52
Although development of PI3Ki resistance or upregulation of alternate signaling pathways are plausible potential explanations for incomplete therapeutic efficacy, the partial treatment responses we observed with BYL719 and idelalisib during relatively short-duration animal experiments likely reflect, at least in part, known solubility issues for oral gavage and poor pharmacokinetic properties of PI3K isoform-selective inhibitors in mice.38,51,53 Similarly, the TORC1/TORC2i AZD2014 moderately inhibited leukemia proliferation in CRLF2/JAK-mutant ALL PDX models, particularly in 2 aggressive relapsed ALL models (ALL121, ALL4364), which may correlate with basal activation of pAktS473. Interestingly, no appreciable reduction in leukemia burden was observed with AZD2014 treatment in the tested BCR-JAK2 fusion model (NL482A) or ABL/PDGFR-mutant models (NL432, NH011, PHL3) despite constitutive mTORC1 activation (pS6 and p4EBP1). However, minimal basal TORC2 signaling activation (pAktS473) was detected in these 4 models. These data suggest biologic differences and signaling dependencies among the genetically heterogeneous Ph-like ALL subgroups, as well as highlight potential limitations of our pharmacodynamic assays with respect to measurement of target inhibition of multiple phosphoproteins at a single time point in animals treated with various inhibitors. Additional studies are indicated to assess such response differences.
As anticipated, we observed augmented antileukemic efficacy with simultaneous targeting of 2 discrete oncogenic pathways in Ph-like ALL. First, gedatolisib monotherapy demonstrated markedly greater antileukemia efficacy in these studies in comparison with prior studies testing single-agent ruxolitinib or dasatinib in Ph-like ALL models.12,14,27,32 Interestingly, although highly effective in all tested models, gedatolisib more potently inhibited ALL proliferation in CRLF2/JAK models than in ABL/PDFGR models, highlighting potential differential signaling dependencies driven by underlying Ph-like genetic alterations. In addition, combined gedatolisib and ruxolitinib or gedatolisib and dasatinib treatment of CRLF2/JAK or ABL/PDGFR models, respectively, resulted in superior therapeutic efficacy, pharmacodynamic phosphoprotein inhibition, and survival benefit. Such dual pathway-targeting approaches may also minimize potential for compensatory signaling upregulation that is a common mechanism of treatment resistance.38,39
In pharmacodynamic studies, we observed few increased signaling sequelae in animals treated with PI3Ki. It is plausible that dynamic signaling changes occur over a longer treatment time period than 4 weeks or in other phosphoproteins than were evaluated in our studies. However, use of next-generation inhibitors that target proximal PI3K pathway proteins or “vertically” inhibit multiple PI3K pathway nodes may more potently shut down aberrant PI3K signaling in Ph-like ALL and perhaps ultimately evoke superior therapeutic efficacy.
Our results are consistent with studies by Badura and colleagues, who assessed effects of various PI3K pathway inhibitors incubated in vitro with primary BCR-ABL1–rearranged and nonrearranged B-ALL cells.54 They also observed greatest antiproliferative and proapoptotic activity of dual PI3K/mTOR inhibition (BGT-226, BEZ-235) in ALL cells vs pan-PI3K (BKM-120), mTOR (everolimus), or TORC1/TORC2 (Torin 1, PP242, KU-0063794) inhibition. Cytotoxic effects of the various inhibitors were similar among Ph+ and non-Ph+ ALL specimens in their study, suggesting that PI3Ki efficacy may not be restricted to specific ALL subsets. Similarly, Kharas et al and Wong et al demonstrated potent in vitro cytotoxicity of PI3K/mTORi incubated with murine or human ALL cell lines, as well as reduction in leukemia burden and prolonged survival in murine and human ALL cell line-engrafted mice treated with PI3K/mTORi.55,56 We also observed greatest phosphosignaling inhibition with PI3K/mTORi (gedatolisib) without upregulation of pAktS473, suggesting potent blockade of the PI3K pathway and consistent with preclinical data from Mallon et al.57
Unlike the Badura study, we did not observe appreciably increased TORC2-mediated pAktS473 with PI3K isoform-selective or TORC1/TORC2 inhibitor treatment, possibly due to differences in specific PI3Ki drugs tested and in vivo vs in vitro systems analyzed. The lack of observed pS6 and p4EBP1 inhibition in a few PDX models may also reflect characteristics of used phosphoantibody reagents, mechanism(s) of action of each PI3Ki, and dynamic signaling networks evaluated in vivo at a single time point. Although we did not specifically investigate selective Akt inhibition given mixed preclinical and clinical data to date,50 our observed basal activation of pAkt in some Ph-like ALL models and recent data demonstrating reversal of steroid resistance in T-ALL with newer Akt inhibitors may warrant future study of these agents.58
We thus demonstrate that activated PI3K pathway signaling is prevalent in Ph-like ALL and can be effectively targeted in vivo using clinically available kinase inhibitors. We report particularly potent efficacy of the PI3K/mTORi gedatolisib as monotherapy and enhanced efficacy in combination with ruxolitinib or dasatinib in specific genetic subsets of Ph-like ALL. Our pharmacodynamic assays also identified constitutive basal activation of PI3K and JAK/STAT phosphoproteins in Ph-like ALL, which may be informative biomarkers to guide precision medicine-focused clinical selection of specific kinase inhibitors for individual leukemias. However, significant and/or dose-limiting toxicities of various PI3K pathway inhibitors in adults with cancer have been reported, as well as modest clinical activity of some inhibitor monotherapies.38,59,60 Careful evaluation of the biochemical mechanisms and antileukemia efficacy of PI3K/mTORi and their toxicity profiles, particularly in combination with JAKi or ABLi, is warranted to facilitate rapid clinical translation of nove kinase inhibitor-based therapies for children and adults with Ph-like ALL.
Presented in part at the 55th annual meeting of the American Society of Hematology, New Orleans, LA, 7-10 December, 2013; the 46th congress of the International Society of Paediatric Oncology, Toronto, ON, Canada, 22-25 October, 2014; and at the 57th annual meeting of the American Society of Hematology, Orlando, FL, 5-8 December, 2015.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Miles Pufall (University of Iowa) and David Fruman (University of California, Irvine) for thoughtful discussions about this work and Shannon Maude (Children’s Hospital of Philadelphia) and Elliot Stieglitz (University of California, San Francisco) for assistance with experimental reagents.
This work was supported by a National Institutes of Health, National Cancer Institute (NIH/NCI) Mentored Career Development Award K08CA184418 (S.K.T.), an NIH/NCI Paul Calabresi Career Development Award for Clinical Oncology to the University of Pennsylvania K12CA076931 (S.K.T.), an NIH/National Center for Research Resources award to the University of Pennsylvania Institute for Translational Medicine and Therapeutics UL1RR024134 (S.K.T.), Alex’s Lemonade Stand Foundation (ALSF) Young Investigator and Center of Excellence awards (S.K.T.), the Rally Foundation for Childhood Cancer Research (S.K.T.), and the Leukemia & Lymphoma Society Specialized Center of Research Program (D.T.T. and S.A.G.). This work was also supported by grants to the Children’s Oncology Group, specifically NCI U10 CA98543 (Chair’s grant and supplement to support the COG High Risk ALL TARGET Project), NCI U10 CA98413 (Statistical Center), and NCI U24 CA114766 (specimen banking). S.K.T. was an ALSF Scholar in Developmental Therapeutics. D.T.T. is supported by a Research Scholar Grant (RSG-14-022-01-CDD) from the American Cancer Society. C.L.W. is the Maurice and Marguerite Liberman Distinguished Chair in Cancer Research Professor of Pathology and Medicine. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics. M.L.L. is the Deborah and Arthur Ablin Chair in Molecular Pediatric Oncology. M.C. is supported by Veterans Administration Merit Award 1I01BX000918. S.A.G. is the Yetta Deitch Novotny Endowed Chair in Pediatric Oncology.
Authorship
Contribution: S.K.T. designed, directed, and performed research, analyzed data, and wrote the manuscript; D.T.T. and C.L.W. provided key reagents and interpreted data; Y.L., F.S., R.C.H., T.R., T.L.V., and I.-M.C. performed research and analyzed and interpreted data; A.E.P., S.P.H., M.L.L., M.C., and S.A.G. contributed to research design and data interpretation; S.P.H. and M.C. edited the manuscript; and all authors critically reviewed the manuscript prior to submission.
Conflict-of-interest disclosure: S.K.T. and M.C. have a research collaboration with Incyte Corporation without financial support. R.C.H., I.-M.C., C.L.W., and S.P.H. are coinventors on US Patent No. 8 568 974 B2 “Identification of novel subgroups of high-risk pediatric precursor-B acute lymphoblastic leukemia, outcome correlations and diagnostic and therapeutic methods related to same.” This technology has not been licensed, and there is no income. The remaining authors declare no competing financial interests.
Correspondence: Sarah K. Tasian, Division of Oncology, Children’s Hospital of Philadelphia, 3501 Civic Center Blvd, CTRB 3010, Philadelphia, PA 19104; e-mail: tasians@e-mail.chop.edu.