Key Points
MDSCs are expanded in AML and contribute to tumor-related immune suppression.
MUC1 mediates MDSC expansion via the promotion of c-myc expression in secreted extracellular vesicles.
Abstract
Myeloid-derived suppressor cells (MDSCs) play a critical role in promoting immune tolerance and disease growth. The mechanism by which tumor cells evoke the expansion of MDSCs in acute myeloid leukemia (AML) has not been well described. We have demonstrated that patients with AML exhibit increased presence of MDSCs in their peripheral blood, in comparison with normal controls. Cytogenetic studies demonstrated that MDSCs in patients with AML may be derived from leukemic or apparently normal progenitors. Engraftment of C57BL/6 mice with TIB-49 AML led to an expansion of CD11b+ Gr1+ MDSCs in bone marrow and spleen. Coculture of the AML cell lines MOLM-4, THP-1 or primary AML cells with donor peripheral blood mononuclear cells elicited a cell contact–dependent expansion of MDSCs. MDSCs were suppressive of autologous T-cell responses as evidenced by reduced T-cell proliferation and a switch from a Th1 to a Th2 phenotype. We hypothesized that the expansion of MDSCs in AML is accomplished by tumor-derived extracellular vesicles (EVs). Using tracking studies, we demonstrated that AML EVs are taken-up myeloid progenitor cells, resulting in the selective proliferation of MDSCs in comparison with functionally competent antigen-presenting cells. The MUC1 oncoprotein was subsequently identified as the critical driver of EV-mediated MDSC expansion. MUC1 induces increased expression of c-myc in EVs that induces proliferation in the target MDSC population via downstream effects on cell cycle proteins. Moreover, we demonstrate that the microRNA miR34a acts as the regulatory mechanism by which MUC1 drives c-myc expression in AML cells and EVs.
Introduction
Acute myeloid leukemia (AML) is a lethal hematologic malignancy affecting over 21 380 people in the United States every year.1 AML arises in the context of a bone marrow microenvironment characterized by an immunosuppressive milieu that fosters tumor growth and immune escape.2 Critical elements of this environment include increased presence of accessory cells with an inhibitory phenotype that polarizes cells toward a tolerizing phenotype.3
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous group of immature myeloid cells with potent immune-suppressing activity.4 Increased presence of MDSCs is associated with tumor progression,5 poor outcomes,6 and decreased effectiveness of immunotherapeutic strategies.7 MDSCs are characterized by the expression of the myeloid markers CD11b and CD33 and absent HLA-DR.8 Two distinct subsets have been further characterized: monocytic MDSCs, with the phenotype CD15−, and granulocytic MDSCs, that are CD15+.4 Although both subtypes have been identified in healthy patients,9 levels are increased in patients with solid malignancies10 and premalignant conditions.11,12 MDSCs exert diverse effects in modulating the interactions between immune effector cells and the malignant cells. MDSCs directly suppress effector CD8+ T cells via T-cell receptor downregulation, mediated by the expression of the enzymes arginase-1 and inducible nitric oxide synthase and by the production of reactive oxygen species.4,13
Although increased numbers of clonally distinct MDSCs have been reported in patients with myelodysplastic syndrome,12 the role of MDSC populations or their function in AML has not been well elucidated. Of note, immature myeloid cells such as MDSCs share common characteristics with myeloid leukemia cells because of the early maturation arrest of leukemic cells. For example, it has been suggested that AML blasts exert their suppressive effects on T cells via a similar arginase-1–dependent mechanism to MDSCs.14-17 These observations lead us to investigate the presence and importance of MDSCs in AML and the critical pathways underlying their accumulation and function. In particular, we investigated the mechanisms of intercellular signaling between the AML tumor cell and the surrounding cells of the immune microenvironment, including MDSCs.
The primary mediator of MDSC expansion in the setting of malignancy is thought to be tumor secretion of inflammatory cytokines such as tumor necrosis factor alpha,18,19 interleukin-1B (IL-1B),20 IL-12,21 IL-18,22 and IL-6.9 More recently, tumor-secreted extracellular vesicles have been demonstrated to be an important mediator of MDSC expansion.23,24 Extracellular vesicles (EVs) are membrane-bound vesicles released ubiquitously by cells and are thought to be important mediators of intercellular communication.25 EVs have a complex nomenclature, which includes the terms exosomes, microvesicles, and oncosomes, defined by size and ranging from 40 to 1000 nM.26-28 Although their biological relevance in cancer has yet to be fully elucidated, it is generally agreed that they carry biologically relevant proteins, messenger RNAs (mRNAs), and microRNAs.28 It has been demonstrated that AML cells release membrane-bound extracellular vesicles,29-32 which transport microRNAs (miRNAs),33 mRNAs,31 cytokines,30 and tumor-derived proteins29 to surrounding cells. Of relevance, the tumor-suppressing microRNA miR34a, a target of p53, has been shown to be crucially involved in regulating the expansion of MDSCs.34
In the present study we demonstrate that patients with AML exhibit increased presence of MDSCs in their peripheral blood in comparison with normal controls. Of note, we demonstrate that MDSCs in patients with AML may be derived from leukemic or apparently normal progenitor populations, suggesting an effect of the tumor on the surrounding myeloid populations irrespective of their clonal derivation. We report on the novel observation that expansion of MDSCs in AML is accomplished by tumor-derived EVs that are shed into the microenvironment and taken up myeloid progenitor cells, resulting in the selective proliferation of MDSCs in comparison with functionally competent antigen-presenting cells.
MUC135 is a uniquely important oncoprotein critically involved in the self-renewal, cell proliferation, and resistance to apoptosis of leukemic cells.36 Although previous reports suggest that MUC1 signaling may exert immunosuppressive effects,37 the nature and mechanism mediating this effect have not been fully elucidated. In the present report, we have identified the MUC1 oncoprotein as a critical driver of the EV-mediated expansion of MDSCs in AML. Specifically, MUC1 induces increased expression of the c-myc in EVs that induce proliferation in the target MDSC population via downstream effects on cell cycle proteins. We subsequently noted that MUC1 appears to mediate its effect on c-myc expression in AML and EVs via posttranscriptional mechanisms. Noncoding RNAs have emerged as critical regulators of biologic pathways and oncogenesis because of their selective binding to target mRNAs and disruption of protein translation.38 MiR34a has been previously described as impacting MDSC expansion in solid tumor models.34 In the present report, we demonstrate that miR34a acts as the regulatory mechanism by which MUC1 drives c-myc expression in AML cells and EVs. Accordingly, silencing of MUC1 results in increased levels of miR34a, a resultant decrease in c-myc expression in the EV population, and a corresponding decrease in AML-mediated expansion of MDSCs.
Materials and methods
For additional information, see the supplemental Methods on the Blood Web site.
Cells and cell lines
Peripheral blood and bone marrow (BM) aspirates were obtained from AML patients in accordance with a protocol approved by the institutional review board. Samples were subjected to Ficoll density gradient centrifugation (Histopaque-1077, Sigma, St. Louis, MO), washed with phosphate-buffered saline (PBS), and used fresh. For experiments using primary AML cells, diagnostic marrows were used with a minimum of 90% blast involvement. For experiments using healthy donor peripheral blood mononuclear cell (PBMCs), total cell fractions were used.
Detailed information regarding cell lines and generation of MUC1 silenced, miR34a overexpressing, and miR-34a silenced cell lines can be found in the supplemental Methods.
Coculture of AML cells and PBMCs
PMBCs from healthy donors were seeded in 6-well plates (Corning Inc., Corning, NY) at 1.5 × 106 cells per well. AML cells from cell lines or fluorescence-activated cell sorter primary samples were irradiated at 7500 rad to prevent proliferation and fluorescently labeled with GranToxiLux dye (OncoImmunin, Gaithersburg, MD). We added 1.5 × 104 AML cells to test wells (ratio of 100:1). Cells were cultured at 37°C in a humidified 5% CO2 incubator and maintained in RPMI 1640 medium (Cellgro, Manassas, VA), supplemented with heat-inactivated 10% human serum albumin (Sigma, St. Louis, MO) and 100 IU/mL of penicillin and 100 µg/mL of streptomycin (Cellgro, Manassas, VA). After 5 days, cells were collected, rinsed twice with PBS, and phenotypically analyzed as described, using the GranToxiLux dye to exclude tumor cells.
Statistical analysis
The Student t test was used to assess statistical significance.
Results
MDSCs are expanded in patients with AML
MDSCs were quantified in peripheral blood samples from patients with active AML and compared with results from healthy controls. Gating strategy is shown in Figure 1A, whereby AML blasts are gated out on the basis of previously clinically defined phenotypes. In this representative example (patient 6), HLA-DR positive blasts are gated out, before CD11b+/HLA−DR−/CD33+ MDSCs are gated on and quantified first as a percentage of total cells (tumor excluded) (Figure 1B) (patients 1–8) and (as in all subsequent figures) as a percentage of immature CD11b+/HLA-DR− myeloid cells (Figure 1C). Disease characteristics of patients from whom samples were obtained are outlined in supplemental Table 1. The mean level of circulating MDSCs in patients with AML in comparison with normal controls was 7.94% (range, 1.70–17.0) and 0.2% (range, 0.02–0.88), respectively (P < .05) (Figure 1B). MDSCs comprised both monocytic and granulocytic fractions (Figure 1D) and were predominantly lineage negative (supplemental Figure 1A). Of note, patients with active AML also demonstrated high levels of MDSCs in bone marrow aspirates (24.2%; range, 0–82) (data not shown).
MDSCs are expanded in patients with AML and can be cytogenetically related to the malignant clone. PBMCs were isolated by Ficoll density-gradient centrifugation and stained with antibodies for CD11b, HLA-DR, CD14, CD15, and CD33 expression. The cells were then analyzed with flow cytometry. (A) Representative example of patient 2 is shown. CD11b+ HLA-DR+ CD14− blasts are shown in gate I (light blue). Monocytic MDSCs (CD11b+ HLA-DR− CD14−/+ CD33+ CD15−) are shown in gate E (purple), granulocytic MDSCs (CD11b+ HLA-DR− CD14−CD33+CD15+) are shown in gate F (orange). PBMCs from AML patients and healthy controls were isolated by Ficoll density-gradient centrifugation and stained with antibodies for CD11b, HLA-DR, CD14, CD15, and CD33 expression. The cells were then analyzed with flow cytometry. If present, tumor cells were gated out on the basis of forward scatter/side scatter and known blast phenotype, and total MDSCs (CD33+CD15− or CD33+CD15+) were quantified as a percentage of total cells (n = 8; P < .05) (B) and as a percentage of gated immature CD11b+/HLA-DR− myeloid cells (n = 7; P < .05) (C). MDSCs were further characterized as granulocytic, by the presence of CD15+, or monocytic, by CD15− and side scatter. (D) Granulocytic and monocytic MDSCs in AML (n = 7) versus healthy donors (n = 9) are shown. MDSCs are shown as a percentage of gated immature CD11b+/HLA-DR− myeloid cells. *P < .05 for both monocytic and granulocytic MDSCs in AML versus healthy donors. SSC, side scatter.
MDSCs are expanded in patients with AML and can be cytogenetically related to the malignant clone. PBMCs were isolated by Ficoll density-gradient centrifugation and stained with antibodies for CD11b, HLA-DR, CD14, CD15, and CD33 expression. The cells were then analyzed with flow cytometry. (A) Representative example of patient 2 is shown. CD11b+ HLA-DR+ CD14− blasts are shown in gate I (light blue). Monocytic MDSCs (CD11b+ HLA-DR− CD14−/+ CD33+ CD15−) are shown in gate E (purple), granulocytic MDSCs (CD11b+ HLA-DR− CD14−CD33+CD15+) are shown in gate F (orange). PBMCs from AML patients and healthy controls were isolated by Ficoll density-gradient centrifugation and stained with antibodies for CD11b, HLA-DR, CD14, CD15, and CD33 expression. The cells were then analyzed with flow cytometry. If present, tumor cells were gated out on the basis of forward scatter/side scatter and known blast phenotype, and total MDSCs (CD33+CD15− or CD33+CD15+) were quantified as a percentage of total cells (n = 8; P < .05) (B) and as a percentage of gated immature CD11b+/HLA-DR− myeloid cells (n = 7; P < .05) (C). MDSCs were further characterized as granulocytic, by the presence of CD15+, or monocytic, by CD15− and side scatter. (D) Granulocytic and monocytic MDSCs in AML (n = 7) versus healthy donors (n = 9) are shown. MDSCs are shown as a percentage of gated immature CD11b+/HLA-DR− myeloid cells. *P < .05 for both monocytic and granulocytic MDSCs in AML versus healthy donors. SSC, side scatter.
MDSCs expand in the presence of AML blasts
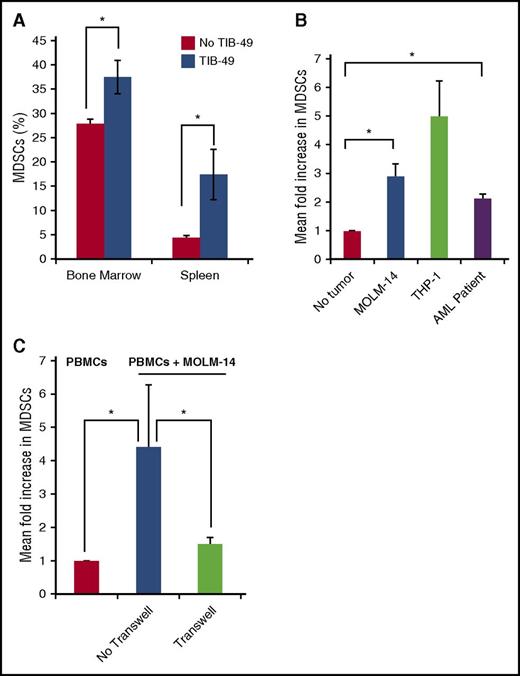
Given these findings, we sought to further characterize the nature of MDSC expansion in AML. In a murine model, leukemic engraftment was associated with increased presence of MDSCs in the spleen and marrow. Mice underwent retro-orbital injection with 1 × 105 cells of the syngeneic murine AML cell line TIB-49. After 3 weeks, at the onset of symptomatic disease, mice were killed and assessed for engraftment (as defined by >1% green fluorescent protein positive [GFP+] cells in the bone marrow) and the presence of murine MDSCs. All TIB-49 injected mice engrafted with leukemia with a mean of 8% GFP+ AML cells in BM and 2.5% in spleen (P < .05) (data not shown). A significant expansion of MDSCs was noted in bone marrow (P < .05) and splenic MDSCs (P < .05), in comparison with a control cohort that had not been challenged with TIB-49 cells (Figure 2A).
MDSCs are expanded in the presence of AML blasts. C57BL/6 mice were inoculated using retro-orbital injections, with 1 × 105 GFP stably transduced murine syngeneic AML TIB-49 cells. At the onset of symptomatic disease at 21 days, mice were analyzed. (A) Bone marrow and splenocytes were analyzed by flow cytometry for the murine MDSC markers mCD11b and mGr-1 (n = 5; *P < .05). (B) PMBCs from healthy donors were cocultured in direct contact with irradiated, fluorescently labeled AML cells at a ratio of 100:1 (PBMCs:AML). After 5 days, cells were analyzed with flow cytometry. Labeled tumor cells were excluded, and total MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells (n = 3; *P < .05 for MOLM-14 and patient AML cells). (C) Healthy donor PBMCs and AML cells were cocultured in direct contact or with 0.4-μM Transwell insert, and MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells (n = 3; *P < .05).
MDSCs are expanded in the presence of AML blasts. C57BL/6 mice were inoculated using retro-orbital injections, with 1 × 105 GFP stably transduced murine syngeneic AML TIB-49 cells. At the onset of symptomatic disease at 21 days, mice were analyzed. (A) Bone marrow and splenocytes were analyzed by flow cytometry for the murine MDSC markers mCD11b and mGr-1 (n = 5; *P < .05). (B) PMBCs from healthy donors were cocultured in direct contact with irradiated, fluorescently labeled AML cells at a ratio of 100:1 (PBMCs:AML). After 5 days, cells were analyzed with flow cytometry. Labeled tumor cells were excluded, and total MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells (n = 3; *P < .05 for MOLM-14 and patient AML cells). (C) Healthy donor PBMCs and AML cells were cocultured in direct contact or with 0.4-μM Transwell insert, and MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells (n = 3; *P < .05).
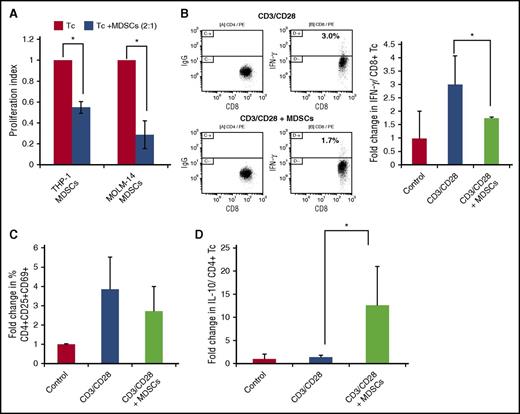
We next sought to further characterize the effect of human AML cells on MDSC expansion using in vitro studies. Coculture of donor PBMCs with MOLM-14 and THP-1 AML cell lines for 5 days resulted in significant fold expansion of cells exhibiting an MDSC phenotype. Similar findings were observed using human AML cell lines and primary patient- derived AML cells in which patients 2 to 4 were used (as per supplemental Table 1) (Figure 2B). An expansion of MDSCs was also seen in an autologous system, whereby patient 7 blasts were cultured with patient 7 PBMCs (once remission was achieved) (supplemental Figure 2). Of note, MOLM-14–mediated expansion of MDSCs was abrogated in a 0.4-μM Transwell system, consistent with the requirement for direct cell contact as opposed to an effect mediated by soluble factors derived from the tumor (Figure 2C). MOLM-14– and THP-1–expanded MDSCs exhibited characteristic immunosuppressive functional properties. Coculture of MDSCs with T cells undergoing activation by CD3/CD28 ligation resulted in blunting of proliferative response (Figure 3A), a 40% reduction in the expression of the markers of T-cell activation (CD69, CD25, CD4) (Figure 3C), a 46% reduction in intracellular expression of interferon-γ (IFN-γ) (Figure 3B), and a 13-fold concurrent increase in CD4 intracellular expression of the inhibitory cytokine IL-10 (Figure 3D). Absolute frequencies of cells are shown in supplemental Table 2).
MDSCs are suppressive of T-cell activation and proliferation. Healthy PBMCs and AML cells were cocultured for 5 days, and MDSCs were isolated by using flow cytometric sorting. T cells autologous to the MDSCs were isolated and stimulated with anti-CD3/CD28 ligation. MDSCs were added at a 2:1 ratio (Tc:MDSCs). (A) After 3 days of culture, T cells were analyzed for proliferation using CellTitreGlo cell luminescence assay (n = 3). After 3 days of culture cells were analyzed for (B) the expression of intracellular IFN-γ by flow cytometry as presented in a representative experiment (left) and summary of three independent experiments (right) and (C) markers of T-cell activation CD4+/CD25+/CD69+ (n = 3) and (D) the expression of intracellular IL-10 by flow cytometry, presented as a summary of three independent experiments. *P < .05. IgG, immunoglobin G.
MDSCs are suppressive of T-cell activation and proliferation. Healthy PBMCs and AML cells were cocultured for 5 days, and MDSCs were isolated by using flow cytometric sorting. T cells autologous to the MDSCs were isolated and stimulated with anti-CD3/CD28 ligation. MDSCs were added at a 2:1 ratio (Tc:MDSCs). (A) After 3 days of culture, T cells were analyzed for proliferation using CellTitreGlo cell luminescence assay (n = 3). After 3 days of culture cells were analyzed for (B) the expression of intracellular IFN-γ by flow cytometry as presented in a representative experiment (left) and summary of three independent experiments (right) and (C) markers of T-cell activation CD4+/CD25+/CD69+ (n = 3) and (D) the expression of intracellular IL-10 by flow cytometry, presented as a summary of three independent experiments. *P < .05. IgG, immunoglobin G.
AML cells release EVs that traffic to surrounding cells, altering the tumor microenvironment
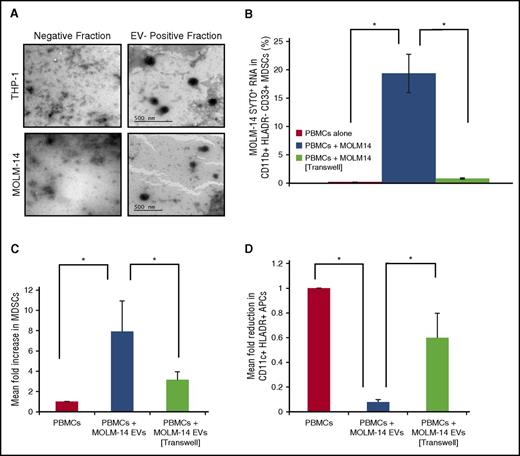
Having demonstrated that MDSC expansion required direct cellular interaction between AML cells and the myeloid compartment, we hypothesized that AML may exert this effect via extracellular vesicles trafficked from AML cells to myeloid accessory cells of the bone marrow microenvironment. The release of EVs by MOLM-14 and THP-1 cells into the surrounding microenvironment was visualized using transmission electron microscopy, demonstrating 100-nm diameter circular structures containing darkly staining protein (Figure 4A). To confirm EV size and purity after isolation, we subjected vesicles to high-sensitivity flow cytometry and compared them with standardized beads, demonstrating minimal debris and vesicles of 200 nm (supplemental Figure 3B). Taken together, we estimate the size of AML EVs to be around 200 nm, because the dehydration step prior to electron microscopy may shrink EVs artificially,39 accounting for the difference seen between these modalities.
AML cells release EVs that traffic to surrounding cells, altering the tumor microenvironment. (A) AML EVs were isolated using a spin-column and visualized using transmission electron microscopy (×1000). The negative spin-column fraction was used as a control. To determine whether AML EVs traffic to surrounding cells, we cocultured PBMCs in direct contact or in a 0.4-μM Transwell, with MOLM-14 AML cells pretreated with SYTO RNA dye (530 nm). (B) After 6 hours, MDSCs were quantified for AML SYTO RNA dye using flow cytometry. Healthy donor PBMCs were cultured for 3 days with AML EVs in direct contact or in a 0.4-μM Transwell and then quantified for (C) CD11b+/HLA-DR−/CD33+ MDSCs (expressed as a percentage of immature CD11b+/HLA-DR− myeloid cells) and (D) HLA-DR+/CD11c+ myeloid APCs by flow cytometry (n = 3). *P < .05.
AML cells release EVs that traffic to surrounding cells, altering the tumor microenvironment. (A) AML EVs were isolated using a spin-column and visualized using transmission electron microscopy (×1000). The negative spin-column fraction was used as a control. To determine whether AML EVs traffic to surrounding cells, we cocultured PBMCs in direct contact or in a 0.4-μM Transwell, with MOLM-14 AML cells pretreated with SYTO RNA dye (530 nm). (B) After 6 hours, MDSCs were quantified for AML SYTO RNA dye using flow cytometry. Healthy donor PBMCs were cultured for 3 days with AML EVs in direct contact or in a 0.4-μM Transwell and then quantified for (C) CD11b+/HLA-DR−/CD33+ MDSCs (expressed as a percentage of immature CD11b+/HLA-DR− myeloid cells) and (D) HLA-DR+/CD11c+ myeloid APCs by flow cytometry (n = 3). *P < .05.
To determine whether AML EVs traffic to surrounding cells, were cocultured PBMCs with MOLM-14 AML cells pretreated with an RNA dye that concentrates in the secreted extracellular vesicles. After 6 hours, MDSCs demonstrated transfer of the tumor-derived RNA-specific dye by flow cytometric analysis, consistent with trafficking of EVs from the MOLM-14 cells into the MDSC population, which was abrogated in the 0.4-μM Transwell (Figure 4B). To confirm that EV trafficking is abrogated in a 0.4-μM Transwell, we placed purified EVs in the top compartment of a 0.4-μM or 3-μM Transwell well with sterile RPMI in the bottom chamber. After 3 hours, the RPMI in the bottom chamber was collected and subjected to high-sensitivity flow cytometry, demonstrating no EVs in the bottom compartment of the 0.4-μM Transwell, but EV trafficking was seen in the 3-μM Transwell (supplemental Figure 3A).
To assess the effect of AML derived on EVs on the phenotypic characteristics and polarization of the myeloid population, we cultured PBMCs for 3 days with MOLM-14 EVs, and the resulting levels of CD11b+/HLADR−/CD33+ MDSCs and HLADR+/CD11c+ myeloid- derived antigen-presenting cells were quantified by multichannel flow cytometry. PBMCs treated with EVs demonstrated an 8-fold increase in MDSCs (Figure 4C) and a corresponding 90% reduction in cells exhibiting antigen-presenting cell phenotypes, both of which were abrogated in a 0.4-μM Transwell system (Figure 4D), and indicating a skew of the tumor microenvironment away from antigen presentation and toward immature immune-suppressive MDSCs. Importantly, a larger Transwell pore size (3 μM) permitting EV trafficking (supplemental Figure 3A) allowed EV-mediated MDSC expansion (supplemental Figure 3C). Absolute frequencies of cells are shown in supplemental Table 2.
MUC1 signaling is critical for the expansion of MDSCs
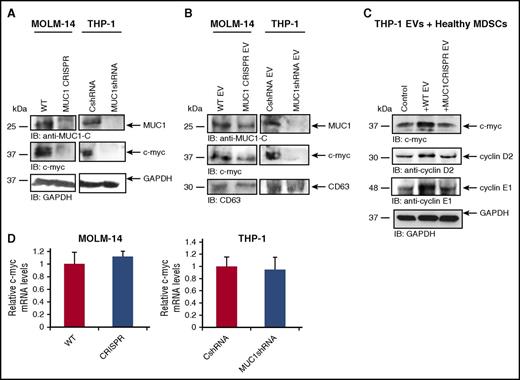
To further elucidate the mechanism by which AML-derived EVs modulate MDSC expansion in the tumor microenvironment, we explored the role of oncogenic drivers in this process. The MUC1 oncogene is aberrantly expressed in solid tumors40 and hematologic malignancies including AML41 and plays a critical role in maintaining the malignant phenotype.42-44 Signaling via the MUC1-C subunit supports tumor cell proliferation and resistance to apoptosis.45 Recent studies have suggested that MUC1 demonstrates immunoregulatory properties.37 As such, we investigated whether MUC1 exerts an immunosuppressive effect on the tumor microenvironment by inducing the expansion of MDSCs. To address this question, we generated human AML cell lines (MOLM-14 and THP-1) in which MUC1 expression was silenced via lentiviral transduction of short hairpin RNA (shRNA) or gene deletion via CRISPR/Cas9 technology (Figure 5A-B). Of note, PBMCs cocultured in direct contact with irradiated MUC1-silenced AML cells resulted in a mean 60% reduction in expansion of MDSCs in comparison with control AML cells in both MOLM-14 and THP-1 cell lines (Figure 5B-C). Absolute frequencies of cells are shown in supplemental Table 2.
MUC1 is critical in the expansion of MDSCs. Stable MOLM-14 and THP-1 AML cell lines silenced for the expression of MUC1 protein were generated by lentiviral transduction of shRNA or control vector. (A) Lysates were prepared and cells analyzed for MUC1 expression using Western blotting. MOLM-14 cells were silenced for MUC1 expression using CRISPR/Cas9 technology to validate the silencing. (B) Lysates were prepared and cells analyzed for MUC1 expression using Western blotting using β-actin as a loading control. Healthy PBMCs and irradiated, fluorescently labeled MUC1-silenced and control AML cells were cocultured for 5 days at a ratio of 100:1 (PBMC:AML). After coculture, cells were analyzed with flow cytometry, and fluorescently labeled blast cells were excluded; CD11b+/HLA-DR−/CD33+ MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells. Summary of three independent experiments for (C) MUC1- silenced MOLM-14 and THP-1 AML cells and (D) CRISPR/Cas9 MUC1-silenced MOLM-14 cells are shown (*P < .05). CRISPR, clustered regularly interspaced short palindromic repeats; IB, immunoblot; WT, wild-type.
MUC1 is critical in the expansion of MDSCs. Stable MOLM-14 and THP-1 AML cell lines silenced for the expression of MUC1 protein were generated by lentiviral transduction of shRNA or control vector. (A) Lysates were prepared and cells analyzed for MUC1 expression using Western blotting. MOLM-14 cells were silenced for MUC1 expression using CRISPR/Cas9 technology to validate the silencing. (B) Lysates were prepared and cells analyzed for MUC1 expression using Western blotting using β-actin as a loading control. Healthy PBMCs and irradiated, fluorescently labeled MUC1-silenced and control AML cells were cocultured for 5 days at a ratio of 100:1 (PBMC:AML). After coculture, cells were analyzed with flow cytometry, and fluorescently labeled blast cells were excluded; CD11b+/HLA-DR−/CD33+ MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells. Summary of three independent experiments for (C) MUC1- silenced MOLM-14 and THP-1 AML cells and (D) CRISPR/Cas9 MUC1-silenced MOLM-14 cells are shown (*P < .05). CRISPR, clustered regularly interspaced short palindromic repeats; IB, immunoblot; WT, wild-type.
Consistent with the requirement for direct cell contact to mediate AML cell expansion of MDSCs, MUC1 does not impact AML secretion of cytokines associated with the recruitment of MDSCs. No significant difference was seen in levels of granulocyte-macrophage colony-stimulating factor, IL-1β, and IL-6 in the cell supernatant following silencing of MUC1 via lentiviral transduction in MOLM-14 or THP-1 cells (supplemental Figure 4). Importantly, MUC1 silencing did not significantly alter the capacity of MOLM-14 or THP-1 cells to secrete EVs, as demonstrated by high-sensitivity flow cytometry on equivalent numbers of MUC1-silenced and control AML cell supernatants (supplemental Figure 3B).
MUC1 promotes c-myc expression in extracellular vesicles, which leads to upregulation of cyclin D2 and E1 in cocultured MDSCs
MUC1 mediates tumor cell proliferation via downstream effectors, including prominently the oncoprotein c-myc that regulates expression of cell cycle proteins.42,43 However, the role of oncoproteins as immunoregulatory agents that mediate MDSC expansion in the tumor microenvironment has not been elucidated. We postulated that MUC1-mediated expression of c-myc in AML cells would potentially impact proliferation of MDSCs in the tumor microenvironment through its transfer via AML-derived EVs. We first demonstrated that silencing of MUC1 in the parent AML cell lines MOLM-14 and THP-1 led to a reduction of MUC1 and c-myc expression (Figure 6A). Moreover, MUC1 silencing in parental AML cells led to a reduction in MUC1 and c-myc expression in EVs derived from those cells, although interestingly, MUC1 abrogation was not as profound in MUC1-silenced EVs from MOLM-14 in comparison with parental MOLM-14 cells (Figure 6B). This difference could hypothetically be due to a preferential loading and export of proteins, such as MUC1, in EVs, as has been suggested.46
MUC1 promotes c-myc expression in EVs, which leads to upregulation of cyclin D2 and E1 in cocultured MDSCs. Stably transduced cell lines silenced for the expression of MUC1 protein were generated by lentiviral transduction of shRNA or CRISPR/Cas9 technology. Control shRNA or wild-type cells were used as controls. Lysates were prepared, and MUC1 and c-myc expression was assessed using Western blot analysis (A) in AMLs cells and (B) in isolated secreted EVs generated from MOLM-14 and THP-1 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and CD63 were used as loading controls. CD11b+/HLA-DR−/CD33+ MDSCs were isolated from healthy donor PBMCs and cultured for 48 hours with EVs isolated from the culture medium of THP-1 cells. (C) PBMCs were lysed and subjected to immunoblot for c-myc and cyclins D2 and E1. GAPDH was used as a loading control. RNA was isolated from MUC1-silenced MOLM-14 and THP-1 cells and subjected to qPCR with primers against c-myc. (D) The c-myc expression in MUC1-silenced MOLM-14 and THP-1 is quantified in relation to control cells; a summary of three experiments is shown.
MUC1 promotes c-myc expression in EVs, which leads to upregulation of cyclin D2 and E1 in cocultured MDSCs. Stably transduced cell lines silenced for the expression of MUC1 protein were generated by lentiviral transduction of shRNA or CRISPR/Cas9 technology. Control shRNA or wild-type cells were used as controls. Lysates were prepared, and MUC1 and c-myc expression was assessed using Western blot analysis (A) in AMLs cells and (B) in isolated secreted EVs generated from MOLM-14 and THP-1 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and CD63 were used as loading controls. CD11b+/HLA-DR−/CD33+ MDSCs were isolated from healthy donor PBMCs and cultured for 48 hours with EVs isolated from the culture medium of THP-1 cells. (C) PBMCs were lysed and subjected to immunoblot for c-myc and cyclins D2 and E1. GAPDH was used as a loading control. RNA was isolated from MUC1-silenced MOLM-14 and THP-1 cells and subjected to qPCR with primers against c-myc. (D) The c-myc expression in MUC1-silenced MOLM-14 and THP-1 is quantified in relation to control cells; a summary of three experiments is shown.
To determine whether the trafficking to myeloid cells of c-myc–containing EVs in the microenvironment resulted in MDSC expansion, we pulsed MDSCs with EVs from wild-type or MUC1-silenced THP-1 cells. Figure 6C shows that control EV-treated MDSCs contained increased levels of c-myc and increased expression of cyclin E1 and cyclin D2, downstream of proproliferative targets of c-myc. MDSCs exposed to MUC1-silenced THP-1 EVs did not demonstrate an increase in c-myc, cyclin E1, or D2 (Figure 6C), suggesting that MUC1 and c-myc are the critical mediators of AML-EV–induced MDSC proliferation.
MUC1 promotes c-myc expression in AML by the suppression of miRNA34a
Having defined MUC1-mediated expansion of MDSCs via increased expression of c-myc and corresponding downstream effectors and their traffic to the surrounding cells via EVs, we subsequently investigated the mechanism by which MUC1 regulates c-myc expression in AML. Of note, silencing of MUC1 in MOLM-14 and THP-1 did not result in decreased levels of c-myc mRNA, suggesting that posttranscriptional regulation was responsible for this effect (Figure 6D).
Noncoding RNAs have been identified as critical mediators of oncogenesis because of their posttranscriptional regulation of protein translation via their binding to target mRNA populations.38 Using TargetScan prediction software, we identified a list of miRNAs with putative binding sites in the c-myc 3′UTR, which included miR34a (data not shown). It has been previously reported that the miR34a family is associated with the modulation of levels of MDSCs in malignancy. As such, we first sought to validate that miR34a regulates c-myc expression in AML. Indeed, overexpression of miR34a using lentiviral transduction of miR34a mimic led to a reduction in c-myc expression in MOLM-14 and THP-1 (Figure 7A). Alternatively, we sought to evaluate whether silencing miR34a could recapitulate the c-myc expression seen in MUC1 wild-type AML cells. MOLM-14 cells were overexpressed for miR34a, in order to obtain sufficient miR34a levels for silencing. Cells were confirmed to be overexpressing miR34a by quantitative polymerase chain reaction (qPCR) for miR34a (supplemental Figure 5). Subsequently, miR34a was silenced in these cells using lentiviral transduction of miR34a-ZIP, demonstrating a dramatic increase in c-myc expression (Figure 7B). Furthermore, in MUC1-silenced THP-1 AML cells, previously shown to have low c-myc expression (Figure 6A), silencing of miR34a demonstrated similar results, with a significant increase in c-myc levels (Figure 7C). Cells were confirmed to be silenced for miR34a by qPCR (supplemental Figure 5).
MUC1 regulates c-myc expression via miR34a in AML cells. MOLM-14 and THP-1 cells were transduced with a miR34a mimic or control, using lentiviral transduction. (A) Lysates were prepared, and c-myc expression was assessed using Western blot analysis. MOLM-14 AML cells overexpressing miR34a were then silenced for miR34a, by lentiviral transduction of miR34a-ZIP or control. (B) Lysates were prepared, and c-myc expression was assessed using Western blot analysis. THP-1 AML cells silenced for MUC1 expression using specific MUC1 shRNA were silenced for miR34a, using lentiviral transduction of miR34a-ZIP or control. (C) Lysates were prepared and c-myc expression was assessed using Western blot analysis. Healthy PBMCs were cocultured with irradiated, fluorescently labeled AML cells with overexpressed miR34a levels, for 5 days at a ratio of 100:1 (PBMC:AML). After coculture, cells were analyzed with flow cytometry, and fluorescently labeled blast cells were excluded; CD11b+/HLA-DR−/CD33+ MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells. (D) A summary of three independent experiments is shown for MOLM-14 and THP-1. Similarly, MDSCs were detected in coculture of PBMCs with miR34a-silenced AML cells MOLM-14 (n = 3) (E) and THP-1 AML cells (n = 3) (F). RNA was isolated from miR34a-overexpressing or miR34a-silenced AML cells and subjected to qPCR with primers against miR34a. (G) The miR34a expression in MUC1-silenced MOLM-14 and THP-1 is quantified in relation to control cells; a summary of three experiments is shown. *P < .05.
MUC1 regulates c-myc expression via miR34a in AML cells. MOLM-14 and THP-1 cells were transduced with a miR34a mimic or control, using lentiviral transduction. (A) Lysates were prepared, and c-myc expression was assessed using Western blot analysis. MOLM-14 AML cells overexpressing miR34a were then silenced for miR34a, by lentiviral transduction of miR34a-ZIP or control. (B) Lysates were prepared, and c-myc expression was assessed using Western blot analysis. THP-1 AML cells silenced for MUC1 expression using specific MUC1 shRNA were silenced for miR34a, using lentiviral transduction of miR34a-ZIP or control. (C) Lysates were prepared and c-myc expression was assessed using Western blot analysis. Healthy PBMCs were cocultured with irradiated, fluorescently labeled AML cells with overexpressed miR34a levels, for 5 days at a ratio of 100:1 (PBMC:AML). After coculture, cells were analyzed with flow cytometry, and fluorescently labeled blast cells were excluded; CD11b+/HLA-DR−/CD33+ MDSCs were quantified as a percentage of immature CD11b+/HLA-DR− myeloid cells. (D) A summary of three independent experiments is shown for MOLM-14 and THP-1. Similarly, MDSCs were detected in coculture of PBMCs with miR34a-silenced AML cells MOLM-14 (n = 3) (E) and THP-1 AML cells (n = 3) (F). RNA was isolated from miR34a-overexpressing or miR34a-silenced AML cells and subjected to qPCR with primers against miR34a. (G) The miR34a expression in MUC1-silenced MOLM-14 and THP-1 is quantified in relation to control cells; a summary of three experiments is shown. *P < .05.
We next confirmed the critical role of miR34a in regulating MDSC expansion in AML. Overexpression of miR34a in wild-type MOLM-14 and THP-1 cells by lentiviral transduction resulted in decreased capacity of the AML cells to induce expansion of MDSCs when cocultured with normal PBMCs (Figure 7D). Furthermore, a coculture of healthy PBMCs with MOLM-14 (Figure 7E) and THP-1 cells (Figure 7F) silenced for miR34a expression resulted in a corresponding increase in MDSCs. Importantly, altering miR34a levels in MOLM-14 or THP-1 was not associated with changes in the rate of apoptosis, which might have otherwise accounted for this change in MDSC expansion (supplemental Figure 6).
Finally, we demonstrated that MUC1 negatively regulates levels of miR34a in AML cells. MUC1-silenced MOLM-14 cells and THP-1 AML cells demonstrated a 13-fold increase in miR34a expression in comparison with the wild-type cell lines as quantified by qPCR analysis (Figure 7G). These findings collectively demonstrate that MUC1 negatively regulates miR34a levels, thereby modulating c-myc expression in AML cells and their resulting EVs, which are shed into the tumor microenvironment.
The clonal relationship between AML and the associated MDSC population
Given the observed increase in MDSCs in the peripheral blood and bone marrow of AML patients, we sought to examine the clonal relationship of the MDSC population with the AML cells as determined by cytogenetic and molecular abnormalities expressed by the dominant malignant clonal population. MDSCs isolated by flow cytometric sorting were interrogated for the presence of cytogenetic or molecular abnormalities that had previously been identified in the leukemic clone. Three patients were examined, all of whom demonstrated that the MDSC phenotype was not uniformly identical with the dominant AML clone. For example, in a patient with AML cells exhibiting three cytogenetic abnormalities, (del7, del20, and trisomy 8) (patient 6, supplemental Table 1), MDSCs expressed only del7 and del20, suggesting a common clonal origin with leukemic precursor prior to the attainment of the trisomy 8 mutation (supplemental Figure 7A). In a second patient with del7 cytogenetic abnormality, only 50% of the MDSCs exhibited the cytogenetic abnormality, consistent with only partial derivation from the dominant AML clone (patient 7, supplemental Table 1; supplemental Figure 7B). In a third patient with AML cells characterized by a NPM1 mutation (patient 8, supplemental Table 1), the MDSC population was found to have the wild-type form of NPM1 (supplemental Figure 7C). These studies suggest that in some cases, the identity of the MDSC could be distinct from the dominant AML clone and is consistent with the observation that AML cells may promote MDSC expansion irrespective of their derivation.
Discussion
MDSCs are a critical component of the tumor microenvironment that modulate interactions between immune effector cells and malignant cells.47 In the present study, we demonstrated increased levels of MDSCs in the peripheral blood of patients with active AML. Coculture of AML cells with healthy PBMCs resulted in the expansion of cells with a characteristic immunosuppressive MDSC phenotype. Consistent with these findings, engraftment of AML in an immune-competent murine model was associated with the significant expansion of MDSCs in the bone marrow and spleen.
AML-mediated expansion of MDSCs is dependent on cell contact and was not observed in a 0.4-μM Transwell model. We hypothesized that AML cells alter the immunologic milieu of the tumor microenvironment through direct communication with accessory cell populations. Extracellular vesicles (EVs) are lipid membrane-bound vesicles released by cells and mediating intercellular communication. In the present study, we demonstrate the release of EVs by AML cells into the microenvironment and demonstrate that their passage is abrogated in a 0.4-μM Transwell, but permitted in a larger-pore Transwell, in keeping with previous studies,48,49 and hypothetically due to EVs aggregating into larger particles that are sterically hindered from traversing the smaller Transwell pores. Importantly, pulsing of PBMCs with AML-derived EVs results in the expansion of MDSCs and the relative suppression of functionally potent antigen-presenting cells.
We subsequently examined which oncogenic drivers are responsible for the capacity of AML cells to alter the microenvironment via the export of EVs. We have previously identified MUC1 as a uniquely important oncoprotein in solid tumors and hematologic malignancies that supports critical aspects of the malignant phenotype, including cell proliferation, resistance to apoptosis, and autonomous self-renewal.41 Accordingly, we have demonstrated that MUC1 is selectively expressed on AML stem cells in comparison with normal hematopoietic stem cells.41 The role of MUC1 in mediating the immunosuppressive milieu in the tumor microenvironment has not been well elucidated. In the present study we have shown that MUC1 is critical for AML expansion of MDSCs. We subsequently investigated how MUC1 signaling, necessary for the expansion of MDSCs, might alter AML extracellular vesicle composition. We demonstrated that AML cells secrete EVs containing c-myc in a MUC1-dependent mechanism. Furthermore, EVs containing c-myc led to an upregulation of the c-myc downstream targets cyclin D2 and cyclin E1 in cocultured MDSCs, indicating that c-myc-containing EVs may drive MDSC proliferation. Critically, EVs from MUC1-silenced AML cells failed to elicit this increase in c-myc and cyclin D2 and E1 expression in EV-exposed MDSCs.
We then sought to determine how MUC1 signaling promotes c-myc signaling in AML. MicroRNAs are small nonencoding RNA molecules involved in posttranslational regulation of gene expression.38 MiR34a, a known p53 inhibitor, has been implicated in regulating the expansion of MDSCs,34 and it is known that tumor cells suppress miR34a expression as part of their self-protective armoury.50 MiR34a is a predicted negative regulator of c-myc, due to a complementary sequence for miR34a in the c-myc 3′UTRr region. We have demonstrated that MUC1 silencing results in increased expression of miRNA34a. Furthermore, overexpression of miR34a in AML cells led to a dramatic downregulation of c-myc. Conversely, silencing of miR34a led to a significant upregulation of c-myc expression, confirming that miR34a regulates c-myc expression in AML.
To confirm miR34a as a critical negative regulator of MDSC expansion, we interrogated miR34a altered cells for their ability to elicit an expansion of MDSCs in cocultured PBMCs. Overexpression of miR34a in AML cells partially abrogated their ability to induce MDSCs from cocultured donor PBMCs. In concert, silencing of miR34a in MUC1-silenced AML cells recapitulated their ability to induce MDSCs in this model.
Previous studies have demonstrated that myeloid cells in the bone marrow microenvironment express heterogeneity with respect to their derivation from leukemic and nonmalignant precursor populations.51 To explore the clonal relationship of the expanded MDSC population to AML, we interrogated isolated MDSCs for the presence of cytogenetic or molecular abnormalities that had previously been identified in the leukemic clone. These studies demonstrated diversity with respect to the origin of the MDSC population. Specifically, there was a lack of uniform identity between the MDSC population and the pattern of cytogenetic or molecular abnormalities that defined the dominant AML clone. It is possible that MDSCs may be derived from a distinct AML subclone, as illustrated by the patient in whom MDSCs expressed two of the three defining cytogenetic abnormalities of the AML cells. MDSCs derived from other patients lack cytogenetic or molecular abnormalities found in the dominant AML clone, consistent with their potential derivation from cytogenetically or NPM1 mutation–negative AML subclones or, alternatively, from normal myeloid cells. A definitive effort to define patterns of clonal derivation of MDSCs would require a larger study cohort. Genomic sequencing may better differentiate MDSCs of normal and malignant derivation52 ; however, the present study suggests that AML-mediated MDSC expansion may occur in the bone marrow niche irrespective of their derivation.
In conclusion, we have demonstrated that MDSCs are expanded in patients with AML and contribute to tumor-related immune suppression. We have shown that the MUC1 oncoprotein is a critical mediator of MDSC expansion, via the promotion of c-myc expression in secreted EVs. Targeting MDSCs in AML, using specific MDSC inhibitors or via MUC1 inhibition, could pave the way for improved responses to immune therapies in AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Judy Lieberman for providing the pLL3.7_hsa-miR-34a plasmid (Addgene plasmid 25791). The authors thank Vasilis Toxavidis and John Tigges for their kind help in providing flow cytometry support.
Authorship
Contribution: A.R.P. designed and performed the research, analyzed data, and wrote the paper; D.S. analyzed data and wrote the paper; H.R., A.W., A.T., M.C., and J.F. performed the research; M.P.B. analyzed data; L.C. and K.P. performed the research; P.S., R.K.L., M.N., and A.A. analyzed data; S.J., M.M., L.M., J.L., R.J., and J.A. provided patient samples; P.P.P. and D.K. designed the research; J.R. and D.A. designed the research and wrote the paper.
Conflict-of-interest disclosure: D.K. holds equity in Genus Oncology and is a consultant to the company. The remaining authors declare no competing financial interests.
Correspondence: Athalia Rachel Pyzer, Department of Bone Marrow Transplantation, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215; e-mail: apyzer@bidmc.harvard.edu.