Abstract
One of the most studied transcription factors in hematopoiesis is the leucine zipper CCAAT-enhancer binding protein α (C/EBPα), which is mainly involved in cell fate decisions for myeloid differentiation. Its involvement in acute myeloid leukemia (AML) is diverse, with patients frequently exhibiting mutations, deregulation of gene expression, or alterations in the function of C/EBPα. In this review, we emphasize the importance of C/EBPα for neutrophil maturation, its role in myeloid priming of hematopoietic stem and progenitor cells, and its indispensable requirement for AML development. We discuss that mutations in the open reading frame of CEBPA lead to an altered C/EBPα function, affecting the expression of downstream genes and consequently deregulating myelopoiesis. The emerging transcriptional mechanisms of CEBPA are discussed based on recent studies. Novel insights on how these mechanisms may be deregulated by oncoproteins or mutations/variants in CEBPA enhancers are suggested in principal to reveal novel mechanisms of how CEBPA is deregulated at the transcriptional level.
Introduction
Lineage-specific transcription factors (TFs) prime chromatin states of hematopoietic stem/progenitor cells (HSPCs) to drive commitment and differentiation of specific cell types in the bone marrow.1 One of the most studied lineage-specific TFs involved in hematopoietic development is CCAAT-enhancer binding protein α (C/EBPα), a leucine zipper TF mainly involved in myeloid development. C/EBPα has been reported to be involved in monopoieisis and granulopoiesis. The mechanism of action of C/EBPα and the interaction with other leucine zipper proteins driving monopoiesis has been extensively reviewed previously.2 Here, we particularly focus on the role of C/EBPα in early hematopoietic development, granulopoiesis, and malignant transformation of myeloid progenitor cells. C/EBPα is the founder of the C/EBP family of TFs, consisting of C/EBPα, C/EBPβ, C/EBPδ, C/EBPε, C/EBPγ, and C/EBPζ, which are named according to their order of discovery.3-12 All CEBP members share a similar C-terminal domain for DNA binding and dimerization, but differ in the N-terminal domain, with CEBPA possessing two transactivation domains for transcription control and protein interactions. The CEBP family of TFs are involved in many different biological pathways, which has been discussed elsewhere.13
Here, we focus on the importance of C/EBPα as a major TF of the neutrophilic differentiation program and how it is recognized as an indispensable factor for the initiation of acute myeloid leukemia (AML). At the same time, frequent aberrations deregulating C/EBPα function or expression are observed in different AML subtypes. Based on recent findings, we will discuss the transcriptional control driven by a specific enhancer regulating CEBPA in the bone marrow, followed by the potential role of this enhancer to be hijacked by different AML-related oncoproteins to deregulate CEBPA expression. By placing C/EBPα at the center of the myeloid lineage hierarchy, this review offers a perspective on C/EBPα as a target of diverse physiological and oncogenic events, which ultimately contribute to the onset or development of AML.
C/EBPα in normal and malignant myeloid priming and development
The nonredundant role of C/EBPα for neutrophil development
CEBPA is an intronless gene located on chromosome 19q in humans and on chromosome 7 in mice, which encodes a 42-kD and a 30-kD DNA-binding protein, both derived from the same gene, but translated from 2 distinct AUG translational start sites (Figure 1). Besides its role in the bone marrow, C/EBPα is also essential for the development of other organs, such as the lungs, liver, intestines, and female reproductive organs.14-21 In the hematopoietic system, C/EBPα is primarily expressed in cells of the myeloid lineage. Zhang and colleagues19 were the first to show that germ-line deletion of Cebpa in mice causes a block in neutrophil differentiation in the bone marrow. This model limited the study of C/EBPα in adult hematopoiesis, because mice died of lung and liver complications shortly after birth. By generating Mx1-Cre–driven conditional knockout mice, it was demonstrated that the excision of Cebpa in the bone marrow of adult mice failed to generate granulocyte/monocyte progenitors and resulted in a complete block of neutrophilic development at the common myeloid progenitor (CMP) stage.22
CEBPA is located on chromosome 19q.32. From the CEBPA mRNA, 2 major proteins are generated from 2 distinct AUG start sites, namely, a p42 and a p30 isoform (A). CEBPA may be transcribed from the 2 alleles (allele A: biallelic mutations in CEBPA may occur at the N-terminus or at the C-terminus [C/N mutation]). Patients with a C/N double mutation generate a p30 isoform only from 1 allele (termed allele 1) and basic leucine zipper (bZIP) domain–mutated p42 and p30 isoforms (red) from the other allele (B). Biallelic mutations at the N-terminal (N/N) generate p30 isoforms only and not p42 isoforms (C). Biallelic mutations at the C-terminus (C/C) generate p42/p30 C-terminally mutated isoforms from both alleles, all defective in the bZIP domain (D). CEBPA double mutant leukemias never express a wild-type p42 protein.
CEBPA is located on chromosome 19q.32. From the CEBPA mRNA, 2 major proteins are generated from 2 distinct AUG start sites, namely, a p42 and a p30 isoform (A). CEBPA may be transcribed from the 2 alleles (allele A: biallelic mutations in CEBPA may occur at the N-terminus or at the C-terminus [C/N mutation]). Patients with a C/N double mutation generate a p30 isoform only from 1 allele (termed allele 1) and basic leucine zipper (bZIP) domain–mutated p42 and p30 isoforms (red) from the other allele (B). Biallelic mutations at the N-terminal (N/N) generate p30 isoforms only and not p42 isoforms (C). Biallelic mutations at the C-terminus (C/C) generate p42/p30 C-terminally mutated isoforms from both alleles, all defective in the bZIP domain (D). CEBPA double mutant leukemias never express a wild-type p42 protein.
During cell fate decisions, C/EBPα primes and activates the myeloid gene expression program by binding to promoters or enhancers of myeloid-related genes, such as CSF3R, IL-6R, CEBPE, GFI-1, or KLF5,23-26 in mouse models as well as in human CD34+ HSPCs of either cord blood or leukemic origin.27,28 C/EBPα competes with other TFs to attenuate the expression of nonmyeloid lineage genes in progenitors of multilineage potential.29-31 In support of this latter observation, murine HSPCs isolated shortly after Cebpa deletion lose the expression of certain myeloid genes and recapitulate the expression of T-cell genes, such as Cd7 or Lck, suggesting a switch toward a myeloid/T-lymphoid phenotype. This mixed myeloid/T-lymphoid phenotype is also observed in a rare CD34+ leukemia subtype in humans, in which CEBPA is silenced by DNA hypermethylation.32
This early phenotype observed upon CEBPA deactivation in murine models and human leukemia, indicates that C/EBPα (1) has a critical role in the regulation of myeloid gene expression at an early hematopoietic stage, and (2) acts as a repressor of nonmyeloid genes. Both observations are discussed further in the coming sections and are supported by in vitro and in vivo models.
Myeloid priming of hematopoietic stem cells by C/EBPα
Given its probable function as a pioneer TF,33,34 at which stage of differentiation does C/EBPα prime the myeloid genome? Porse and colleagues34 conducted chromatin studies in murine cKit+ CD150+ long-term hematopoietic stem cells (LT-HSCs) to show that C/ebpα binds to chromatin at loci of myeloid-associated genes before they are marked by active chromatin modifications. This finding suggests that C/ebpα binds and primes genes for myeloid commitment. A number of studies suggest that C/ebpα may act as a pioneer TF in cooperation with other TFs, such as Pu.1 or Runx1, to prime the myeloid gene expression program at very early stages of hematopoiesis.33
Wolfler and colleagues35 generated a Cebpa-Cre/YFP reporter mouse and reported that C/ebpα is expressed in <10% of the cKIT+CD150+ defined LT-HSC population in the bone marrow. These findings are in line with recent published data from the groups of Grimes36 and Amit,37 who conducted single-cell RNA sequencing and found that a small proportion of bone marrow HSPCs expresses Cebpa.
In addition to the block in myeloid differentiation, the conditional Cebpa-knockout mouse model by Tenen and colleagues22,38 shows that LT-HSCs exit their quiescent state and enter the G1 phase of the cell cycle, leading to expansion. On the other hand, the conditional Cebpa-knockout mouse model by Porse and colleagues34 showed that LT-HSCs undergo exhaustion and increase in apoptosis. These studies may seem conflicting, but these differences are most probably due to the dissimilarity in timing of the analysis of these mice. In parallel with these findings, a severe quantitative loss of LT-HSCs has also been reported in 2 separate studies in which a myeloid-specific (+37 kb) Cebpa enhancer was deleted.39,40 Therefore, phenotypically, it is very clear that reduced Cebpa expression, either by deleting the Cebpa gene or the Cebpa-enhancer, exerts detrimental effects on the frequency of the LT-HSC population. Interestingly, to investigate whether reduced Cebpa levels also influence the function of hematopoietic stem cells (HSCs), transplantation experiments of Cebpa enhancer-deleted bone marrow cells in recipient mice showed no difference in the number of LT-HSCs when compared with controls at 19 weeks posttransplantation, which can be explained by the presence of residual Cebpa levels after enhancer deletion. In fact, transplantation of bone marrow cells exhibiting complete ablation of Cebpa expression from Cebpa-knockout mice showed a 20-fold reduction in LT-HSC numbers at 16 weeks posttransplantation.34 Altogether, these findings suggest that C/EBPα is required to maintain the integrity of HSCs and, hence, the hematopoietic system.
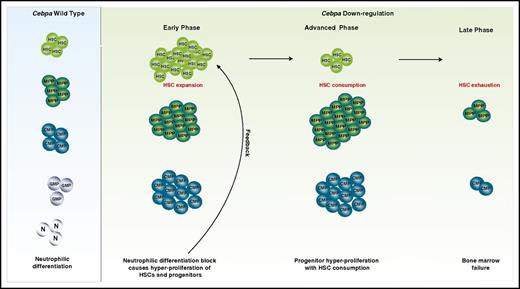
Together, these results all point toward the importance of C/ebpα for the maintenance of bone marrow HSC integrity and raise further an important question: why does the loss of Cebpa in mice in only a subset of stem/progenitor cells influence the behavior of the whole HSPC population, including the Cebpa-negative HSPCs? Possibly, a feedback mechanism is activated to stimulate the generation of additional myeloid-primed progenitors and compensate for the loss of the granulocytic lineage. This would lead to HSC exhaustion caused by an induced HSC response to exit from the quiescence state and undergo cell division to generate sufficient myeloid progenitors (Figure 2). It could also be that the loss of such a critical TF might alter the cell to cell communication between the Cebpa+ HSPC compartment and the bone marrow microenvironment, which further imposes a global effect on the HSC population.41 To understand the mechanism of action of the above-proposed ideas, further experimental investigations are required. Knocking out Cebpa at a later stage of myeloid commitment, such as in CMPs, would provide a sufficient time frame to monitor the biological response of the HSCs to compensate for the loss of granulopoiesis in a time-dependent manner. Transplantation of Cebpa-knockout HSPCs into sublethally irradiated wild-type recipient mice opens new opportunities to study whether these Cebpa-knockout cells stimulate HSC exhaustion, resulting in bone marrow failure. These models should also address the involvement of the microenvironment and whether stromal and endothelial cells from the transplanted host become susceptible to alterations that ultimately influence the HSC population. Whether the proposed events are also a hallmark in human disease is yet to be investigated.
C/EBPα primes HSPCs for neutrophilic differentiation. We hypothesize that downregulation of C/EBPα in the bone marrow in mice results in the following phases. The early phase: a block of neutrophilic differentiation causes a feedback mechanism on HSCs and progenitors to stimulate more differentiation. This results in an increase in cycling of HSCs and expansion of downstream progenitors. Advanced phase: HSCs are consumed, while progenitors still show expansion. Late phase: HSC and progenitor exhaustion leading to bone marrow failure. GMP, granulocytic/monocytic progenitor; MPP, multipotent progenitor; N, neutrophils.
C/EBPα primes HSPCs for neutrophilic differentiation. We hypothesize that downregulation of C/EBPα in the bone marrow in mice results in the following phases. The early phase: a block of neutrophilic differentiation causes a feedback mechanism on HSCs and progenitors to stimulate more differentiation. This results in an increase in cycling of HSCs and expansion of downstream progenitors. Advanced phase: HSCs are consumed, while progenitors still show expansion. Late phase: HSC and progenitor exhaustion leading to bone marrow failure. GMP, granulocytic/monocytic progenitor; MPP, multipotent progenitor; N, neutrophils.
C/EBPα in myeloid reprograming and cell fate decisions
Transdifferentiation studies by Graf and colleagues42,43 previously showed that when an estrogen-inducible C/EBPα-ER (CEBPA fused to the estrogen receptor) is expressed in a nonmyeloid bone marrow cell, it acts in synergy with other TFs to induce myeloid differentiation. They showed that C/EBPα, in collaboration with PU.1 or C/EBPβ, reverses the lymphoid phenotype of B-cell or T-cell progenitors into myeloid progenitors to eventually differentiate into monocytes/granulocytes in vitro. Whether the cell commits to one cell type or another depends on the TF network that is available within the cell under investigation. In the presence of the Yamanaka factors (Oct4, cMYC, KLF4, and SOX2),44 C/EBPα poises change to transdifferentiate B cells rapidly into induced pluripotent stem cells by first generating GMPs.33,45-48 Other studies showed that C/EBPα is also capable of transdifferentiating nonhematopoietic cells into various cell types. For example, fibroblasts can be transformed into myeloid progenitors by C/EBPα in the presence of PU.1 or into adipocytes in the presence of PPAR-γ and SREBP-1.49-52
The repressive role of C/EBPα in hematopoiesis is less understood. Previous studies have shown that C/EBPα induces cell cycle exit coupled with differentiation, by repressing cell cycle–related TFs, including Myc53 and c-Jun.54,55 A recent study showed that C/EBPα represses the B-cell lineage genes by forming a complex with chromatin-modifying proteins, including LSD1 and HDAC1.46 Such a mechanism might also be applied in other cell types because progenitors from Cebpa-knockout mice exhibit upregulation of T-cell–related genes, such as Cd7 and Lck, whereas C/EBPα retroviral reintroduction in these progenitors attenuated the expression of these genes to drive myeloid differentiation.32
During cell fate decisions, Cebpa is negatively regulated by other nonmyeloid factors to exclude the myeloid differentiation program. De Obaldia et al56 reported that in mouse bone marrow, C/ebpα expression is repressed by the Notch1 target, Hes1. Interestingly, they showed that T-cell development in Hes1–deficient progenitors was restored upon C/ebpα deletion, indicating that C/ebpα acts as a main repressor of T-cell development.56 Moreover, Rothenberg and colleagues57 have shown that Cebpa expression in the thymus is lost during T-cell commitment by an extensive increase of a repressive histone mark deposition in the Cebpa locus.
In summary, C/ebpα is a potent differentiation mediator in different cell types, especially for the myeloid lineage. Its importance in activating the myeloid program is indispensable to generate GMP from any cell type, which occurs only in the presence of other pioneer myeloid factors, such as Pu.1. In addition, Cebpa is one of the primary targets to be shut down by other lineage-specific TFs in order to exclude myelopoiesis, emphasizing its important role as a granulocytic differentiation TF.
C/EBPα in AML initiation and development
It has long been observed that AML is initiated in myeloid committed progenitors.58,59 Cozzio and colleagues60 demonstrated that murine CMPs and GMPs that were transduced with an MLL-ENL fusion gene acquired leukemogenic potential in vitro and in vivo. Based on the myeloid progenitor potential of generating in vitro serial replating and serial transplantation with minimal cell numbers in vivo, these cells were termed leukemic stem cells. Such leukemic stem cells with a strong leukemogenic potential were also later confirmed when the MLL-AF9 fusion oncoprotein was retrovirally transduced in mouse GMPs.61 Interestingly, leukemic GMPs shared identical gene expression profiles with wild-type GMPs, but acquired a self-renewal gene expression program to propagate in vitro and in vivo.61 Leukemic blasts from Cebpa biallelic mutant knockin mice share an identical gene expression program with these MLL-AF9–transduced leukemic GMPs.62 Collectively, these findings indicate that a myeloid differentiation program is required to initiate myeloid leukemia.
Moreover, such findings raise paradoxical questions as to whether the expression of C/EBPα, which may act as a tumor suppressor protein, is also required for the initiation of leukemia. Gene expression profiles of AML patient cells showed that CEBPA is sufficiently expressed at the messenger RNA (mRNA) level in all AML subtypes, except for 1 subgroup with a myeloid/T-lymphoid immunophenotype.32 Although this finding needs confirmation at the protein level, such observations indicate that AML requires an adequate degree of differentiation by involving C/EBPα, at least at the early stages of leukemic initiation. Porse and colleagues34 used the MLL-ENL and MLL-AF9 mouse model to test this hypothesis. They compared the degree of leukemogenesis exhibited by these 2 fusion oncoproteins in a Cebpa wild-type and Cebpa-knockout background. The latter failed to generate any leukemia both in vitro and in vivo.34 They confirmed that once leukemia was propagated in the Cebpa wild-type mice, it did not matter anymore whether Cebpa was expressed or not, thus confirming the role of C/EBPα in AML initiation as a differentiation mediator, but not in AML maintenance.
Using similar mouse models, a recent study by Ye et al63 confirms these findings. In addition, they show that by administering the growth factors interleukin-3 or granulocyte-macrophage colony-stimulating factor, the MLL fusion oncoproteins induce myeloid leukemia even in the absence of C/ebpα.63 Thus, a divergent pathway may take over the C/ebpα pathway when enhanced signaling is introduced by the ectopic addition of myeloid growth factors, even when C/ebpα is not expressed. Possibly, this divergence is via a redundant pathway that takes place in cases of “emergency,” and the main factor known to be responsible for similar events in granulopoiesis is the other CEBP family member, C/ebpβ.64 Thus, these findings wrap up the whole concept that leukemia develops only if a sufficient degree of differentiation is present, with C/EBPα being one of the important factors for AML initiation.
Mutant C/EBPα as a driver of AML
CEBPA mutations in AML: biological function and clinical implications
The strong myeloid phenotype observed in Cebpa-knockout mice prompted the Tenen group to investigate AML patients for mutations in the open reading frame (ORF) of CEBPA.34,65-70 Mutations in the CEBPA ORF in human AML occur in ∼7% to 15% of cases.34,65-70 Of these CEBPA-mutant AMLs, 30% exhibit mutations on 1 allele, termed here as CEBPA single mutants (CEBPAsm), with the majority occurring at the N-terminus as out-of-frame mutations. The other 70% of CEBPA mutant AMLs have both alleles affected and are usually termed as CEBPA double mutant (CEBPAdm) AMLs (Figure 1). Usually, 1 allele carries an out-of-frame N-terminal mutation and the other carries an in-frame C-terminal mutation (referred to as an N/C mutant).65 Very rare combinations of biallelic mutations, such as N/N or C/C, also occur in human AMLs and may either be derived from same C or N mutations in the different alleles, or from mitotic recombination of the long arm of chromosome 19.71,72
The N-terminal out-of-frame mutations induce a stop codon after the first ATG site, leading to translation initiation from the third ATG site and generating the shorter p30 isoform in excess, which has been reported to act as a dominant negative of the p42 isoform.34,65-70 An imbalance in the p42:p30 ratio caused by N-terminal mutations is associated with increased proliferation and minimal differentiation of myeloid progenitor cells.73,74 The C-terminal in-frame mutations on the other allele cause a defect in the bZIP domain. However, the C-terminal mutations still generate a balanced ratio of p42:p30 isoforms. Depending on the position of the mutation at the bZIP domain, the C/EBPα p42 is defective either in (1) the DNA-binding domain, which disables the DNA-binding property of C/EBPα, or (2) at the leucine zipper domain, which interferes with the formation of homo- and heterodimers.75-77 This implies that CEBPAdm cases lack the wild-type p42 isoform, thus translating only defective C/EBPα isoforms, which support the onset of leukemia (see the next section).
Those with CEBPAdm are among the AML patients with a favorable prognosis.34,65-70 CEBPA mutation analysis is now routinely carried out at AML diagnosis in many academic centers and is applied as a prognostic marker. A single CEBPA mutation has no predictive value on treatment response or survival in AML. The treatment of choice and outcome for CEBPAsm AML is determined by mutations in other genes, such as in NPM1 and/or FLT3.65,78,79
CEBPAdm but not CEBPAsm drives the onset of AML in humans and in mice
All CEBPAdm AMLs carry a very similar gene expression profile that distinguishes them from other AML subtypes, including CEBPAsm leukemias.65,67,80 In fact, a gene expression signature of ∼20 genes only has been defined, which predicts the presence of a CEBPAdm in AML. These results demonstrate that CEBPAdm AML should be considered as a unique AML subtype that is biologically different from CEBPAsm leukemias.
Mutations in the ORF deregulate the function of C/EBPα, but are they sufficient to drive leukemogenesis? Nerlov and colleagues73,81 addressed this question by generating mutant mice that carried either an N-terminal or a C-terminal mutation in Cebpa. Single-mutant animals did not develop AML, suggesting that other mutations are indeed required. As mentioned above, CEBPAsm AMLs frequently harbor mutations in genes, such as NPM1, FLT3, or ASXL1.78,79 Thus, these mutant genes seem to collaborate with single mutations in CEBPA (mostly N-terminal) to drive AML. An additional line of evidence to this statement is observed in families with germline N-terminal CEBPAsm. Individuals from these families only develop AML if a second mutation is acquired on the other allele (usually as a C-terminal mutant), or else an additional mutation occurs in other AML-related genes.82 This is in line with the mouse reports by Nerlov and colleagues,73,81 demonstrating that AML arises upon transplantation of fetal liver cells carrying combinations of knockin CEBPAdm mutations, namely, N/N, C/N, or C/C mutations, with the C/N combination driving the most aggressive form of AML. The latency of 9 to 14 months for these mice to develop AML suggests that other cooperative mutations are acquired in time, which might decrease the latency of AML in these mice when introduced.
Gene mutations of high interest, frequently observed in human CEBPAdm AML, are the ones that occur in the TF, GATA2. In ∼30% to 40% of CEBPAdm AMLs, mutations occur in GATA2, most frequently in the DNA-binding zinc-finger domain.83,84 In addition, 2 recent studies also presented, for the first time, mutations co-ccurring in the CSF3R gene with CEBPAdm AML.85,86 The generation of models by cross-breeding Cebpa C/N with Gata2 zinc finger mutant mice or with Csf3R mutant mice would aid our understanding of the biological role of these mutations in CEBPAdm-driven human leukemia development.
Transcriptional control of CEBPA
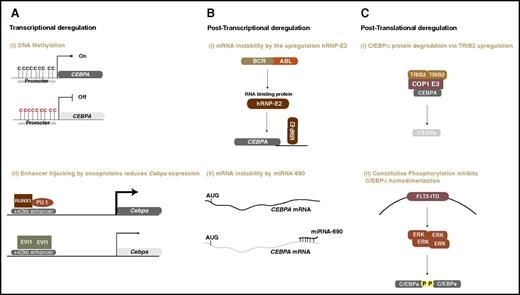
Aberrant mechanisms deregulating CEBPA function or expression occur at the transcriptional, posttranscriptional, translational, and posttranslational level in myeloid malignancies, as shown in Figure 3.87-95 Studies reported transcriptional deregulation of CEBPA either by DNA methylation or by the recruitment of transcriptional repressor complexes to regulatory elements in the CEBPA locus.96 Posttranscriptional deregulation of CEBPA was shown by the BCR-ABL fusion oncoprotein and by micro-RNA-690.93,97 In both scenarios, the CEBPA transcripts are more susceptible to instability. Protein degradation mediated by Tribbles2 and altered phosphorylation through increased FLT3-ITD signaling are examples of aberrant posttranslational control in leukemia.98,99 These mechanisms have previously been reviewed in detail by others,2,100 thus, in this review, we focus more on the emerging roles of chromatin configuration and enhancers of CEBPA in myeloid development and disease.
Oncogenic mechanisms of C/EBPα deregulation in myeloid malignancies. (A) Transcriptional deregulation: (i) promoter DNA methylation silences CEBPA ; and (ii) oncogenic transcription regulators (eg, AML1-ETO or EVI1) bind to CEBPA enhancer and downregulate mRNA expression. (B) Posttranscriptional deregulation: (i) the BCR-ABL oncoprotein upregulates an RNA-binding protein that destabilizes CEBPA transcripts; (ii) upregulation of microRNA-690 destabilizes CEBPA transcripts and alters granulocytic development. (C) Posttranslational deregulation: (i) an increase in tribbles pseudokinase 2 (TRIB2) levels degrades the CEBPA protein by the recruitment of COP1 ubiquitin ligase117 ; (ii) internal tandem duplication in the FLT3 receptor constitutively activates the signaling molecule ERK, which inhibits homodimerization of C/EBPα and, hence, interferes with its function by phosphorylation on serine 21.
Oncogenic mechanisms of C/EBPα deregulation in myeloid malignancies. (A) Transcriptional deregulation: (i) promoter DNA methylation silences CEBPA ; and (ii) oncogenic transcription regulators (eg, AML1-ETO or EVI1) bind to CEBPA enhancer and downregulate mRNA expression. (B) Posttranscriptional deregulation: (i) the BCR-ABL oncoprotein upregulates an RNA-binding protein that destabilizes CEBPA transcripts; (ii) upregulation of microRNA-690 destabilizes CEBPA transcripts and alters granulocytic development. (C) Posttranslational deregulation: (i) an increase in tribbles pseudokinase 2 (TRIB2) levels degrades the CEBPA protein by the recruitment of COP1 ubiquitin ligase117 ; (ii) internal tandem duplication in the FLT3 receptor constitutively activates the signaling molecule ERK, which inhibits homodimerization of C/EBPα and, hence, interferes with its function by phosphorylation on serine 21.
The topology of the CEBPA locus in transcription regulation
Studies are revealing the role of enhancers on CEBPA transcriptional output during neutrophilic maturation in the bone marrow40,101 and how they are susceptible to deregulation by oncogenic mechanisms in AML. CEBPA is located in a topological-associated domain (TAD) of 170 kb on the long arm of chromosome 19, with borders demarcated by the architectural protein, CTCF.39,102,103 These borders restrict interactions to occur between genes and their corresponding enhancers within the TAD. Genes that occur within the same TAD are usually coregulated.104,105 In fact, the CEBPA-TAD also includes another CEBP family member, CEBPG, which is located 5′ of CEBPA. Both genes are located at close proximity in the nuclear 3-dimensional space and form loop interactions with other 14 potential enhancers located 5′and 3′ of CEBPA. The enhancers within the CEBPA TAD are active in different CEBPA-expressing tissues in combinatorial patterns and in a cell type–specific manner. In the bone marrow, myeloid-primed progenitors and mature myeloid cells exhibit a combinatorial pattern of 8 active enhancers from the 14 found in the whole locus. Of these 8 enhancers, 1 located at +42 kb has been studied extensively.39,40,101,106,107 This enhancer is highly conserved between humans and mice (located at +37 kb in mice) and is active in the fetal liver and the dorsal aorta during early fetal development.108 At later developmental stages and in adult life, it’s main function is to prime the myeloid gene expression program for neutrophilic differentiation by forming chromatin complexes, involving HSC TFs, such as PU.1, ERG, RUNX1, and C/EBPα itself to modulate CEBPA expression. In 2 separate studies, deletion of this enhancer in mice downregulated the expression of Cebpa, which ultimately reduced GMP formation and neutrophilic differentiation. The bone marrow cells lacking the +37 kb enhancer showed characteristics of myeloid transformation, namely, they could indefinitely be replated with no evidence of neutrophil development in vitro, similar to C/ebpα knockout bone marrow cells. Other enhancers seem to be important for neutrophil development as well, but at later stages of maturation. Multiple enhancer knockouts of these enhancers, either alone or in combination, should shed light on their function in the regulation of Cebpa expression and myeloid development.
Hijacking of the CEBPA locus by potential oncoproteins
Given that the +37 kb enhancer acts autonomously to regulate Cebpa expression in early HSPCs and later in CMPs and GMPs, it is possible that changes in enhancer function, possibly in combination with the other enhancers, might have potential leukemiogenic implications in humans. Genome-wide investigations of oncogenic TF binding show that leukemia-associated oncoproteins physically interact with several genomic loci to deregulate the expression of protooncogenes and tumor suppressor genes.109 The +42 kb enhancer (human +37 homolog) is a common target for the fusion oncoproteins known as RUNX1-RUNX1T1 (previous called AML1-ETO) and CBFB-MYH11 (unpublished data). Similarly, EVI1, also called MECOM or PRDM3, binds the +42 Kb enhancer in human EVI1-transformed leukemias (R.D., Eric Bindels, and Marije Havermans, personal observations). Interestingly, CEBPA expression in these subtypes of AML is often downregulated, and the direct binding of these oncoproteins to the enhancer could explain this effect. In fact, Perkins and colleagues94 demonstrated that Evi1 binding to the murine +37 Kb enhancer is associated with a reduction of Cebpa expression in Evi1-expressing mouse myeloid leukemia lines. Potentially, the oncoproteins may alter active chromatin states at the enhancer(s), leading to inactivation of CEBPA. Whether this mechanism is also the case in human AML needs further investigation.
The oncoprotein RUNX1-RUNX1T1 has been long associated with negative regulation of CEBPA expression in AML,110-113 but the mechanism underlying this interesting finding is poorly understood. RUNX1-RUNX1T1 is known to recruit histone deacetylases via the nervy homologs of RUNX1T1 and binds to DNA on promoters and enhancers via the DNA-binding domain of RUNX1. Because RUNX1-RUNX1T1 binds the +42 kb enhancer at RUNX1 motifs, as demonstrated in cell lines as well as in patient samples,112 we hypothesize that RUNX1-RUNX1T1 recruits histone deacetylases, causing deacetylation, and disturbs the enhancer-promoter interaction leading to the downregulation of CEBPA expression.
Unexplained low CEBPA levels in AML: do mutations or nucleotide variants in the regulatory domains play a role?
Low CEBPA expression without any evidence of known underlying AML-related abnormalities accounts for 10% to 30% of AMLs. Unknown oncoproteins that may deregulate CEBPA expression cannot be excluded. However, other possible oncogenic mechanisms, such as deletions or mutations causing alterations in TF-binding sites, should be further investigated. The presence of single nucleotide variants occurring in TF consensus sequences within enhancers might be another reasonable explanation for decreased CEBPA levels. Genome-wide association studies showed several events of single nucleotide variants (SNVs) occurring at enhancers of disease-causing genes.114,115 Using the knowledge revealed so far about the role of SNVs or mutations in gene deregulation and disease susceptibility, it could be hypothesized that such alterations may be causative for low CEBPA expression in these CEBPAlow AML subgroups. The application of DNA custom capture sequencing in large AML cohorts would be a potential tool to reveal mutations or SNVs that may be occurring in the CEBPA locus. Such studies could aid our understanding of deregulated expression of CEBPA in myeloid malignancies. These potential mutations or SNVs could be used as susceptibility markers for myeloid-related disorders in the normal population as well as in preleukemic conditions.
Concluding remarks and future perspectives
This review provides a comprehensive perspective of indispensable role of C/EBPα in neutrophilic differentiation. The emerging dual function of C/EBPα in AML received particular attention in this review; adequate CEBPA levels are required for the initiation of AML, whereas mutations in its ORF or deregulated expression are key for the leukemic state. The role of C/EBPα in HSPC biology is still not well understood. This is largely masked by the heterogeneity of bone marrow samples obtained from mouse models or patients, which makes it a challenge to define clearly its function within the HSPC population. Single-cell RNA sequencing is an emerging technology that overcomes the problem of heterogeneity. The application of this technology, in combination with in vitro and in vivo functional studies, provides a sensitive platform to delineate the function of CEBPA in HSPCs. In this review, we also discussed the potential of bypassing the C/EBPα-differentiation pathway by the administration of growth factors63,64 Otherwise, it has been proposed that C/EBPβ could take over and drive neutrophil development in the absence of functional C/EBPα via emergency hematopoiesis.101 In cases where C/EBPα is expressed but functionally defective, such an approach can open a new window for drug administration to induce differentiation in a C/EBPα-independent pathway. Another alternative is to reactivate C/EBPα expression by small molecules.116 These approaches offer a therapeutic alternative for a wide range of patients with low CEBPA expression that might be predisposed to preleukemic conditions and, hence, overcome any differentiation-related aberrations in combination with other currently available therapy.
Acknowledgments
The authors thank Bas Wouters for critically reading the manuscript.
This work was supported by the Dutch Cancer Foundation “Konining Wilhelmina Fonds” and the Ladt Tata Memorial Trust Foundation.
Authorship
Contribution: R.A. and R.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Department of Hematology, Erasmus University Medical Center, Doctor Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.

![Figure 1. CEBPA is located on chromosome 19q.32. From the CEBPA mRNA, 2 major proteins are generated from 2 distinct AUG start sites, namely, a p42 and a p30 isoform (A). CEBPA may be transcribed from the 2 alleles (allele A: biallelic mutations in CEBPA may occur at the N-terminus or at the C-terminus [C/N mutation]). Patients with a C/N double mutation generate a p30 isoform only from 1 allele (termed allele 1) and basic leucine zipper (bZIP) domain–mutated p42 and p30 isoforms (red) from the other allele (B). Biallelic mutations at the N-terminal (N/N) generate p30 isoforms only and not p42 isoforms (C). Biallelic mutations at the C-terminus (C/C) generate p42/p30 C-terminally mutated isoforms from both alleles, all defective in the bZIP domain (D). CEBPA double mutant leukemias never express a wild-type p42 protein.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/15/10.1182_blood-2016-09-687822/4/m_blood687822f1.jpeg?Expires=1767732291&Signature=A2KwT6qj3SWbO1UPneZXigcFTNAmjm58scKFjggI~Z07loawBKQ7vxpktefo7dKtSfOp1JHxbHLcJWejmRBA-BnDo7s3YyjqCuwXtIfQoHyGf2ayyMyLWi6NNZO6ItslBDggpCM75MaurhicIk4nTDDBrqTV89XnXOnulG0vBIvUHn1kuevN4s-XH1NrA6JN3MIKL3i4h486cBDMWMMdS85VzhQkRFeD2dm3equkK0G4vTSKlrmHqwf-Kgvss4mvZXuyO7UELqZPDWzID3AGQKy05t3cQ-lOC6LtBNOv0Xhqszeln9qzo8DXovQgqxWNprjAW8IglGEJuxjgwhC2kA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal