Key Points
Capture sequencing reveals that PDL SRs cluster into 2 discrete breakpoint regions.
PDL SRs are significantly associated with increased protein expression and limit T-cell activation.
Abstract
Programmed death ligands (PDLs) are immune-regulatory molecules that are frequently affected by chromosomal alterations in B-cell lymphomas. Although PDL copy-number variations are well characterized, a detailed and comprehensive analysis of structural rearrangements (SRs) and associated phenotypic consequences is largely lacking. Here, we used oligonucleotide capture sequencing of 67 formalin-fixed paraffin-embedded tissues derived from primary B-cell lymphomas and 1 cell line to detect and characterize, at base-pair resolution, SRs of the PDL locus (9p24.1; harboring PDL1/CD274 and PDL2/PDCD1LG2). We describe 36 novel PDL SRs, including 17 intrachromosomal events (inversions, duplications, deletions) and 19 translocations involving BZRAP-AS1, CD44, GET4, IL4R, KIAA0226L, MID1, RCC1, PTPN1 and segments of the immunoglobulin loci. Moreover, analysis of the precise chromosomal breakpoints reveals 2 distinct cluster breakpoint regions (CBRs) within either CD274 (CBR1) or PDCD1LG2 (CBR2). To determine the phenotypic consequences of these SRs, we performed immunohistochemistry for CD274 and PDCD1LG2 on primary pretreatment biopsies and found that PDL SRs are significantly associated with PDL protein expression. Finally, stable ectopic expression of wild-type PDCD1LG2 and the PDCD1LG2-IGHV7-81 fusion showed, in coculture, significantly reduced T-cell activation. Taken together, our data demonstrate the complementary utility of fluorescence in situ hybridization and capture sequencing approaches and provide a classification scheme for PDL SRs with implications for future studies using PDL immune-checkpoint inhibitors in B-cell lymphomas.
Introduction
As with other B7 family members, programmed death ligands (PDL) 1 (CD274) and 2 (PDCD1LG2) maintain peripheral tolerance by engaging their cognate receptor PD1 (PDCD1) on target cells.1-3 Several lymphomas, including diffuse large B-cell lymphoma (DLBCL), primary mediastinal large B-cell lymphoma (PMBCL), and classical Hodgkin lymphoma (cHL), exploit the PDL axis to suppress the antitumor response.4-7 PDL expression is aberrated in malignant B cells through a combination of altered gene and transcriptional regulation, in addition to somatically acquired copy-number variations (CNV) and chromosomal structural rearrangements (SRs).4,6-8 As assessed by fluorescence in situ hybridization (FISH), CNVs appear most frequently in cHL (50% to 70%), whereas SRs are common to PMBCL (20%) and primary testicular lymphoma (7%).6,9 Previously reported recurrent PDL SR partners include CIITA and immunoglobulin heavy chain segments, which drive PDL expression through juxtaposition of a highly active promoter and enhancing elements, respectively.5,6
Although studies have established PDL SRs are frequent, few primary cases have been characterized at base-pair resolution, and therefore, the partner genes and patterns of rearrangement have not thus far been appreciated.7 Furthermore, no studies have empirically evaluated downstream associations of the various types of PDL SRs with their phenotypic or functional consequences.
To inform more comprehensively on the landscape and functionality of PDL SRs, we performed custom oligonucleotide capture sequencing of the PDL locus on 67 primary B-cell lymphoma specimens and 1 cHL-derived cell line (L-1236). Capture sequencing using frozen or formalin-fixed paraffin-embedded tissue (FFPET) has proven sensitive for resolving single base-pair somatic mutations and SRs in multiple tumor types.10-14 However, this approach remains unexplored for large-scale SR detection in lymphoma FFPET. Here, we report specific breakpoint anatomy for 36 novel PDL SRs, including 19 previously undescribed translocations and 17 intrachromosomal SRs. We classify PDL SRs into distinct subgroups according to cluster breakpoint regions (CBRs), CBR1 in CD274 and CBR2 in PDCD1LG2, and firmly establish that, together, FISH and capture sequencing identify a previously underestimated SR frequency of PDLs. Subsequently, we show that PDL SRs, as with CNVs, are significantly associated with protein expression (P < .05) in primary lymphoma specimens. Finally, using stably transduced DOHH-2 cells with constructs representative of PDL CNVs and 3′ SRs, we demonstrate that wild-type and chimeric PDCD1LG2 protein localizes to the cell surface and limits activation of T cells in vitro (P < .01). As an increasing number of immune-checkpoint inhibitors targeting the PDL axis become available, knowledge of underlying genomic events will contribute to informed selection of immunotherapeutic agents and might provide a framework for studying resistance to these therapies.
Materials and methods
Cohort selection and DNA extraction
Cases were selected based on prior knowledge of PDL rearrangement status as assessed by FISH.6 To expand on our previously published lymphoma case cohorts, we performed FISH on an additional 217 DLBCL specimens, which yielded 5 break-apart cases with material available for capture sequencing (see supplemental Results, available on the Blood Web site). In total, we selected 68 cases for investigation of which 41% (28 of 68) contained a PDL rearrangement. As capture sequencing of FFPET has not previously been used to identify SRs in lymphomas, nonstructurally rearranged cases, for example, cases with normal signal patterns or CNVs, were included in the sequencing cohort to assess capture sequencing sensitivity and specificity. These cases were also used to determine the proportion of cases for which capture sequencing could identify SRs not detectable by FISH, so-called FISH-silent SRs.
Our cohort of 68 cases consisted of the cHL-derived cell line L-1236, 19 DLBCLs, 4 primary central nervous system lymphomas (PCNSLs), 8 primary testicular lymphomas (PTLs), 1 follicular lymphoma, and 35 PMBCLs (supplemental Table 1). Genomic material was extracted using a Qiagen AllPrep DNA/RNA FFPE Kit, and DNA was quantified using an Invitrogen Qubit system. This study abided by the principles set forth by the Declaration of Helsinki and was approved by the BC Cancer Agency (REB: H11-00684).
Capture sequencing and analysis
Prior to sequencing, genomic libraries were enriched for the region harboring CD274 and PDCD1LG2 using an Agilent SureSelect custom oligonucleotide design targeting chr9:5 449 434-5 573 579 with 5× probe tiling, standard boosting, and no masking. The genomic coordinates were selected based on the location of previously described rearrangements and included all previously reported translocation breakpoints.5-7 The 16 cases with the highest library yield were first used as a pilot and were pooled and sequenced on an Illumina HiSeq 2500 (v3 chemistry, 100 bp paired-end reads). Subsequently, the remaining 52 cases were divided into 2 pools of 17 and 1 pool of 18 cases and sequenced on an Illumina HiSeq 2500 (v4 chemistry, 125 bp paired-end reads). Each pool of libraries was sequenced in 1 lane of a flow cell. Reads were aligned to GRCh37, and predictions were generated using the deStruct (v0.1.3; https://bitbucket.org/dranew/destruct) and DELLY (v0.5.5) tools.7 Select predictions were subsequently Sanger validated (see supplemental Methods for details).
Immunohistochemistry
Sections of 2 tissue microarrays encompassing previously published PMBCL cases were cut (4 μm) and deparaffinized followed by antigen retrieval in a decloaking chamber with Diva Decloaker solution.5,15 Automated immunohistochemical staining was performed on the Biocare Intellipath FLX autostainer; all reagents (with the exception of the antibodies) were purchased from Biocare. Sections were blocked with peroxidase-1 and Background Sniper. CD274 (PDL1) rabbit monoclonal antibody (clone SP142; Spring Bioscience) was applied at a concentration of 1:100 in Da Vinci Green Diluent for 30 minutes at room temperature. PDCD1LG2 (PDL2) mouse monoclonal antibody (clone 366C.9E5; Gordon J. Freeman, Dana-Farber Cancer Institute) was used at a concentration of 0.07 μg/mL in Renaissance background reducing diluent for 30 minutes at room temperature. MACH 2 rabbit-horseradish peroxidase and mouse-horseradish peroxidase polymers were applied for 30 minutes and visualized using intelliPATH FLX diaminobenzidine chromogen. Staining was reported as a histoscore, the product of the percentage of positive tumor cells (scale 0-100, increments of 10) and staining intensity (scale 0-3, increments of 1). A Nikon Eclipse 80i microscope equipped with a Nikon DS-Ri1 camera was used for taking representative pictures. P values were calculated using an exact Wilcoxon-Mann-Whitney test.
Retroviral transduction
Wild-type PDCD1LG2 and PDCD1LG2-IGHV7-81 alleles were amplified using the Phusion DNA polymerase (New England Biolabs) from the PMBCL cell line U2940 and the cHL cell line L-1236, respectively. cDNAs were ligated into the tetracycline-inducible pRetroCMV/TO/GFP vector (kindly provided by Louis M. Staudt, National Cancer Institute), as previously described.15 HEK cells were transfected (lipofectamine 2000; ThermoFisher), and 48 hours later, supernatant containing viral particles was harvested and subsequently used for B-cell line transduction, as previously described.15 The DLBCL-derived (germinal center B-cell type) cell line DOHH-2 (obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ]) was selected for manipulation, as it expresses neither CD274 nor PDCD1LG2.6 Transduced DOHH-2 cells underwent puromycin selection for 4 days prior to validating ectopic expression of PDCD1LG2 (clone HIH18; BD Biosciences) and GFP via flow cytometry.
Coculturing and flow cytometry
Jurkat T cells (kindly provided by Andrew P. Weng, BC Cancer Agency) were stimulated concurrently with 1× phorbol myristate acetate/Ionomycin (eBioscience) with the addition of either empty vector, PDCD1LG2 wild-type, or PDCD1LG2-IGHV7-81-engineered DOHH-2 cells at T:B ratios of 1:1 or 1:5. Cells were cocultured for 28 hours in accordance with the DSMZ repository directions (https://www.dsmz.de), subsequently blocked with 1× phosphate-buffered saline/1% heat-inactivated fetal bovine serum (ThermoFisher), and stained with anti-CD5 (clone L17F12; BD Biosciences) and anti-CD69 (clone FN50; BioLegend) for 30 minutes. CD69 was evaluated within the CD5+/GFP-negative cell population, for which at least 10 000 events were acquired, on an LSRFortessa cytometer (BD Biosciences). Replicate experiments were assessed for significance using a paired, 2-tailed Student t test.
Results
Capture sequencing characterizes 36 novel PDL SRs at base-pair resolution
To identify PDL rearrangement partners, we performed capture sequencing of the PDL locus for 67 clinical specimens and 1 cHL-derived cell line (L-1236) for which FISH break-apart status was known (28/68 positive). After filtering and merging the predicted SRs (see supplemental Methods), we retained 86 SRs in 30 cases (supplemental Tables 2 and 3). Of these, we selected 36 events observed in 25 cases (Table 1, supplemental Table 4) for validation and further analysis. Fourteen events involved CD274; 19 involved PDCD1LG2, and 3 involved both PDLs concurrently. Twenty-seven of 31 (87%) predictions were validated using Sanger sequencing (supplemental Table 5); 4 predictions could only be validated via polymerase chain reaction (PCR) due to repetitive base stretches embedded within partner loci. The 5 remaining events could not be validated due to tissue exhaustion. However, based on the breakpoint anatomy and number of reads supporting the variant, we consider these events to be high-confidence predictions (supplemental Figure 1). The congruency of SR findings between primary specimens, as evaluated by FISH and capture sequencing, revealed an overlap of 81%. However, 13 of 36 (36%) reported PDL SRs were observed exclusively with capture sequencing, so-called FISH-silent SRs. See supplemental Results for details.
SRs identified by oligonucleotide capture sequencing of primary B-cell lymphoma tissue specimens
| Case . | Path . | Event . | Gene 1 . | Gene 2 . | Break 1 . | Break 2 . | FISH . | Sanger . | Subtype . |
|---|---|---|---|---|---|---|---|---|---|
| CS01 | DLBCL | DEL | CD274 | MLANA | Chr9:5467908 | Chr9:5889215 | CNV | Y | Intrachromosomal |
| CS02 | PMBCL | TRA | CTSL1P2 | PDCD1LG2 | Chr10:48174662 | Chr9:5526439 | BA | Y | Other TRA (5′) |
| CS08 | PMBCL | DUP | CD274 | PDCD1LG2 | Chr9:5447066 | Chr9:5570695 | Normal | Y | Intrachromosomal |
| CS09 | PTL | INV | CD274 | GRHPR | Chr9:5477220 | Chr9:37409812 | BA | Y | Intrachromosomal |
| CS10 | PTL | TRA | PTPN1 | CD274 | Chr20:49127848 | Chr9:5450396 | BA | Y | CBR1 |
| DUP | CD274 | CD274 | Chr9:5451269 | Chr9:5467963 | Y | Intrachromosomal | |||
| INV | CD274 | RIC1 | Chr9:5467981 | Chr9:5627175 | Y | Intrachromosomal | |||
| DEL | CD274 | RIC1 | Chr9:5466595 | Chr9:5775524 | Y | Intrachromosomal | |||
| DEL | CD274 | CD274 | Chr9:5451258 | Chr9:5451585 | Y | Intrachromosomal | |||
| CS11 | PCNSL | TRA | CD44 | CD274 | Chr11:35161226 | Chr9:5452813 | BA | N* | CBR1 |
| DEL | PDCD1LG2 | IL33 | Chr9:5491418 | Chr9:6135848 | N* | Intrachromosomal | |||
| CS12 | PMBCL | TRA | BZRAP1-AS1 | PDCD1LG2 | Chr17:56409009 | Chr9:5518645 | BA | Y | CBR2 |
| TRA | AK309109 | PDCD1LG2 | Chr10:47272696 | Chr9:5518489 | Y | CBR2 | |||
| CS21 | PMBCL | DEL | CD274 | PDCD1LG2 | Chr9:5467992 | Chr9:5508323 | Normal | N# | Intrachromosomal |
| CS25 | PMBCL | TRA | RCC1 | PDCD1LG2 | Chr1:28833535 | Chr9:5518615 | BA | Y | CBR2 |
| CS29 | PMBCL | DUP | PDCD1LG2 | PDCD1LG2 | Chr9:5500059 | Chr9:5570090 | Normal | Y | Intrachromosomal |
| CS33 | PMBCL | TRA | BZRAP1-AS1 | PDCD1LG2 | Chr17:56409576 | Chr9:5517129 | BA | Y | CBR2 |
| CS36 | PMBCL | TRA | IGK | PDCD1LG2 | Chr2:89159416 | Chr9:5510982 | BA | Y | CBR2 |
| CS38 | PMBCL | DEL | CD274 | CD274 | Chr9:5467983 | Chr9:5470588 | BA | Y | Intrachromosomal |
| CS39 | DLBCL | TRA | KIAA0226L | CD274 | Chr13:46946572 | Chr9:5451322 | BA | Y | CBR1 |
| CS42 | DLBCL | TRA | CD274 | MCM3 | Chr9:5466034 | Chr6:52178267 | BA | Y | Other TRA (3′) |
| CS43 | PTL | INV | CD274 | GRHPR | Chr9:5480091 | Chr9:37381934 | BA | Y | Intrachromosomal |
| CS44 | PMBCL | DEL | PDCD1LG2 | KIAA2026 | Chr9:5563053 | Chr9:5984410 | BA | N# | Intrachromosomal |
| CS47 | PMBCL | TRA | IL4R | PDCD1LG2 | Chr16:27326593 | Chr9:5511678 | BA | Y | CBR2 |
| TRA | GET4 | PDCD1LG2 | Chr7:932289 | Chr9:5513369 | Y | CBR2 | |||
| TRA | PDCD1LG2 | MID1 | Chr9:5565388 | ChrX:10651175 | N# | Other TRA (3′) | |||
| CS48 | PMBCL | TRA | OCLN | PDCD1LG2 | Chr5:68784424 | Chr9:5491406 | BA | N# | Other TRA (intergenic) |
| CS49 | PMBCL | TRA | IGLL5 | PDCD1LG2 | Chr22:23231719 | Chr9:5510728 | BA | Y | CBR2 |
| CS51 | PMBCL | TRA | IL4R | PDCD1LG2 | Chr16:27326880 | Chr9:5518313 | BA | Y | CBR2 |
| CS52 | DLBCL | INV | PDCD1LG2 | POLR1E | Chr9:5527098 | Chr9:37498687 | BA | Y | Intrachromosomal |
| CS54 | PMBCL | INV | CD274 | PDCD1LG2 | Chr9:5469428 | Chr9:5561319 | Normal | Y | Intrachromosomal |
| CS63 | DLBCL | TRA | IGK | CD274 | Chr2:89157495 | Chr9:5450316 | BA | N* | CBR1 |
| TRA | RPIA | PDCD1LG2 | Chr2:89051353 | Chr9:5502180 | N* | Other TRA (intergenic) | |||
| DEL | CD274 | UHRF2 | Chr9:5470453 | Chr9:6440816 | N* | Intrachromosomal | |||
| CS65 | PMBCL | DEL | PDCD1LG2 | PDCD1LG2 | Chr9:5518701 | Chr9:5520276 | BA | Y | Intrachromosomal |
| TRA | IGLL5 | PDCD1LG2 | Chr22:23235076 | Chr9:5511361 | Y | CBR2 |
| Case . | Path . | Event . | Gene 1 . | Gene 2 . | Break 1 . | Break 2 . | FISH . | Sanger . | Subtype . |
|---|---|---|---|---|---|---|---|---|---|
| CS01 | DLBCL | DEL | CD274 | MLANA | Chr9:5467908 | Chr9:5889215 | CNV | Y | Intrachromosomal |
| CS02 | PMBCL | TRA | CTSL1P2 | PDCD1LG2 | Chr10:48174662 | Chr9:5526439 | BA | Y | Other TRA (5′) |
| CS08 | PMBCL | DUP | CD274 | PDCD1LG2 | Chr9:5447066 | Chr9:5570695 | Normal | Y | Intrachromosomal |
| CS09 | PTL | INV | CD274 | GRHPR | Chr9:5477220 | Chr9:37409812 | BA | Y | Intrachromosomal |
| CS10 | PTL | TRA | PTPN1 | CD274 | Chr20:49127848 | Chr9:5450396 | BA | Y | CBR1 |
| DUP | CD274 | CD274 | Chr9:5451269 | Chr9:5467963 | Y | Intrachromosomal | |||
| INV | CD274 | RIC1 | Chr9:5467981 | Chr9:5627175 | Y | Intrachromosomal | |||
| DEL | CD274 | RIC1 | Chr9:5466595 | Chr9:5775524 | Y | Intrachromosomal | |||
| DEL | CD274 | CD274 | Chr9:5451258 | Chr9:5451585 | Y | Intrachromosomal | |||
| CS11 | PCNSL | TRA | CD44 | CD274 | Chr11:35161226 | Chr9:5452813 | BA | N* | CBR1 |
| DEL | PDCD1LG2 | IL33 | Chr9:5491418 | Chr9:6135848 | N* | Intrachromosomal | |||
| CS12 | PMBCL | TRA | BZRAP1-AS1 | PDCD1LG2 | Chr17:56409009 | Chr9:5518645 | BA | Y | CBR2 |
| TRA | AK309109 | PDCD1LG2 | Chr10:47272696 | Chr9:5518489 | Y | CBR2 | |||
| CS21 | PMBCL | DEL | CD274 | PDCD1LG2 | Chr9:5467992 | Chr9:5508323 | Normal | N# | Intrachromosomal |
| CS25 | PMBCL | TRA | RCC1 | PDCD1LG2 | Chr1:28833535 | Chr9:5518615 | BA | Y | CBR2 |
| CS29 | PMBCL | DUP | PDCD1LG2 | PDCD1LG2 | Chr9:5500059 | Chr9:5570090 | Normal | Y | Intrachromosomal |
| CS33 | PMBCL | TRA | BZRAP1-AS1 | PDCD1LG2 | Chr17:56409576 | Chr9:5517129 | BA | Y | CBR2 |
| CS36 | PMBCL | TRA | IGK | PDCD1LG2 | Chr2:89159416 | Chr9:5510982 | BA | Y | CBR2 |
| CS38 | PMBCL | DEL | CD274 | CD274 | Chr9:5467983 | Chr9:5470588 | BA | Y | Intrachromosomal |
| CS39 | DLBCL | TRA | KIAA0226L | CD274 | Chr13:46946572 | Chr9:5451322 | BA | Y | CBR1 |
| CS42 | DLBCL | TRA | CD274 | MCM3 | Chr9:5466034 | Chr6:52178267 | BA | Y | Other TRA (3′) |
| CS43 | PTL | INV | CD274 | GRHPR | Chr9:5480091 | Chr9:37381934 | BA | Y | Intrachromosomal |
| CS44 | PMBCL | DEL | PDCD1LG2 | KIAA2026 | Chr9:5563053 | Chr9:5984410 | BA | N# | Intrachromosomal |
| CS47 | PMBCL | TRA | IL4R | PDCD1LG2 | Chr16:27326593 | Chr9:5511678 | BA | Y | CBR2 |
| TRA | GET4 | PDCD1LG2 | Chr7:932289 | Chr9:5513369 | Y | CBR2 | |||
| TRA | PDCD1LG2 | MID1 | Chr9:5565388 | ChrX:10651175 | N# | Other TRA (3′) | |||
| CS48 | PMBCL | TRA | OCLN | PDCD1LG2 | Chr5:68784424 | Chr9:5491406 | BA | N# | Other TRA (intergenic) |
| CS49 | PMBCL | TRA | IGLL5 | PDCD1LG2 | Chr22:23231719 | Chr9:5510728 | BA | Y | CBR2 |
| CS51 | PMBCL | TRA | IL4R | PDCD1LG2 | Chr16:27326880 | Chr9:5518313 | BA | Y | CBR2 |
| CS52 | DLBCL | INV | PDCD1LG2 | POLR1E | Chr9:5527098 | Chr9:37498687 | BA | Y | Intrachromosomal |
| CS54 | PMBCL | INV | CD274 | PDCD1LG2 | Chr9:5469428 | Chr9:5561319 | Normal | Y | Intrachromosomal |
| CS63 | DLBCL | TRA | IGK | CD274 | Chr2:89157495 | Chr9:5450316 | BA | N* | CBR1 |
| TRA | RPIA | PDCD1LG2 | Chr2:89051353 | Chr9:5502180 | N* | Other TRA (intergenic) | |||
| DEL | CD274 | UHRF2 | Chr9:5470453 | Chr9:6440816 | N* | Intrachromosomal | |||
| CS65 | PMBCL | DEL | PDCD1LG2 | PDCD1LG2 | Chr9:5518701 | Chr9:5520276 | BA | Y | Intrachromosomal |
| TRA | IGLL5 | PDCD1LG2 | Chr22:23235076 | Chr9:5511361 | Y | CBR2 |
Rearrangement partner genes listed are those that are either the closest to the breakpoint or most biologically plausible based on chromosomal strand orientation. Coordinates are based on the GRCh37 genome build.
BA, break-apart positive; DEL, deletion; DUP, duplication/eversion; INV, inversion; N*, not validated due to tissue unavailability; N#, PCR but not Sanger sequencing validated; Path, pathology; TRA, translocation; Y, yes.
PDL translocations occur in well-defined cluster regions CBR1 and CBR2
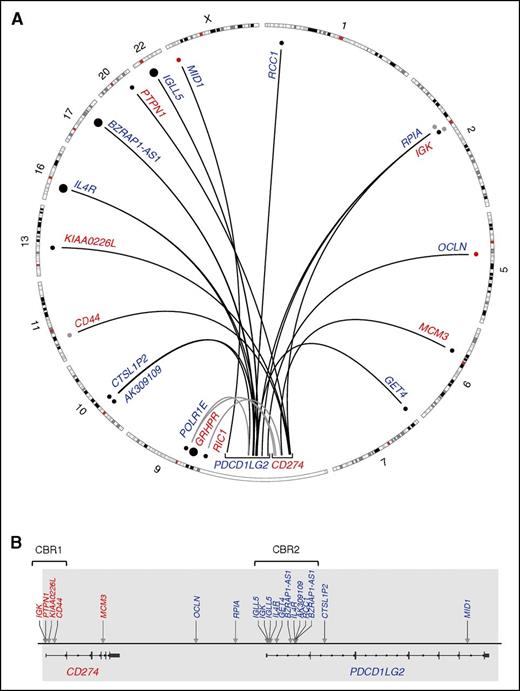
In total, 19 novel translocation partners were identified: 13 in PMBCLs, 4 in DLBCLs, 1 in PTL, and 1 in PCNSL (Table 1). Fourteen translocations involved PDCD1LG2 and 5 translocations involved CD274. No exact translocation breakpoints were conserved, but rearrangements involving the partners BZRAP-AS1, IGK, IGLL5, and IL4R were recurrent, with IGK involving both CD274 and PDCD1LG2.
Translocation-specific CBRs were observed within the first introns of both CD274 and PDCD1LG2. Here, we define these regions as CBR1 and CBR2, respectively. Four CD274 translocations were observed in the region spanning 300 bp upstream of exon 1 to the first 2.5 kb of intron 1, and 10 translocations were found within the first 8 kb of intron 1 of PDCD1LG2 (Figure 1). Reading frame was assessed for the CBR rearrangements and the CD44-CD274 translocation was the only SR to encode a putative chimeric protein. In this case (CS11), a protein is predicted to exist consisting of the first 21 amino acids of CD44 encoded by exon 1, followed by 5 amino acids of the untranslated region (UTR) of CD274 exon 2, and subsequently, the remainder of the normal CD274 amino acid sequence. As genetic material for this case was exhausted, we were unable to validate this hypothesis.
The structural rearrangement landscape of the PDL locus (9p24.1) as determined by capture sequencing. (A) A circos plot representing translocation (black arcs) and large inversion (gray arcs) partners of CD274 (red) and PDCD1LG2 (blue). Smaller dots denote single reported partners, whereas large dots denote partners that were observed twice within our cohort. Black dots were Sanger validated; red dots were PCR validated only, and gray dots were not validated due to exhaustion of clinical material. (B) A linear plot denoting the location of the translocation breakpoints in the PDL region (arrows) with partners labeled above. Newly annotated CBRs, CBR1 and CBR2, are indicated. Gray shading represents the captured region.
The structural rearrangement landscape of the PDL locus (9p24.1) as determined by capture sequencing. (A) A circos plot representing translocation (black arcs) and large inversion (gray arcs) partners of CD274 (red) and PDCD1LG2 (blue). Smaller dots denote single reported partners, whereas large dots denote partners that were observed twice within our cohort. Black dots were Sanger validated; red dots were PCR validated only, and gray dots were not validated due to exhaustion of clinical material. (B) A linear plot denoting the location of the translocation breakpoints in the PDL region (arrows) with partners labeled above. Newly annotated CBRs, CBR1 and CBR2, are indicated. Gray shading represents the captured region.
In addition to translocations occurring in CBR1 and CBR2, we also define a third subcategory of SRs consisting of rearrangements outside the CBRs, so-called nonclustered translocations. This category includes 2 rearrangements that were observed near the 3′ end of each gene: those that involved MCM3 and MID1 (Figure 1). It also encompasses the 2 intergenic breakpoints upstream of PDCD1LG2 and the PDCD1LG2 intron 2 translocation.
Intrachromosomal PDL SRs show recurrent patterns and define a fourth subgroup of rearrangements
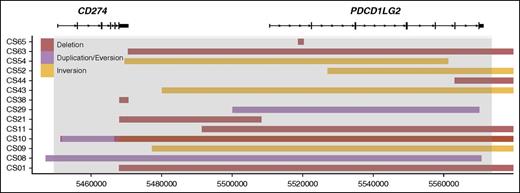
Intrachromosomal rearrangement predictions included 3 duplications, 9 deletions, and 5 inversions (Figure 2). Duplications involved the PDL gene regions, with 1 of the duplications involving both ligands and the other 2 duplicating CD274 and PDCD1LG2 individually. Deletions commonly resulted in the loss of the CD274 3′ UTR (5 SRs) or the loss of the entire PDCD1LG2 coding region (4 SRs), and these losses were seen concurrently in 3 cases. Deletion of the PDCD1LG2 3′ UTR was also observed in 1 case. Inversions could be subcategorized into 2 entities: microinversions confined to the region of the CD274 and PDCD1LG2 coding space and pericentric macroinversions that spanned >100 kb. Three cases had single macroinversions and no other SR identified. In all 3 cases, the partner region was the area surrounding the POLR1E/GRHPR genes (9p13.2). Together, the intrachromosomal events define a fourth subgroup of SRs.
A linear map of intrachromosomal PDL SRs. The deletions, duplications, and inversions identified in the PDL region are shown. The x-axis represents genomic coordinates, and individual cases are shown on the y-axis. Boxes represent the genomic span of the SRs. Gray shading represents the captured region, and RefSeq gene models are shown to scale for reference.
A linear map of intrachromosomal PDL SRs. The deletions, duplications, and inversions identified in the PDL region are shown. The x-axis represents genomic coordinates, and individual cases are shown on the y-axis. Boxes represent the genomic span of the SRs. Gray shading represents the captured region, and RefSeq gene models are shown to scale for reference.
SRs are significantly associated with increased PDL protein expression in primary lymphoma specimens
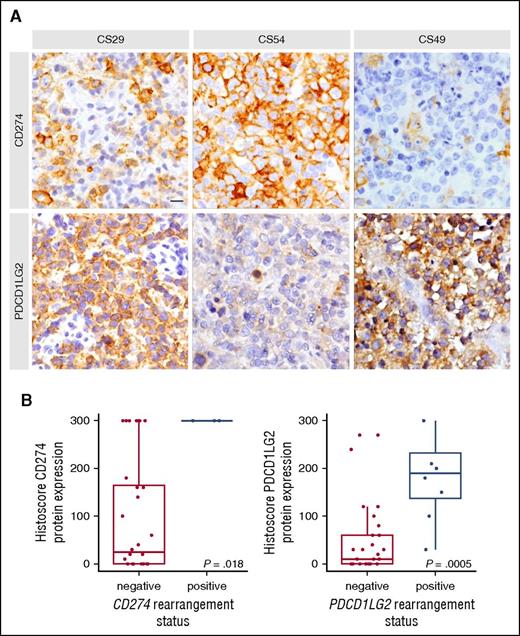
To assess the phenotypic impact of different SR subtypes, immunohistochemistry (IHC) was used to characterize expression of the CD274 and PDCD1LG2 proteins (supplemental Table 6). IHC data were available for 2 of 4 cases harboring a CBR1 rearrangement, and both showed positivity for CD274 in 100% of cells. Of the 8 cases with a CBR2 rearrangement, 6 had IHC data available, and all cases stained positively for PDCD1LG2 (≥20% positive cells). Figure 3 highlights a representative case (CS49) in which a CBR2 rearrangement was associated with specific expression of PDCD1LG2.
Representative IHC staining and protein level associations based on capture sequencing rearrangement status for CD274 (PDL1) and PDCD1LG2 (PDL2). (A) In case CS29, only macrophages stained for CD274, whereas the majority of tumor cells express PDCD1LG2 due to a focal amplification of the PDCD1LG2 gene locus. CS54 harbored an inversion displacing the 3′ end of CD274 and disrupting the coding sequence of PDCD1LG2. Concordantly, the tumor cells display strong positivity for CD274 and remain negative for PDCD1LG2. CS49 harbors a CBR2 translocation and shows a similar staining pattern as described for CS29. Scale bar, 25 μm. (B) Cases were considered rearranged if they had a CBR translocation or a focal amplification of the gene region. The bold line represents the median, and the box denotes the interquartile range. Individual cases are represented by points. P value was calculated using an exact Wilcoxon-Mann-Whitney test.
Representative IHC staining and protein level associations based on capture sequencing rearrangement status for CD274 (PDL1) and PDCD1LG2 (PDL2). (A) In case CS29, only macrophages stained for CD274, whereas the majority of tumor cells express PDCD1LG2 due to a focal amplification of the PDCD1LG2 gene locus. CS54 harbored an inversion displacing the 3′ end of CD274 and disrupting the coding sequence of PDCD1LG2. Concordantly, the tumor cells display strong positivity for CD274 and remain negative for PDCD1LG2. CS49 harbors a CBR2 translocation and shows a similar staining pattern as described for CS29. Scale bar, 25 μm. (B) Cases were considered rearranged if they had a CBR translocation or a focal amplification of the gene region. The bold line represents the median, and the box denotes the interquartile range. Individual cases are represented by points. P value was calculated using an exact Wilcoxon-Mann-Whitney test.
Moreover, cases with focal amplifications showed CD274 and PDCD1LG2 positivity in the case where both loci were duplicated (CS08; supplemental Figure 2), whereas positivity of only 1 duplicated ligand was observed in the other cases (CS10 and CS29; Figure 3). Taken together, cases harboring a CBR translocation or a focal gene amplification were significantly associated with increased PDL protein expression (Wilcoxon-Mann-Whitney P = .018 for CD274, P < .001 for PDCD1LG2; Figure 3). In addition, 3 cases with deletions of the 3′ UTR in either CD274 or PDCD1LG2 corresponded to increased protein levels (CS10, CS38, CS44), and a microinversion displacing the CD274 3′ UTR and disrupting the PDCD1LG2 coding sequence was IHC positive for CD274 and negative for PDCD1LG2 (CS54; Figure 3).
PDL translocations resulting in increased surface protein expression significantly inhibit T-cell activation in vitro
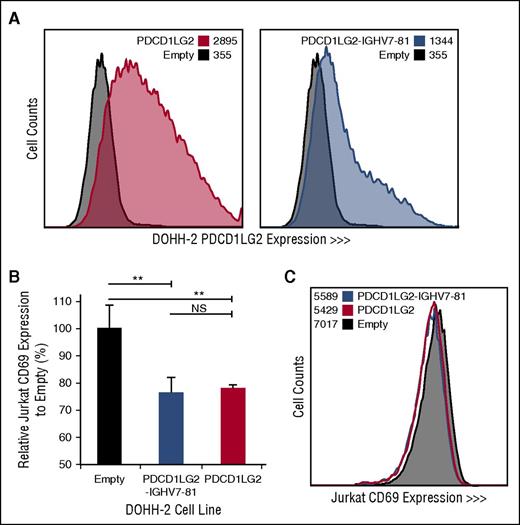
To determine the cellular localization and functional effect of specific PDL SRs, we retrovirally transduced DOHH-2, a non-PDL expressing DLBCL-derived cell line with wild-type PDCD1LG2 and the PDCD1LG2-IGHV7-81 3′ fusion, originally observed in the cHL-derived cell line L-1236. PDCD1LG2 surface expression demonstrates that the chimeric PDCD1LG2 protein, although lacking exons 6 and 7, is expressed on the cell membrane of the engineered cells (Figure 4). Coculturing phorbol myristate acetate/ionomycin-activated Jurkat T cells with engineered DOHH-2 cells expressing either wild-type PDCD1LG2 or PDCD1LG2-IGHV7-81 revealed a significant decrease (Student t test, P < .01) in the lymphocyte early activation marker, CD69, on Jurkat cells compared with the empty vector control (Figure 4; supplemental Figure 3). This observation was dose-dependent between the 1:1 and 1:5 T:B cell ratios.
Engineered DOHH-2 cell lines and Jurkat CD69 expression following coculturing. (A) PDCD1LG2 surface expression on DOHH-2 cell lines stably transduced with wild-type PDCD1LG2 or PDCD1LG2-IGHV7-81 is provided compared to DOHH-2 cells transduced with empty vector. (B) CD69 expression in Jurkat cells following stimulation and coculturing for 28 hours with transduced DOHH-2 cell lines. Coculturing was performed in triplicate, using a ratio of 1:5 T:B cells. Significance was assessed using a 2-tailed, paired Student t test. **P < .01. NS, not significant. (C) Representative distribution of CD69 among Jurkat cells cocultured with empty vector, PDCD1LG2 wild-type expressing, or PDCD1LG2-IGHV7-81 expressing DOHH-2.
Engineered DOHH-2 cell lines and Jurkat CD69 expression following coculturing. (A) PDCD1LG2 surface expression on DOHH-2 cell lines stably transduced with wild-type PDCD1LG2 or PDCD1LG2-IGHV7-81 is provided compared to DOHH-2 cells transduced with empty vector. (B) CD69 expression in Jurkat cells following stimulation and coculturing for 28 hours with transduced DOHH-2 cell lines. Coculturing was performed in triplicate, using a ratio of 1:5 T:B cells. Significance was assessed using a 2-tailed, paired Student t test. **P < .01. NS, not significant. (C) Representative distribution of CD69 among Jurkat cells cocultured with empty vector, PDCD1LG2 wild-type expressing, or PDCD1LG2-IGHV7-81 expressing DOHH-2.
Discussion
Our study provides the first comprehensive characterization of PDL rearrangement breakpoint anatomy and establishes that FFPET-derived genomic material can be used effectively for capture sequencing to identify intra- and interchromosomal SRs in lymphomas. Furthermore, we establish that capture sequencing and FISH are complementary techniques that are best used in conjunction to identify the true frequency of PDL SRs. In aggregate, these new data shed light on the genetic complexity of the PDL loci and their likely important role in B-cell malignancies and acquired immune escape.
To date (including our present study), 29 PDL translocation partners have been observed; other previously reported partners include BCNP1, CIITA, IGHV7-81, IGHG4, IGL, NRG1, and TBL1XR1.5-7,16 Collectively, these data provide the basis for categorizing such translocations and intrachromosomal SRs into 4 distinct subgroups: CBR1, CBR2, nonclustered translocations, and intrachromosomal SRs. Here, we show that translocations arising in CBR2 are the most common (13 total reported events) followed by those arising in CBR1 (7 total reported events). In maintaining the complete coding sequence of both ligands, CBR1 and CBR2 translocations are shown to increase ligand expression as confirmed by IHC in this study and in agreement with published literature.6,7,17,18 PDL overexpression is achieved through a combination of promoter swaps with highly expressed genes in B cells (eg, CD44 and CIITA) and introduction of enhancer elements proximal to the second coding exon of either CD274 or PDCD1LG2. These translocations might also serve a dual purpose to abrogate expression of partner genes with tumor suppressor function (GET4, KIAA0226L, PTPN1) similarly to CIITA 5′ SRs (Figure 5).5,19-23 Furthermore, 5′ PDL translocations have been previously demonstrated to confer an immune-privilege phenotype in the context of T-cell activation.5
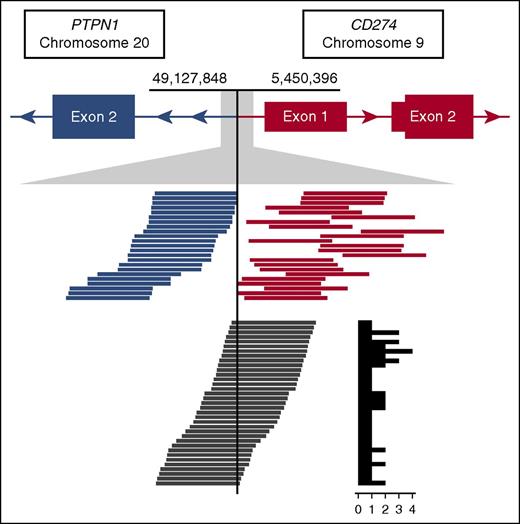
Read support for the CBR1 t(9;20) identified in case CS10. The top track represents gene models in der9, where PTPN1 is joined from intron 1 onward in opposite orientation upstream of CD274. Tracks below show an expansion of the breakpoint region. Blue and orange lines represent the paired spanning reads supporting the rearrangement, and dark gray lines represent the split reads with the supporting count summarized in a histogram to the right.
Read support for the CBR1 t(9;20) identified in case CS10. The top track represents gene models in der9, where PTPN1 is joined from intron 1 onward in opposite orientation upstream of CD274. Tracks below show an expansion of the breakpoint region. Blue and orange lines represent the paired spanning reads supporting the rearrangement, and dark gray lines represent the split reads with the supporting count summarized in a histogram to the right.
The 9 translocations occurring outside of CBR1 and CBR2 (5 of which are newly reported here) are a mixture of 5′ and 3′ events distributed across the PDL region, which we define here as a third category of translocations. The 3′ translocations are predicted to result in the loss of exon 7 and/or 6, which may remove microRNA target sites, increase transcript half-life, and improve translational efficiency due to altered secondary mRNA structures.24-28 The pathophysiological effect of these 3′ translocations is supported by the presence of PDCD1LG2 expression in the PDCD1LG2 3′ rearranged case. As with CBR1 and CBR2 translocations, regulatory elements from partner genes are involved in subgroup 3 translocations (eg, IGHV7-81 and IGHG4).6,7 We also demonstrate for the first time that exons 6 and 7 of PDCD1LG2 are not necessary for localization to the cell surface and that subgroup 3 and 4 SRs (including nonclustered 3′ translocations and 3′ deletions) can abrogate T-cell activity in a similar fashion to CBR1 and CBR2 rearrangements.5
In addition, we identified 17 intrachromosomal events that include deletions, macro- and microinversions, and duplications. These events add to 6 intrachromosomal rearrangements that have been previously observed in immune-privilege lymphomas.7,16,23 The functional effects of these rearrangements on both PDLs by IHC suggest that an increase in ligand expression is achieved through deletion of 3′ UTRs and duplications of 1 or both PDL genes.7 Our coculturing experiments using engineered cells, which are representative of duplications (wild-type PDCD1LG2-expressing DOHH-2) and 3′ deletions (PDCD1LG2-IGHV7-81-expressing DOHH-2 cells, because this fusion results in the loss of the 2 terminal 3′ PDCD1LG2 exons), provide additional support for this observation. An additional feature of several deletions includes the loss of the entire PDCD1LG2 coding sequence. We postulate that these deletions might serve as allelic exclusion-like events, whereby tumor cells preferentially select for CD274 expression, or alternatively, select for PDCD1LG2 harboring polymorphisms that may change the biological activity of ligand-receptor binding.29 Specific advantages of selecting for CD274 over PDCD1LG2 expression include smaller 5′ UTR in conjunction with smaller introns allowing increased translational efficiency and reduced microRNA degradation, more transient ligand-receptor binding state, inducibility of the ligand by interferon-γ, and ability to interact with the receptor CD80 in addition to PD1.30
As an increasing number of immune-checkpoint inhibitor therapies specifically targeting the PDLs become available, we anticipate clinical utility of biomarkers related to PDL CNVs, SRs, and expression. However, the exact biomarker approaches and methodologies to guide clinical decision making with respect to the rational use of immune checkpoint inhibition need to be explored. Specifically, a recent phase I clinical trial in Hodgkin lymphoma suggested that a high objective response rate in relapsed disease was linked to amplification of the PDL locus in a substantial proportion of Hodgkin lymphoma cases, indicating biomarker potential of determining PDL copy-number status.9 Our data strongly support the added value of capture sequencing to our understanding of the disparate action of PDL SRs, as they relate to heterogeneous PDL expression: either CD274 or PDCD1LG2. Our description of distinct CBRs might explain variable clinical courses under checkpoint inhibitor treatment and potential development of treatment resistance. Therefore, the exact knowledge of breakpoint anatomy and FISH-silent SRs might add to information derived from commonly used methodologies such as FISH and IHC. Furthermore, the clinical impact of PDL rearrangement partners, such as response to checkpoint inhibition or conventional therapies, still needs to be determined. Of relevance, observations in DLBCL suggest that varying MYC translocation partners (ie, immunoglobulin versus nonimmunoglobulin) influence prognosis after cyclophosphamide, doxorubicin, vincristine, and prednisone, plus rituximab therapy.31 Taken together, understanding the underlying SRs leading to altered PDL expression may be critical for future approaches matching specific lymphoma phenotypes to immunotherapies, as well as anticipating therapeutic resistance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the technical development and production groups of Canada’s Michael Smith Genome Sciences Centre for data generation. The authors thank Andrew McPherson for helpful discussions and feedback regarding methodology and Adele Telenius for assistance with functional experiments. The authors also thank Katy Milne and Brad H. Nelson for performing IHC staining, Andrew P. Weng for the Jurkat cell line, and Gordon J. Freeman for providing the PDCD1LG2 antibody.
This work was supported by the Canadian Cancer Society (grant 702519 to C.S.) and the Terry Fox Research Institute (grant 1023 to C.S. and R.D.G.). L.C.C. is supported by the Canadian Institutes of Health Research (CIHR) Bioinformatics Training Program. D.D.W.T. is supported by a BC Cancer Agency–CIHR–University of British Columbia (UBC) MD/PhD Studentship, a UBC Four-Year Fellowship, a Killam Doctoral Scholarship, and a CIHR Vanier Scholarship. A.M. is supported by postdoctoral fellowship awards from the Mildred Scheel Cancer Foundation (Deutsche Krebshilfe) and the Michael Smith Foundation for Health Research. M.A.M. is the UBC Canada Research Chair in Genome Science. C.S. is the recipient of a Career Investigator Award from the Michael Smith Foundation for Health Research.
Authorship
Contribution: The project was designed by A.J.M., A.M., B.W.W., C.S., D.D.W.T., D.W.S., K.J.S., R.D.G., and S.B.-N.; D.D.W.T. and S.B.-N. performed FISH analyses; A.M., C.S., D.D.W.T., and S.B.-N. performed cohort selection; A.M. and S.B.-N. performed nucleic acid extractions; A.J.M., M.A.M., and Y.Z. designed and performed the capture sequencing protocols; L.C.C. performed sequencing and bioinformatics analysis; D.D.W.T. completed validations of the predicted variants; A.M. performed IHC analysis; A.M. and D.D.W.T. performed the functional experiments; A.M., C.S., D.D.W.T., and L.C.C. interpreted the results; D.D.W.T. and L.C.C. wrote the manuscript; and all authors edited and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Steidl, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: csteidl@bccancer.bc.ca.
References
Author notes
L.C.C., D.D.W.T., and A.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal