Key Points
BTKCys481 mutations, including multiple mutated variants within individual patients are common in ibrutinib-progressing WM patients.
BTKCys481 mutations were associated with mutated CXCR4 in WM patients progressing on ibrutinib.
Abstract
Ibrutinib produces high response rates and durable remissions in Waldenström macroglobulinemia (WM) that are impacted by MYD88 and CXCR4WHIM mutations. Disease progression can develop on ibrutinib, although the molecular basis remains to be clarified. We sequenced sorted CD19+ lymphoplasmacytic cells from 6 WM patients who progressed after achieving major responses on ibrutinib using Sanger, TA cloning and sequencing, and highly sensitive and allele-specific polymerase chain reaction (AS-PCR) assays that we developed for Bruton tyrosine kinase (BTK) mutations. AS-PCR assays were used to screen patients with and without progressive disease on ibrutinib, and ibrutinib-naïve disease. Targeted next-generation sequencing was used to validate AS-PCR findings, assess for other BTK mutations, and other targets in B-cell receptor and MYD88 signaling. Among the 6 progressing patients, 3 had BTKCys481 variants that included BTKCys481Ser(c.1635G>C and c.1634T>A) and BTKCys481Arg(c.1634T>C). Two of these patients had multiple BTK mutations. Screening of 38 additional patients on ibrutinib without clinical progression identified BTKCys481 mutations in 2 (5.1%) individuals, both of whom subsequently progressed. BTKCys481 mutations were not detected in baseline samples or in 100 ibrutinib-naive WM patients. Using mutated MYD88 as a tumor marker, BTKCys481 mutations were subclonal, with a highly variable clonal distribution. Targeted deep-sequencing confirmed AS-PCR findings, and identified an additional BTKCys481Tyr(c.1634G>A) mutation in the 2 patients with multiple other BTKCys481 mutations, as well as CARD11Leu878Phe(c.2632C>T) and PLCγ2Tyr495His(c.1483T>C) mutations. Four of the 5 patients with BTKC481 variants were CXCR4 mutated. BTKCys481 mutations are common in WM patients with clinical progression on ibrutinib, and are associated with mutated CXCR4.
Introduction
Activating somatic mutations in MYD88 and the C-terminal domain of CXCR4 (warts, hypogammaglobulinemia, immunodeficiency, and myelokathexis-like) are present in ∼90% to 95% and 30% to 40% of Waldenström macroglobulinemia (WM) patients, respectively.1-4 MYD88 mutations trigger pro-survival nuclear factor-κB signaling through Bruton tyrosine kinase (BTK), which is a target of ibrutinib. CXCR4 mutations are similar to those found in the germ line warts, hypogammaglobulinemia, immunodeficiency, and myelokathexis syndrome, promote AKT and extracellular signal-regulated kinase 1/2 activation, and are associated with both in vitro and clinical drug resistance to ibrutinib.4-7 These findings prompted clinical investigation of ibrutinib in previously treated WM patients who showed high levels of response activity and durable responses, and supported the regulatory approval of ibrutinib in WM in the United States and Europe.8,9 An important revelation in this study was the role of MYD88 and CXCR4 mutation status as determinants of primary response in WM.10 Patients who lacked MYD88 mutations (ie, were wild-type [WT] for MYD88) had no major responses, whereas those with MYD88 mutations who were CXCR4 mutated had fewer major responses vs those WT for CXCR4 (62% vs 92%). Furthermore, major responses were also delayed by 6 months or more for CXCR4-mutated individuals. High response rates with durable activity were also observed in a multicenter study that administered ibrutinib to heavily pre-treated, rituximab refractory WM patients.11 Patients with CXCR4 mutations also showed delayed responses, and the 1 patient with WT MYD88 included in this study showed no response to ibrutinib. Despite the highly active nature of ibrutinib in WM, clinical progression occurs and mechanistic insights are lacking in WM. Among chronic lymphocytic leukemia (CLL) patients who progressed on ibrutinib, mutations in the ibrutinib binding (BTKCys481), gatekeeper (BTKThr474), and SH2 non-kinase (BTKThr316) domains of BTK have been reported.12-15 Loss of ibrutinib binding due to BTKCys481 mutations permits downstream survival signaling that includes PLCγ2 and CARD11. Activating mutations in PLCγ2 and CARD11 have also been identified in CLL patients progressing on ibrutinib, whereas BTKCys481 and CARD11 mutations have also been identified in mantle cell lymphoma (MCL) patients who progressed on ibrutinib.14-17
We therefore examined sorted lymphoplasmacytic cells from WM patients with progressive disease on ibrutinib, on ibrutinib without clinical progression at time of bone marrow (BM) sampling, as well as untreated and previously treated ibrutinib-naïve patients, for mutations known to be associated with ibrutinib progression. For these efforts, we used Sanger, as well as cloning and sequencing studies to identify BTKCys481 mutations associated with clinical progression in WM, and developed highly sensitive and allele-specific polymerase chain reaction (AS-PCR) assays for their detection to perform screening. Targeted next-generation sequencing (NGS) was also used to validate AS-PCR findings, and to assess for other BTK mutations, as well as MYD88, CXCR4, and other select targets in B-cell receptor (BCR) and MYD88 signaling (PLCγ2, CARD11, HCK, and LYN).
Patients and methods
Patient samples and methods
We identified 6 WM patients who had a major response and subsequently progressed on ibrutinib. All 6 of these patients had relapsed disease, and were symptomatic at time of ibrutinib initiation. Their clinical and laboratory characteristics are shown in Table 1. The median time to progression on ibrutinib for these patients was 16.3 (range, 7.7-37.1) months. We used Sanger, as well as cloning and sequencing studies to screen these patients for BTK mutations. We subsequently developed and validated highly sensitive and specific nested AS-PCR assays for 3 BTK mutations that we identified, and screened 38 patients on ibrutinib without clinical progression at the time of repeat BM sampling, as well as 100 (50 untreated, 50 previously treated) ibrutinib-naïve patients. Targeted, deep NGS was used to confirm findings, to assess MYD88 and CXCR4 mutation status, and assess for other mutations in BTK, as well as MYD88, CXCR4, and other targets in BCR and MYD88 signaling. CD19+ cells from BM aspirates were isolated, and DNA was extracted as previously described and used for sample analysis.18,19 Subject participation was approved by the Harvard Cancer Center/Dana-Farber Cancer Institute’s Institutional Review Board, and all participants provided written consent for use of their samples.
Baseline clinical and laboratory characteristics of WM patients who progressed on ibrutinib
| Baseline characteristics . | WM1 . | WM2 . | WM3 . | WM4 . | WM5 . | WM6 . |
|---|---|---|---|---|---|---|
| Age (y) | 77 | 92 | 61 | 66 | 78 | 73 |
| Sex | Male | Male | Male | Male | Male | Male |
| Serum IgM (mg/dL) | 4130 | 921 | 3630 | 2490 | 1300 | 5790 |
| Serum IgM M-spike (g/dL) | 2.21 | 0.86 | 2.09 | 1.41 | 0.98 | 3.62 |
| Hb (g/dL) | 10.3 | 8.0 | 12.3 | 9.1 | 8.9 | 10.8 |
| Serum β2-microglobulin (mg/L) | 4.5 | N/A | 2.5 | 5.6 | 14.2 | 3.9 |
| BM involvement (%) | 30 | N/A | 70 | 40 | 5 | 30 |
| Prior therapies | Fludarabine, R, CPR, bendamustine-R, bortezomib/dex, tositumomab | R | R, ofatumumab, C, pentostatin | R | Cladribine, C, R, IFN-α, bendamustine, bortezomib/dex | R, chlorambucil, bendamustine |
| Time on ibrutinib (mo) | 9.6 | 7.7 | 37.1 | 36.4 | 9.1 | 23.0 |
| Best response to ibrutinib | PR | PR | PR | VGPR | VGPR | PR |
| Events supporting progressive disease from best response | BM 30%→70% IgM M-spike 0.55→1.12 Hb 11.8→10.5 | IgM M-spike NQ→0.55 Hb 9.5→7.3 | BM 20%→90% Hb 13.4→10.7 | Splenic enlargement Hb 10.6→7.8 New pleural effusion | BM 5%→60% IgM M-spike 0.23→0.81 Hb 9.6→8.4 | BM 15→80% IgM 2647→3970 Hb 15.1→7.7 |
| MYD88 status | L265P | L265P | L265P | S243N | L265P | L265P |
| CXCR4 status | WT | S339fs | S338X | WT | WT | S338fs |
| Baseline characteristics . | WM1 . | WM2 . | WM3 . | WM4 . | WM5 . | WM6 . |
|---|---|---|---|---|---|---|
| Age (y) | 77 | 92 | 61 | 66 | 78 | 73 |
| Sex | Male | Male | Male | Male | Male | Male |
| Serum IgM (mg/dL) | 4130 | 921 | 3630 | 2490 | 1300 | 5790 |
| Serum IgM M-spike (g/dL) | 2.21 | 0.86 | 2.09 | 1.41 | 0.98 | 3.62 |
| Hb (g/dL) | 10.3 | 8.0 | 12.3 | 9.1 | 8.9 | 10.8 |
| Serum β2-microglobulin (mg/L) | 4.5 | N/A | 2.5 | 5.6 | 14.2 | 3.9 |
| BM involvement (%) | 30 | N/A | 70 | 40 | 5 | 30 |
| Prior therapies | Fludarabine, R, CPR, bendamustine-R, bortezomib/dex, tositumomab | R | R, ofatumumab, C, pentostatin | R | Cladribine, C, R, IFN-α, bendamustine, bortezomib/dex | R, chlorambucil, bendamustine |
| Time on ibrutinib (mo) | 9.6 | 7.7 | 37.1 | 36.4 | 9.1 | 23.0 |
| Best response to ibrutinib | PR | PR | PR | VGPR | VGPR | PR |
| Events supporting progressive disease from best response | BM 30%→70% IgM M-spike 0.55→1.12 Hb 11.8→10.5 | IgM M-spike NQ→0.55 Hb 9.5→7.3 | BM 20%→90% Hb 13.4→10.7 | Splenic enlargement Hb 10.6→7.8 New pleural effusion | BM 5%→60% IgM M-spike 0.23→0.81 Hb 9.6→8.4 | BM 15→80% IgM 2647→3970 Hb 15.1→7.7 |
| MYD88 status | L265P | L265P | L265P | S243N | L265P | L265P |
| CXCR4 status | WT | S339fs | S338X | WT | WT | S338fs |
C, cyclophosphamide; dex, dexamethasone; fs, denotes frameshift mutation present at this amino acid site; Hb, hemoglobin; IFN, interferon; IgM, immunoglobulin M; N/A, not available; NQ, faint, not quantifiable; P, prednisone; PR, partial response; R, rituximab; VGPR, very good partial response.
Sanger sequencing and cloning of BTKCys481 mutants
A 382 bp fragment covering the BTKCys481 was amplified by PCR. The forward and reverse PCR primers were 5′-TGAGAAGCTGGTGCAGTTGTATG-3′ and 5′-CTGGAGATATTTGATGGGCTCAG-3′, respectively. The amplified PCR products were isolated by QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), and sequenced using forward and reverse PCR primers. PCR products were cloned into the TA cloning vector, and selected colonies sequenced using the M13 primers (Genewiz, South Plainfield, NJ).
Development of quantitative AS-PCR assays for BTK mutations at Cys481
Highly sensitive nested-PCR assays were developed to detect known BTKCys481 mutations that included BTKCys481Ser(c.1635G>C and c.1634T>A) and BTKCys481Arg(c.1634T>C).
The primers are listed in supplemental Table 1 on the Blood Web site, and the assay conditions are presented in the supplemental Appendix. The amplification plots, dissociation curves, and standard curves for BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C) assays are presented in supplemental Figure 1.
Estimation of WM cell fraction that expressed BTKCys481 mutations
To estimate the proportion of WM cells that expressed BTKCys481 mutations, MYD88L265P was assumed as a tumor marker. The fraction of cells expressive of BTKCys481 and MYD88L265P mutations was determined by ΔCT and standard curves. The ratio of cells expressing BTKCys481/MYD88L265P was calculated. Copy number variants for the MYD88L265P locus were also determined by a TaqMan assay.20 Using targeted deep sequencing analysis, the ratio of mutated BTKCys481/MYD88 allele frequency was determined.
Targeted deep sequencing
Targeted deep NGS was performed for 8 patients that included 6 patients with ibrutinib progression, and 2 others on active ibrutinib without clinical progression at the time of sampling who were positive for BTK mutations by AS-PCR. Tumor samples from the 8 patients, along with germ line (CD19-depleted peripheral blood mononuclear cells) samples available for 6 of these patients were sequenced. The library was generated using HaloPlexHS 1-500kb (Agilent Technologies, Santa Clara, CA), and sequencing data were generated from MiSeq paired-end sequences and aligned to HG19/GRCh37 ensemble genome reference using SureCall (http://www.agilent.com/search/?Ntt=surecall).21 Molecular bar coding was used to identify reads, and reads supporting each call were calculated using the Integrative Genomics Viewer (Broad Institute, Cambridge, MA). The median number of reads per patient was 774 (range, 372-1895). The genes included in the targeted deep sequencing included BTK, PLCγ2, CARD11, LYN, HCK, MYD88, and CXCR4.
Results
Sanger, cloning, and sequencing results for WM patients who progressed on ibrutinib
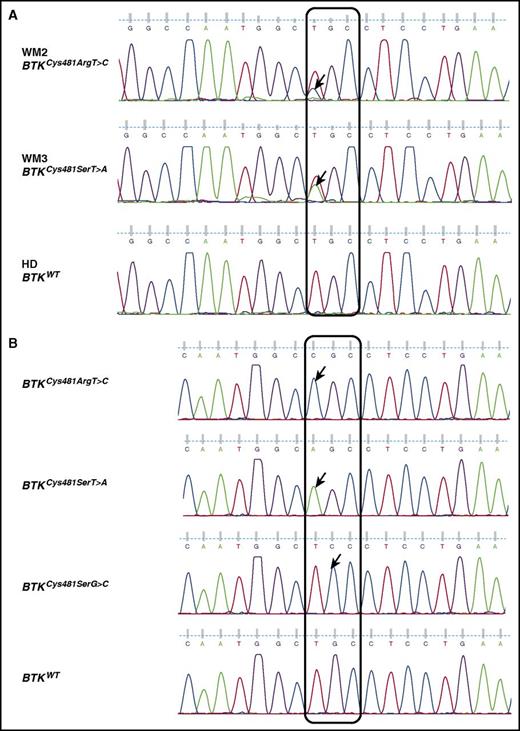
Six WM patients who progressed on active ibrutinib therapy were sequenced for BTKCys481 mutations. Of the 6 patients, 5 had a MYD88L265P and 1 had a MYD88S243N mutation. Three of these 6 patients (50%) also had CXCR4 mutations. By Sanger sequencing, a BTKCys481Arg(c.1634T>C) mutation was identified in patient WM2, whereas a BTKCys481Ser(c.1634T>A) was found in patient WM3 (Figure 1A). Cloning and sequencing analysis confirmed the presence of these mutations, and identified additional BTKCys481 mutations in both patients as follows: 17/107 (15.9%), 21/107 (19.6%), and 7/107 (6.5%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM2; whereas 2/119 (1.7%), 46/119 (38.7%), and 8/119 (6.7%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM3. Baseline samples were available for 5 of the 6 progressing patients (all except WM2). No BTK mutations were detected by Sanger, and cloning and sequencing studies, in these samples. Representative tracings for the cloning and sequencing studies are shown in Figure 1B.
Representative Sanger sequencing traces for patients with BTKCys481variants. (A) Sanger sequencing traces on CD19-sorted BM cells showing BTKCys481Arg(c.1634T>C) and BTKCys481Ser(c.1634T>A) variants in patients WM2 and WM3, respectively. (B) Representative Sanger sequencing traces from cloning and sequencing studies show the presence of multiple BTKCys481 variants in patient WM2. Cloning and sequencing analysis showed 17/107 (15.9%), 21/107 (19.6%), and 7/107 (6.5%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM2; whereas 2/119 (1.7%), 46/119 (38.7%), and 8/119 (6.7%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM3. Arrows denote nucleotide variants. HD, healthy donor.
Representative Sanger sequencing traces for patients with BTKCys481variants. (A) Sanger sequencing traces on CD19-sorted BM cells showing BTKCys481Arg(c.1634T>C) and BTKCys481Ser(c.1634T>A) variants in patients WM2 and WM3, respectively. (B) Representative Sanger sequencing traces from cloning and sequencing studies show the presence of multiple BTKCys481 variants in patient WM2. Cloning and sequencing analysis showed 17/107 (15.9%), 21/107 (19.6%), and 7/107 (6.5%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM2; whereas 2/119 (1.7%), 46/119 (38.7%), and 8/119 (6.7%) clones expressed BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C), respectively, for patient WM3. Arrows denote nucleotide variants. HD, healthy donor.
Detection of BTKCys481 mutations in WM patients who progressed on ibrutinib using nested AS-PCR
The findings from the cloning and sequencing analyses encouraged us to develop more sensitive methods to evaluate for ibrutinib-resistant clones. To ensure high sensitivity and reliability, we developed nested AS-PCR assays for BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C). The AS-PCR assays developed for BTKCys481Ser(c.1635G>C) and BTKCys481Ser(c.1634T>A) detected these mutations at a dilution of 0.1%, and for BTKCys481Arg(c.1634T>C) at a dilution of 0.8% with ≥2-cycle difference from the WT DNA background. The details for the development of these assays are presented in the supplemental Appendix.
We then applied the nested AS-PCR assays to the 6 WM patients who progressed on ibrutinib. The samples from baseline and the time of disease progression were analyzed in parallel. We identified by nested AS-PCR assays the three BTKCys481 mutations, which we found by Sanger, and cloning and sequencing analyses, in patients WM2 and WM3. In addition, a BTKCys481Ser(c.1635G>C) mutation was also identified in patient WM6 that was not detected by Sanger sequencing. Thus, the nested AS-PCR assays were able to identify BTKCys481 mutations in 3 of 6 WM patients who progressed on ibrutinib that included the 2 patients with multiple BTKCys481 mutations. All 3 of these patients also carried MYD88L265P and CXCR4 mutations. Baseline samples were available for 5 of the 6 progressing patients (all except WM2). No BTK mutations were detected by AS-PCR studies in these samples.
To better understand the proportion of tumor cells expressing BTKCys481 mutations in these patients, MYD88L265P was assumed as a tumor marker and the ratio of cells expressing BTKCys481 to MYD88L265P was calculated. No copy number variants in the MYD88L265P locus were detected by TaqMan assays in any of the patients. The fraction of cells expressing BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), and BTKCys481Ser(c.1635G>C) relative to MYD88L265P was 32.3%, 12.9%, and 4.2%, respectively, for patient WM2; and 1.2%, 28.0%, and 2.5%, respectively, for patient WM3. Therefore, the combined fraction of cells expressing the BTKCys481 mutations relative to MYD88L265P were 49.4% and 31.7% for patients WM2 and WM3, respectively. In patient WM6, the fraction of cells expressing BTKCys481Ser(c.1635G>C) relative to MYD88L265P was 3.8%.
Serial samples were only available from patient WM3 to permit multiple time point longitudinal analysis for BTKCys481 mutations by nested AS-PCR assays. This patient had cryoglobulinemia that resulted in erratic IgM readings. Serial BM and Hb levels were therefore tracked. At baseline, the patient had fatigue with an Hb level of 12.3 g/dL and BM tumor involvement of 80%. At month 12, the patient reported a good energy level with an Hb level of 13.4 g/dL and a repeat BM biopsy showed 50% involvement. At month 24, the patient continued to describe a good energy level with an Hb level of 13.3 g/dL, and BM biopsy showed 20% tumor infiltration. By month 36, the patient was again complaining of fatigue, with a decrease in Hb level to 9.3 g/dL, and a repeat BM biopsy showed 90% tumor infiltration.
Coincident with the above time points, no BTKCys481 mutations were detected by AS-PCR assays at baseline or at month 12. At month 24, BTKCys481Ser(c.1634T>A) and BTKCys481Ser(c.1635G>C) mutations were first detected, and constituted 0.71% and 0.19% of the total MYD88L265P clone. By month 36, the BTKCys481Ser(c.1634T>A) and BTKCys481Ser(c.1635G>C) mutations were markedly higher at 26.1% and 3.62% of the total MYD88L265P clone. In addition, the BTKCys481Arg(c.1634T>C) clone was also first detectable at month 36 for this patient, and made up 2.54% of the total MYD88L265P clone.
Screening for BTKCys481 mutations by nested AS-PCR assays in patients on active ibrutinib therapy without clinical progression
We next applied the nested AS-PCR assays to samples from 38 patients on active ibrutinib therapy without clinical progression. The median time on ibrutinib for these patients at last BM sampling was 31.9 (range, 3.9-41.3) months, and the median time on ibrutinib was 45.1 (range, 3.9-not reached) months. Among these patients, 2 (5.1%) expressed BTKCys481Ser(c.1635G>C) variants in samples taken after 24 months on ibrutinib. Baseline samples were available for both of these patients, and no BTK mutations were detected by AS-PCR assays. The estimated fraction of cells expressing BTKCys481Ser(c.1635G>C) relative to MYD88L265P was 2.0% for patient WM7, and 1.0% for patient WM8 at last BM sampling. A CXCR4 mutation was also detected in patient WM7 at time of last BM sampling. Subsequently, 6 patients had progressive disease, including 3 with and 3 without systemic progression (2 amyloidosis and 1 Bing-Neel syndrome). Included among the 3 patients with systemic progression were patients WM7 and WM8, both of whom had refractory disease with 85% and 90% disease involvement, respectively, at time of ibrutinib initiation. The time on ibrutinib for these patients was 21.3 and 21.4 months at time of BM sampling for BTKCys481 variants. Both achieved a very good partial response, and progressed to 18.5 and 9.2 months following detection of BTKCys481 variants.
Screening for BTKCys481 mutations by nested AS-PCR assays in ibrutinib-naïve patients
We next applied the nested AS-PCR assays to samples from 50 untreated and 50 previously treated ibrutinib-naïve patients. No BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), or BTKCys481Ser(c.1635G>C) mutations were detected by the AS-PCR assays in any of these patients.
Targeted deep NGS
To confirm the AS-PCR findings and screen for other potential mutations associated with secondary ibrutinib resistance, targeted deep sequencing for MYD88, CXCR4, BTK, PLCγ2, CARD11, LYN, and HCK was performed for the 6 patients who progressed on ibrutinib, as well as the 2 patients on ibrutinib who were detectable by AS-PCR for BTKCys481Ser(c.1635G>C) mutation. For these 8 patients, 6 (WM1, 3, 4, 5, 7, and 8) had paired CD19-depleted peripheral blood mononuclear cells for germ line comparison. All BTKCys481 mutations that were detected by the AS-PCR assays were confirmed by targeted deep sequencing (Table 2). The paired sample analysis confirmed the somatic nature of the BTKCys481 mutations in all 3 patients for whom a germ line sample was available (WM3, 7, and 8). In addition, targeted deep sequencing detected an additional BTKCys481 mutation, ie, BTKCys481Tyr(c.1634G>A) in patients WM2 and WM3. Thus, patients WM2 and WM3 had four distinct BTKCys481 mutations. The fraction of cells with BTKCys481 mutations relative to MYD88L265P was estimated by the deep targeted sequencing reads. As shown in Table 2, the fraction of cells expressing BTKCys481Arg(c.1634T>C), BTKCys481Ser(c.1634T>A), BTKCys481Ser(c.1635G>C), and BTKCys481Tyr(c.1634G>A) relative to MYD88L265P was 32.4%, 6.6%, 5.8%, and 1.0%, respectively, for patient WM2; and 0.3%, 34.4%, 6.5%, and 0.3%, respectively, for patient WM3. In aggregate, the total fraction of cells expressing any type of BTKCys481 mutation relative to MYD88L265P was 45.8% and 41.5% for patients WM2 and WM3, respectively.
Targeted deep sequencing results for BTK, PLCγ2, and CARD11 mutations in WM patients
| Patient . | BTKCys481Arg(T>C) . | BTKCys481Ser(T>A) . | BTKCys481Ser(G>C) . | BTKCys481Tyr(G>A) . | PLCγ2Tyr495His(T>C) . | CARD11Leu878Phe(C>T) . |
|---|---|---|---|---|---|---|
| WM1 | ND | ND | ND | ND | ND | ND |
| WM2 | 32.4% | 6.6% | 5.8% | 1.0% | ND | ND |
| WM3 | 0.3% | 34.4% | 6.5% | 0.3% | ND | 0.2% |
| WM4 | ND | ND | ND | ND | ND | ND |
| WM5 | ND | ND | ND | ND | ND | ND |
| WM6 | ND | ND | 10.3% | ND | 11.9% | ND |
| WM7 | ND | ND | 1.5% | ND | ND | ND |
| WM8 | ND | ND | 0.7% | ND | ND | ND |
| Patient . | BTKCys481Arg(T>C) . | BTKCys481Ser(T>A) . | BTKCys481Ser(G>C) . | BTKCys481Tyr(G>A) . | PLCγ2Tyr495His(T>C) . | CARD11Leu878Phe(C>T) . |
|---|---|---|---|---|---|---|
| WM1 | ND | ND | ND | ND | ND | ND |
| WM2 | 32.4% | 6.6% | 5.8% | 1.0% | ND | ND |
| WM3 | 0.3% | 34.4% | 6.5% | 0.3% | ND | 0.2% |
| WM4 | ND | ND | ND | ND | ND | ND |
| WM5 | ND | ND | ND | ND | ND | ND |
| WM6 | ND | ND | 10.3% | ND | 11.9% | ND |
| WM7 | ND | ND | 1.5% | ND | ND | ND |
| WM8 | ND | ND | 0.7% | ND | ND | ND |
Results are presented for 6 patients who progressed on ibrutinib (WM1-WM6), and for 2 patients (WM7 and WM8) who were positive for a BTKCys481 variant from screening 38 patients on ibrutinib without clinical progression at time of sequencing.
ND, not detected.
In addition to BTKCys481, targeted deep sequencing identified a PLCγ2Tyr495His(c.1483T>C) variant in patient WM6 who had a BTKCys481Ser(c.1635G>C) mutation (Table 2). The fraction of cells expressing BTKCys481Ser(c.1635G>C) and PLCγ2Tyr495His(c.1483T>C) relative to MYD88L265P was 10.3% and 11.9%, respectively, in this patient. The somatic nature of PLCγ2Tyr495His(c.1483T>C) was inferred for this patient who lacked germ line tissue by its rarity in healthy donors, ie, found in only 1 Asian (n = 12 561), but no European (n = 36 590), African (n = 4881), or Latino (n = 5764) individuals (http://exac.broadinstitute.org).22 Lastly, a novel mutation, ie, CARD11Leu878Phe(c.2632C>T) was identified in patient WM3 who had multiple BTKCys481 variants by targeted deep sequencing (Table 2). The fraction of cells expressing this mutation relative to MYD88L265P was 0.2% (4/1895 reads), and was absent in the germ line sample for this patient.
Discussion
Despite high response rates and durable remissions in WM, disease progression can occur on active ibrutinib therapy. Understanding the molecular mechanism(s) responsible for acquired resistance to ibrutinib may improve treatment strategy, and potentially direct novel drug discovery. We therefore performed a targeted genomic analysis in WM patients who progressed on active ibrutinib therapy using multiple sequencing approaches. Akin to CLL and MCL, BTKCys481 mutations were frequently identified in progressing WM patients, and accounted for half of the progression events.
An important distinction was the multitude of BTKCys481 mutations that were identified within individual WM patients. Using mutated MYD88 as a tumor marker, BTKCys481 mutations appeared to be primarily subclonal, with a highly variable clonal distribution. Among the 3 progressing patients with BTKCys481 mutations, 2 (WM2 and WM3) had four different BTKCys481 mutations, with frequencies that ranged from 0.3% to 34.4% for each mutation. No gatekeeper BTKThr474 or non-Cys481 BTK mutations were observed. Baseline samples were available for 5 of the 6 progressing patients (all but WM2), as well as the 2 patients on active ibrutinib (WM7 and WM8), and were analyzed by Sanger sequencing and AS-PCR assays. No BTK mutations were detected suggesting that these mutations were acquired, although they could have existed at such very low clonal frequencies as to not be detectable by our assays. Similar observations have also been made in CLL patients progressing on ibrutinib with acquired BTK mutations. The highly variable and predominantly subclonal nature of BTKCys481 variants in patients progressing on ibrutinib has also been observed in CLL, raising the possibility that other genomic or epigenomic events may also be contributing to resistance. It also remains possible that although the BTK-mutated clone is subclonal, it could be exerting pro-growth and/or survival effects directly or indirectly through micro-environmental interactions on neighboring non–BTK-mutated clones.
The finding of CXCR4 mutations in 4 of the 5 patients with BTKC481 variants may allude to underlying genomic instability. Similar observations have been made in CLL patients, wherein BTK mutations appear more common in 17p (p53)-deleted patients.12 Although TP53 mutations are rare in WM, and were not observed in any of the ibrutinib-resistant patients in this series by targeted NGS (data not shown), dysregulated RAG1, RAG2, and ATM expression are commonly observed in WM.23 Further insights into the role of these dysregulated genes, as well as other genomic or epigenomic variants that contribute to the acquisition of BTKC481 variants in WM patients on ibrutinib are needed. The findings may also indicate an increased susceptibility of CXCR4-mutated patients to develop BTKCys481-related resistance to ibrutinib therapy. A frontline study of single-agent ibrutinib in WM patients with serial deep whole exome sequencing is now fully enrolled and may help validate these observations (#NCT02604511). CXCR4 mutations are subclonal in most WM patients with a highly variable clonality within the WM clone, as defined by mutated MYD88.20 We were unable to address in these studies whether the BTKCys481 variants occurred within CXCR4-mutated cells, and prospective studies utilizing sorted single tumor cell sequencing are needed to clarify this important point.
The emergence and expansion kinetics of the various BTKCys481 variants in WM patients who developed resistance to ibrutinib was also of interest. Although the underlying genomic background within WM clones could have contributed to the emergence and expansion kinetics of the various BTKCys481 variants, the amino acid substitution itself may also be critical. A recent study used site-directed mutagenesis to examine different amino acid substitutions at BTKCys481.24 Substitutions of cysteine by serine and threonine, but not tyrosine, tryptophan, phenylalanine, glycine, or arginine permitted BTK phosphorylation, and triggered downstream PLCγ2 activation in the presence of ibrutinib. BTKCys481Ser were the predominant or sole BTKCys481 variants generated by either T>A or G>C transversions at n.1634 in 4 of 5 WM patients, and are observed in most CLL and MCL patients carrying a BTKCys481 variant. However, in our series, a BTKCys481Arg constituted the predominant variant in 1 patient (WM3) with multiple BTKCys481 clones including BTKCys481Ser, and was observed in another patient (WM2), whereas BTKCys481Tyr variants were seen in 2 patients (WM2 and WM3). Both BTKCys481Arg and BTKCys481Tyr would not have been predicted as promoting ibrutinib resistance.24 Further mechanistic insights are therefore needed to clarify whether an underlying genomic instability in WM patients may account for the multitude of observed BTKCys481 variants, and their potential functionality in MYD88-mutated patients.
Akin to CLL, we also observed a putative PLCγ2 variant in 1 WM patient who also had a BTKCys481SerG>C mutation (WM6).12,16 The PLCγ2Tyr495His mutation observed in the WM patient has not been previously reported, and is located within the auto-inhibitory domain wherein PLCγ2 mutations have been identified in ibrutinib-resistant CLL or MCL patients.25 In vitro transduction studies have eluded to circumvention of BTK signaling by a PLCγ2 mutation found in the auto-inhibitory domain in ibrutinib-resistant patients, and the potential to abrogate aberrant PLCγ2 signaling by agents that target the BCR members LYN or SYK.25 We also observed a CARD11Leu878Phe mutation at a very low frequency in a patient with multiple BTKCys481Ser variants (WM3). CARD11 mutations in the coil-coiled domain were observed in MYD88-mutated activated B-cell subtype of diffuse large B-cell lymphoma patients, and are associated with in vitro, as well as primary clinical resistance to ibrutinib in activated B-cell diffuse large B-cell lymphoma and MCL.26-28 The variant observed by us has not been previously described and resides outside the coil-coiled domain of CARD11. Functional studies are therefore needed to characterize this novel mutation and its potential contribution to ibrutinib resistance.
In conclusion, the findings of this study provide the first reported insights into the molecular mechanisms associated with ibrutinib resistance in WM, and highlight the emergence of multiple BTK-mutated clones, including non-BTKCys481Ser clones, as well as novel PLCγ2 and CARD11 mutations within individual patients who progressed on active ibrutinib therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Edward and Linda Nelson Fund for WM Research, the Kerry Robertson Fund for WM Research, Peter S. Bing, a translational research grant from the Leukemia and Lymphoma Society, a research grant from the International Waldenström’s Macroglobulinemia Foundation, and support from the Bauman Family Foundation. The study is dedicated in memory of Edward Nelson.
Authorship
Contribution: S.P.T., L.X., and Z.R.H. designed the study; C.J.P., K.M., J.G., T.D., M.L.P., R.A., J.J.C., R.R.F., and S.P.T. collected study samples and data; L.X., N.T., G.Y., J.G.C., X.L., M.D., and A.K. processed tumor samples and performed mutation analysis; L.X., Z.R.H., and S.P.T. analyzed the study data; and L.X. and S.P.T. wrote the manuscript.
Conflict-of-interest disclosure: M.L.P., R.A., R.R.F., and S.P.T. have received research funding, speaker honoraria, and/or consulting fees from Pharmacyclics Inc., and/or Janssen Pharmaceuticals. J.J.C. received honoraria from Alexion, Celgene, Janssen, and Pharmacyclics, and research funding from Millennium, Pharmacyclics, Gilead, and AbbVie. The remaining authors declare no competing financial interests.
Correspondence: Steven P. Treon, Bing Center for Waldenström’s Macroglobulinemia, Dana-Farber Cancer Institute, M548, 450 Brookline Ave, Boston, MA 02215; e-mail: steven_treon@dfci.harvard.edu.