Key Points
Responders to melphalan therapy are characterized by slower rates of NER and DSB/R mechanisms and higher apoptotic rates.
The DSB/R inhibitor SCR7 enhances cytotoxicity of melphalan against myeloma plasma cells.
Abstract
DNA repair activity of malignant cells seems to influence therapeutic outcome and patients’ survival. Herein, we investigated the mechanistic basis for the link between DNA repair efficiency and response to antimyeloma therapy. Nucleotide excision repair (NER), interstrand cross-links repair (ICL/R), double-strand breaks repair (DSB/R), and chromatin structure were evaluated in multiple myeloma (MM) cell lines (melphalan-sensitive RPMI8226; melphalan-resistant LR5) and bone marrow plasma cells (BMPCs) from MM patients who responded (n = 17) or did not respond (n = 9) to subsequent melphalan therapy. The effect of DSB/R inhibition was also evaluated. Responders’ BMPCs showed slower rates of NER and DSB/R (P < .0022), similar rates of ICL/R, and more condensed chromatin structure compared with nonresponders. Moreover, apoptosis rates of BMPCs were inversely correlated with individual DNA repair efficiency and were higher in responders’ cells compared with those of nonresponders (P = .0011). Similarly, RPMI8226 cells showed slower rates of NER and DSB/R, comparable rates of ICL/R, more condensed chromatin structure, and higher sensitivity than LR5 cells. Interestingly, cotreatment of BMPCs or cell lines with DSB/R inhibitors significantly reduced the rates of DSB/R and increased melphalan sensitivity of the cells, with the nonhomologous end-joining inhibitor SCR7 showing the strongest effect. Together, responders’ BMPCs are characterized by lower efficiencies of NER and DSB/R mechanisms, resulting in higher accumulation of the extremely cytotoxic ICLs and DSBs lesions, which in turn triggers the induction of the apoptotic pathway. Moreover, the enhancement of melphalan cytotoxicity by DSB/R inhibition offers a promising strategy toward improvement of existing antimyeloma regimens.
Introduction
Multiple myeloma (MM) is a malignant disorder accounting for ∼10% of hematologic malignancies.1 It is characterized by clonal proliferation of long-lived plasma cells associated with the overproduction of monoclonal immunoglobulin.2 The main clinical manifestations of the disease include anemia, recurrent infections, renal failure, and osteolytic lesions, which lead to devastating complications for the patients and their quality of life. To date, high-dose melphalan (HDM) followed by autologous stem cell transplantation (ASCT) is the gold standard of treatment for eligible patients with MM, whereas melphalan remains the backbone of treatment for elderly patients or the patients who are not eligible for ASCT.3,4 Although advances in the understanding of the biology of the disease have translated into novel therapeutic strategies in the past decade,5,6 the disease remains incurable because of the development of resistance and relapse in almost all patients.7,8
Melphalan is a nitrogen mustard clinically used in the treatment of several cancers.9 This alkylating agent induces N-alkylpurine-monoadducts, with a small fraction of them forming interstrand crosslinks (ICLs).10 Failure to repair ICLs’ lesions before the DNA replication process may induce DNA breaks or chromosomal rearrangements, or lead to cell death.11 N-alkylpurine-monoadducts are exclusively repaired by nucleotide excision repair (NER),12 whereas molecular components of NER, Fanconi anemia repair pathway, homologous recombination (HR), nonhomologous end-joining (NHEJ), and translesion synthesis are all required for adequate ICL repair.13-15
DNA double-strand breaks (DSBs) are indirectly formed as a consequence of melphalan-induced oxidative stress,16 and as intermediates of DNA repair pathways such as base-excision repair and ICL repair.14,17 DSBs are highly toxic lesions that can generate genetic instability and profound genome rearrangements. Once detected, DSBs can be repaired by 2 distinct mechanisms: HR, which acts preferentially in S and G2 phases of the cell cycle, and NHEJ, which is active throughout the whole cell cycle.18,19 The formation of DSBs is always followed by the phosphorylation of the histone H2AX, a variant of the H2A protein family, which is a component of the histone octamer in nucleosomes.20 Previous studies have shown that γH2AX can act as a highly sensitive and general marker of DNA damage induced by various ICL-inducing agents such as nitrogen mustards and platinum-based drugs.21-23
Repair of DNA and, thus, preservation of the genetic code are critical for normal cellular function.24 However, tumor cells use DNA repair pathways to develop resistance to chemotherapy. Therefore, inhibiting DNA repair may override this drug resistance. Many inhibitors targeting DNA repair pathways (PARP1, DNA-PK, ATM, ATR, MGMT, APE) or cell-cycle checkpoints (CHK1, CHK2) have now been developed and might be useful to induce tumor cell apoptosis in combination with DNA damage-inducing drugs.25-30
Herein, to further investigate the mechanistic basis for the link between DNA repair efficiency and response to antimyeloma therapy, we studied major DNA repair mechanisms in MM cell lines and malignant bone marrow plasma cells (BMPCs) from patients with MM before antimyeloma therapy. We found that BMPCs from responders to melphalan therapy are characterized by slower rates of NER and DSBs repair (DSB/R) compared with nonresponders. Moreover, we provide evidence that inhibitors of DSB/R might be proven efficient when they are used in combination with DNA-damaging chemotherapeutic drugs.
Materials and methods
Patients
A total of 26 unselected patients newly diagnosed with MM (12 female/14 male; median age 60 years, range 42-66) were included in the study (Table 1). All patients were staged according to the International Staging System.31 All patients received as first-line treatment HDM supported by ASCT. Response assessment was based on the International Myeloma Working Group criteria.32 The patients were categorized according to their outcome to responders (≥ partial responders, n = 17) and nonresponders (n = 9) to melphalan treatment. None of the patients received any antimyeloma treatment previously, including any supportive treatment (ie, bisphosphonates). All the BMPC patients’ samples were collected at diagnosis before the administration of any antimyeloma or supportive treatment. The study was approved by the institutional review board of Alexandra Hospital, and all subjects provided informed consent. The study was conducted according to the Declaration of Helsinki.
Patients and disease characteristics
| Characteristic . | Patients . | ||
|---|---|---|---|
| N . | Age (y) . | % of total . | |
| Sex | |||
| Female | 12 | 46.2 | |
| Male | 14 | 53.8 | |
| Age, median (range) | 60 (42-66) | ||
| Ig subtype | |||
| IgG | 12 | 46.2 | |
| IgA | 9 | 34.6 | |
| IgM | 0 | 0 | |
| IgE | 0 | 0 | |
| FLCs | 5 | 19.2 | |
| Nonsecretory | 0 | 0 | |
| ISS stage | |||
| I | 5 | 19.2 | |
| II | 8 | 30.8 | |
| III | 13 | 50.0 | |
| Response to HDM | |||
| Responders | 17 | 65.4 | |
| Nonresponders | 9 | 34.6 | |
| High-risk cytogenetics* | |||
| Responders | 6 | 35.3 | |
| Nonresponders | 4 | 44.4 | |
| Characteristic . | Patients . | ||
|---|---|---|---|
| N . | Age (y) . | % of total . | |
| Sex | |||
| Female | 12 | 46.2 | |
| Male | 14 | 53.8 | |
| Age, median (range) | 60 (42-66) | ||
| Ig subtype | |||
| IgG | 12 | 46.2 | |
| IgA | 9 | 34.6 | |
| IgM | 0 | 0 | |
| IgE | 0 | 0 | |
| FLCs | 5 | 19.2 | |
| Nonsecretory | 0 | 0 | |
| ISS stage | |||
| I | 5 | 19.2 | |
| II | 8 | 30.8 | |
| III | 13 | 50.0 | |
| Response to HDM | |||
| Responders | 17 | 65.4 | |
| Nonresponders | 9 | 34.6 | |
| High-risk cytogenetics* | |||
| Responders | 6 | 35.3 | |
| Nonresponders | 4 | 44.4 | |
High-risk cytogenetics are defined as the presence of t(4;14), t(4;20), deletion 17p13, or 1q21 gain.
Mononuclear cell suspensions were prepared from bone marrow aspirates by Ficoll-Paque plus gradient centrifugation (Amersham Pharmacia AB, Uppsala, Sweden), and plasma cell isolation was performed using the CD138 MicroBeads and magnet-assisted cell sorting (MACS; Miltenyi Biotec, GmbH, Germany). The purity of plasma cells was >90% as confirmed by flow cytometry (Becton-Dickinson, San Jose, CA).
Preparation of monohydroxymelphalan
Preparation of the monofunctional derivative of melphalan was performed as previously described.33 Briefly, melphalan (Sigma) was partially hydrolyzed by incubation in HCl, and reaction products were separated by chromatography using a C18 Sep-pak cartridge (Vac 35cm3, 10-g capacity; Waters, Millipore Corp).
Cell lines
Human MM cell lines (melphalan-sensitive, RPMI8226, and melphalan-resistant, LR5) were cultured in RPMI 1640 medium (Sigma-Aldrich) containing 10% (vol/vol) fetal bovine serum (Gibco), 1% (vol/vol) of 200 mM l-glutamine, 1% (vol/vol) of 10 mg/mL gentamicin (Gibco), and 1% (vol/vol) of 250 μg/mL amphotericin B (Sigma-Aldrich). Melphalan-resistant LR5 cells were maintained under constant selection through the addition of 1 μM melphalan (Sigma-Aldrich) twice per week. Subsequent to the addition of the maintenance melphalan, LR5 cells were allowed to grow for at least for 3 days before any additional treatment.
Cell treatment
Cell lines were treated with melphalan or monohydroxymelphalan (100 μg/mL, 5 min, 37°C) in culture medium. BMPCs were treated ex vivo with 100 μg/mL melphalan (5 min, 37°C) in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin (100 U/mL), streptomycin (100 μg/mL) and l-glutamine (2 mmol/L). To specifically inhibit DSB/R, cell lines were treated for 0 to 120 hours at 37°C with various concentrations (0-100 μg/mL) of NU7026 (Sigma, N1537), RI-1 (Calbiochem, 553514), or SCR7 (Access bio, M60082-25), all diluted in dimethyl sulfoxide (DMSO).
Measurement of DNA damage and repair
Monoadducts and ICLs were measured in the N-ras gene using Southern blot analysis as described previously (supplemental Materials and methods, available on the Blood Web site).34
Immunofluorescence antigen staining and confocal laser scanning microscope analysis
Aliquots of 2 × 104cells were adhered to a coverslip, fixed, and stored at −70°C until the analysis of γH2AX and Rad51.22,23 Briefly, cells were incubated with antibodies against γH2AX (serine-139, Cell Signaling) or Rad51 (Santa Cruz Biotechnology), washed, incubated with fluorescent secondary antibodies (Alexa Fluor 488 goat anti-mouse IgG; Abcam), and images were visualized with a Leica TCS SP-1 confocal laser scanning microscope. Foci were manually counted in 200-cells-per-treatment condition, and results are expressed as the mean γH2AX or Rad51 foci per cell (mean ± standard deviation [SD]) from 3 independent experiments.
Micrococcal nuclease digestion-based analysis of chromatin condensation
Nuclei were isolated from untreated cells and digested with 1 unit of micrococcal nuclease (Sigma-Aldrich) for 10 minutes at 37°C. DNA was purified, separated in 1.5% agarose gel, transferred onto nitrocellulose, and hybridized with the appropriate labeled probe.35
Apoptosis and cytotoxicity assays
Cells (2 × 104cells) were treated with various doses of melphalan (0-200 μg/mL) for 5 minutes, followed by 24 hours or 72 hours postincubation time. Then, the Cell Death Detection ELISA-PLUS kit (Roche Diagnostics Corporation) was used to determine apoptosis at 24 hours according to the protocol provided by the manufacturer (supplemental Materials and methods).36 Moreover, ApoTox-Glo Triplex Assay (Promega) was used to measure the cells’ viability, cytotoxicity, and apoptosis at 24 hours and 72 hours according to the manufacturer’s instructions (supplemental Materials and methods).
Cell viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.37 Briefly, cells (8 × 103cells/well) were seeded into 96-well plates and, after 24 hours, exposed to the agents under study. After treatment, the medium was replaced with 100 μL of MTT solution (0.5 mg/mL in cell culture medium) and incubated at 37°C for 2 hours. MTT solution was then removed, and MTT formazan was dissolved in 100 μL DMSO. Absorbance was measured at 570 nm using the Tecan Safire2 reader.
Statistical analysis
All data from DNA repair assessment for cell lines and BMPCs are shown as mean values ± SD. To assess the linear association between DNA damage burden and apoptosis rates, linear regression analysis was performed. Student t test was used to determine differences in cell viability. Comparisons within RPMI 8226 and LR5 cell lines, as well as within BMPCs, among the different melphalan therapy groups (NU7026, SCR7, and R1-1) and the reference group of no inhibitor, were performed using the Wilcoxon rank-sum statistical test. The same statistical test was used for comparisons between the RPMI 8226 and LR5 cell lines, as well as in BMPCs between responders and nonresponders, for each of the melphalan therapy groups (No inhibitor, NU7026, SCR7, and R1-1). P < .05 was considered statistically significant.
Results
Determining dose and duration of exposure
A pair of commonly used MM cell lines sharing several common biological characteristics—the melphalan-sensitive RPMI8226 and the melphalan-resistant LR5—which was derived from RPMI 8226 after exposure to gradually increasing concentrations of melphalan,38 were treated with melphalan in combination with a DSB/R inhibitor. Three repair inhibitors were used: two of them modify NHEJ (NU7026 and SCR7)39-41 and IR-1 disrupts HR (supplemental Materials and methods).42 To investigate the effect of the combination treatment of melphalan with an inhibitor, we first selected the optimal concentrations of the inhibitors that, when administered alone, showed minimal decrease in cell viability. Cell viability was evaluated using the MTT assay, where IC50 values were calculated. There was no effect on the cell viability after treatment with DMSO, thus excluding any potential toxicity from medium, contributing to the effect of melphalan. After treatment with an inhibitor, both cell lines showed a dose-dependent decrease in cell viability (supplemental Figure 1), with the IC50 values being significantly lower in RPMI 8226 than in LR5 cells (Student t test: NU7026, P = .0348; SCR7, P < .001; IR-1, P = .001). Moreover, after treatment of both cell lines with inhibitors (NU7026, 10 μg/mL; SCR7, 20 μg/mL; RI-1, 5 μg/mL) for various time periods (up to 120 hours), a time-dependent decrease in cell viability was observed (supplemental Figure 2).
In addition, to define the optimal concentrations of the inhibitors, which induce maximal accumulation of melphalan DNA damage, cell lines were treated with various concentrations of the inhibitors (0-100 μg/mL) for 24 hours and then with 100 μg/mL melphalan for 5 minutes in the presence of the inhibitor. Cells were subsequently incubated for 48 hours in melphalan-free medium in the presence of the inhibitor, and γH2AX foci were measured. The results were expressed as the area under the curve (AUC) for γH2AX foci during the whole experiment, which reflects the overall DNA damage burden resulting from initial damage formation and DNA repair. Both cell lines showed similar results (supplemental Figure 3). Using NU7026, a dose-dependent increase in the accumulation of γH2AX foci was observed up to 10 μg/mL, with the levels of γH2AX foci decreasing at higher concentrations. The corresponding concentrations of the other inhibitors were 20 μg/mL for SCR7 and 5 μg/mL for RI-1. There was no effect on the phosphorylation of H2AX after treatment of each type of cells with DMSO or an inhibitor alone (data not shown). Thus, the optimal concentrations of the inhibitors, which in combination with melphalan induce maximal inhibition of DSBs repair with minimal decrease in cell viability, were 10 μg/mL NU7026, 20 μg/mL SCR7, and 5 μg/mL RI-1.
Slower rates of NER and DSB/R and higher apoptosis rates in the melphalan-sensitive cell line
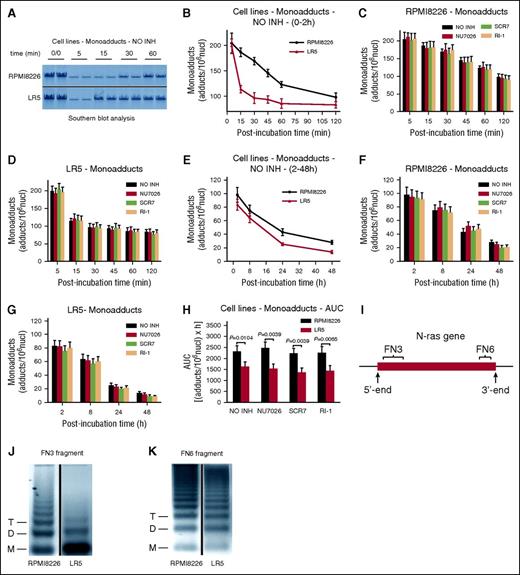
We have shown previously that in both PBMCs and BMPCs from patients with MM, the efficiency of the N-ras–specific NER correlates with the drug sensitivity of these cells and reflects the individualized response to melphalan therapy.43 In the present study, the kinetics of N-ras–specific monoadducts repair were followed for up to 48 hours after treatment of RPMI 8226 and LR5 cells with melphalan by using Southern blot analysis (Figure 1A). Similar formation of monoadducts was observed in both cell types at the end of the 5-minute treatment (Figure 1B). Biphasic repair of DNA damage was observed, with a fast component extending up to 2 hours after treatment (Figure 1B), and a slower progression of repair thereafter (Figure 1E). Interestingly, a much slower early phase of repair was found in RPMI 8226 than in LR5 cells, resulting in significantly higher levels of monoadducts in RPMI 8226 cells (Figure 1B). On the other hand, similar rates of adduct loss were observed in both cell lines during the second phase of repair (Figure 1E). In agreement with the rates of NER mentioned before, significantly higher total amounts of monoadducts, expressed as AUC, were found in RPMI 8226 than in LR5 cells (Figure 1H; supplemental Table 1). The cotreatment of cells with a DSB/R inhibitor had no significant impact on the monoadducts’ levels (Figure 1C-D,F-H). Similar results were obtained by using a monofunctional derivative of melphalan, which induced only monoadducts (supplemental Figure 4).33
Kinetics of monoadducts repair in MM cell lines. (A) Representative autoradiograms for the Southern blot analysis of monoadducts. 0/0, no treatment. A horizontal line has been inserted to indicate a repositioned gel lane. The kinetics of monoadducts repair 0 to 2 hours (B) and 2 to 48 hours (E) after treatment with melphalan alone are shown. NO INH, no inhibitor. The kinetics of monoadducts repair 0 to 2 hours (C-D) and 2 to 48 hours (F-G) after treatment of RPMI 8226 (C,F) and LR5 (D,G) cells with melphalan in the presence or not of an inhibitor are also presented. (H) Total amounts of monoadducts, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (I) The gene locus where the kinetics of DNA repair and chromatin condensation were evaluated. Representative autoradiograms showing chromatin condensation of the untreated RPMI 8226 and LR5 cells in the FN3 (J) and FN6 (K) fragments of the N-ras gene locus. Vertical lines have been inserted to indicate a repositioned gel lane. Symbols M, D, and T represent the positions of nucleosome monomer, dimer, and trimer, respectively. The experiments shown were based on a minimum of 3 independent repeats. Bar graphs and error bars show mean ± standard deviation (SD).
Kinetics of monoadducts repair in MM cell lines. (A) Representative autoradiograms for the Southern blot analysis of monoadducts. 0/0, no treatment. A horizontal line has been inserted to indicate a repositioned gel lane. The kinetics of monoadducts repair 0 to 2 hours (B) and 2 to 48 hours (E) after treatment with melphalan alone are shown. NO INH, no inhibitor. The kinetics of monoadducts repair 0 to 2 hours (C-D) and 2 to 48 hours (F-G) after treatment of RPMI 8226 (C,F) and LR5 (D,G) cells with melphalan in the presence or not of an inhibitor are also presented. (H) Total amounts of monoadducts, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (I) The gene locus where the kinetics of DNA repair and chromatin condensation were evaluated. Representative autoradiograms showing chromatin condensation of the untreated RPMI 8226 and LR5 cells in the FN3 (J) and FN6 (K) fragments of the N-ras gene locus. Vertical lines have been inserted to indicate a repositioned gel lane. Symbols M, D, and T represent the positions of nucleosome monomer, dimer, and trimer, respectively. The experiments shown were based on a minimum of 3 independent repeats. Bar graphs and error bars show mean ± standard deviation (SD).
Furthermore, we analyzed chromatin condensation in different fragments of the N-ras gene locus (Figure 1I). In both cell lines the FN3 fragment (located close to the transcription initiation site of the gene) exhibited greater looseness of chromatin structure than the FN6 fragment (located near the 3′-end) (Figure 1J-K). Interestingly, we found that the looseness of the chromatin structure of the FN3 fragment was much higher in the melphalan-resistant LR5 cell line than in the melphalan-sensitive RPMI 8226 cells (Figure 1J), whereas chromatin condensation of the FN6 fragment was very similar in both MM cell lines (Figure 1K).
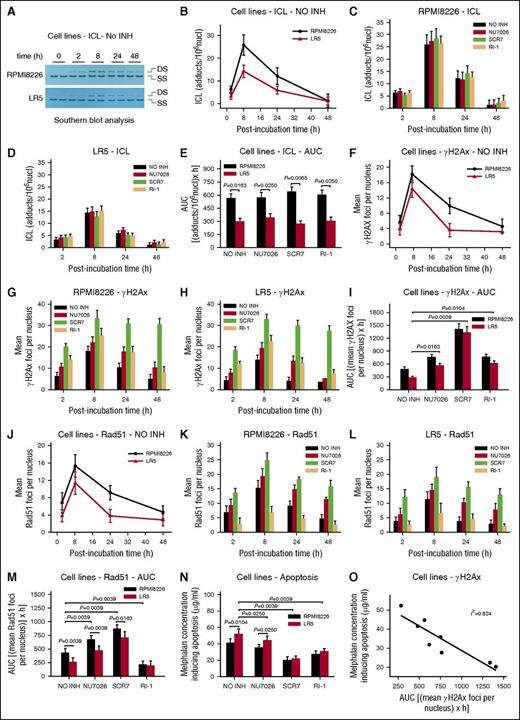
The kinetics of ICL formation and repair were also analyzed; of note, the disappearance of the ICLs (Figure 2A) reflects the excision of one of the crosslinked bases, the “unhooking” step, and not necessarily the complete removal of the damage.44 We found that the accumulation of ICLs was slower compared with monoadducts’ accumulation, reaching maximal levels within 8 hours (Figure 2B). Thereafter, ICLs’ levels were decreased and the rates of adduct loss were similar in the different cell types. Interestingly, the melphalan-sensitive cell line RPMI 8226 exhibited significantly higher ICL burden (expressed as AUC) compared with the melphalan-resistant LR5 cells (Figure 2E; supplemental Table 1). As expected, the combination treatment of melphalan with DNA repair inhibitors had no significant effect on ICL levels (Figure 2C-E).
Interstrand crosslinks formation/repair, γH2AX and Rad51 foci formation, and melphalan toxicity in MM cell lines. (A) Representative autoradiograms for the Southern blot analysis of ICLs 0 to 48 hours after treatment with melphalan alone. DS, double-stranded DNA; SS, denatured, single-stranded DNA. (B) The kinetics of ICL formation and repair 2 to 48 hours after treatment with melphalan alone. NO INH, no inhibitor. The formation and repair of ICLs 2 to 48 hours after treatment of RPMI 8226 (C) or LR5 (D) cells with melphalan in the presence or not of an inhibitor. (E) Total amounts of ICLs, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (F) γH2AX foci formation 2 to 48 hours after treatment with melphalan alone. The formation of γH2AX foci after treatment of RPMI 8226 (G) or LR5 (H) cells with melphalan alone or in combination with an inhibitor. (I) Total amounts of γH2AX foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (J) Rad51 foci formation 2 to 48 hours after treatment with melphalan alone. Formation of Rad51 foci after treatment of RPMI 8226 (K) or LR5 (L) cells with melphalan alone or in combination with an inhibitor. (M) Total amounts of Rad51 foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (N) The induction of apoptosis 24 hours after treatment with melphalan in the presence or not of an inhibitor. (O) Correlation between the melphalan doses required for the induction of apoptosis and the drug-induced γH2AX foci in the same cells. The experiments shown were based on a minimum of 3 independent repeats. Bar graph and error bars show mean ± SD.
Interstrand crosslinks formation/repair, γH2AX and Rad51 foci formation, and melphalan toxicity in MM cell lines. (A) Representative autoradiograms for the Southern blot analysis of ICLs 0 to 48 hours after treatment with melphalan alone. DS, double-stranded DNA; SS, denatured, single-stranded DNA. (B) The kinetics of ICL formation and repair 2 to 48 hours after treatment with melphalan alone. NO INH, no inhibitor. The formation and repair of ICLs 2 to 48 hours after treatment of RPMI 8226 (C) or LR5 (D) cells with melphalan in the presence or not of an inhibitor. (E) Total amounts of ICLs, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (F) γH2AX foci formation 2 to 48 hours after treatment with melphalan alone. The formation of γH2AX foci after treatment of RPMI 8226 (G) or LR5 (H) cells with melphalan alone or in combination with an inhibitor. (I) Total amounts of γH2AX foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (J) Rad51 foci formation 2 to 48 hours after treatment with melphalan alone. Formation of Rad51 foci after treatment of RPMI 8226 (K) or LR5 (L) cells with melphalan alone or in combination with an inhibitor. (M) Total amounts of Rad51 foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. (N) The induction of apoptosis 24 hours after treatment with melphalan in the presence or not of an inhibitor. (O) Correlation between the melphalan doses required for the induction of apoptosis and the drug-induced γH2AX foci in the same cells. The experiments shown were based on a minimum of 3 independent repeats. Bar graph and error bars show mean ± SD.
Moreover, to study the formation and repair of DNA DSBs, γH2AX foci as a marker of DNA damage and the formation of Rad51 foci as a marker of homologous recombination were examined. We found that γH2AX foci reached maximal levels within 8 hours and decreased thereafter. In accordance with previous data showing that melphalan-induced γH2AX foci persist for a shorter time in melphalan-resistant LR5 cells,28 we found that RPMI 8226 cells showed significantly lower γH2AX foci removal capacity and higher γH2AX foci burden than LR5 cells (Figure 2F-I; supplemental Table 1). All DSB/R inhibitors increased the accumulation of γH2AX in both cell lines (Figure 2G-I). It is worth mentioning that, in line with the high levels of ligase IV observed in these cell lines,28 we found that SCR7 showed the strongest effect. In harmony with γH2AX foci formation, Rad51 foci reached maximal levels within 8 hours and decreased thereafter, whereas LR5 cells showed lower levels of Rad51 foci compared with RPMI 8226 cells (Figure 2J-M; supplemental Table 1). In agreement with previous data,45 homologous recombination was increased as a compensatory response to NHEJ inhibition, whereas treatment with IR-1 diminished Rad51 foci (Figure 2K-M).
The induction of apoptosis was also measured 24 hours after melphalan treatment by using the Cell Death Detection ELISA-PLUS assay. We found that the lowest concentrations of melphalan required for the induction of apoptosis were higher in LR5 (52.2 ± 5.1 μg/mL) compared with RPMI 8226 cells (41.5 ± 5.0 μg/mL) (Figure 2N; supplemental Table 1), indicating higher apoptosis rates in the melphalan-sensitive RPMI 8226 cells. Interestingly, we found that although the inhibitors per se did not result in significant cytotoxicity (data not shown), cotreatment of these agents with melphalan sensitized both cell lines (Figure 2N; supplemental Table 1). Similar results were obtained by using the ApoTox-Glo Triplex Assay 24 hours and 72 hours after treatment (supplemental Figures 5 and 6). Thus, increased apoptosis and cytotoxicity as well as decreased cell viability were observed in RPMI 8226 compared with LR5 cells, especially 72 hours post melphalan treatment. Again, in both cell lines, increased sensitivity toward melphalan was observed in the presence of the inhibitors; SCR7 showed the strongest effect. Finally, an inverse correlation was observed between the melphalan concentration required for the induction of apoptosis and the DSBs levels measured in the same cell line (linear regression analysis; r2 = 0.824; Figure 2O).
Responders’ BMPCs are characterized by slower rates of NER and DSB/R and increased melphalan sensitivity
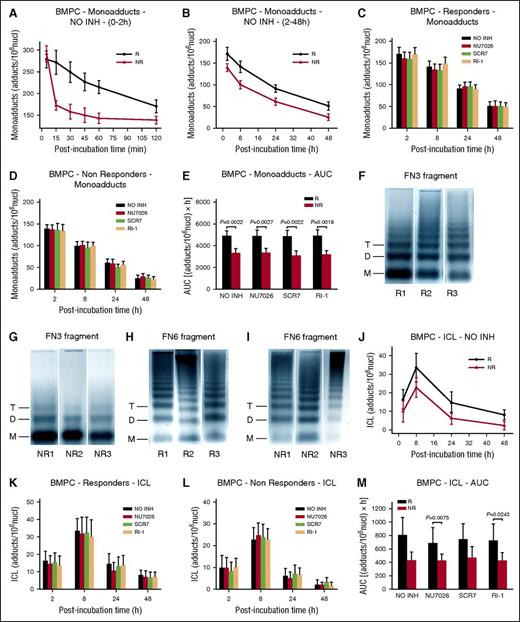
Similar formation of monoadducts was found at the end of the 5-minute ex vivo melphalan exposure in the BMPCs of all individuals tested (Figure 3A). In line with the results from the MM cell lines, monoadducts were diminished over 2 sharply demarcated phases: a rapid first phase (0-2 hours after melphalan treatment) (Figure 3A), and a much slower second phase (2-48 hours after treatment) (Figure 3B). Interestingly, although a much slower early phase of repair was found in responders’ BMPCs, very similar rates of adduct loss were observed in both groups of patients during the second phase of repair. In accordance with these data, the monoadducts’ burden, expressed as AUC for DNA adducts, was significantly higher in responders compared with nonresponders (Figure 3E; supplemental Table 2). DSB/R inhibitors had no effect on the monoadduct levels (Figure 3C-E; supplemental Table 2).
Kinetics of monoadducts and ICLs repair and chromatin condensation in BMPCs. (A) The kinetics of monoadducts repair 0 to 2 hours (A) and 2 to 48 hours (B) after treatment with melphalan alone. R, responders; NR, nonresponders; NO INH, no inhibitor. The kinetics of monoadducts repair 2 to 48 hours after treatment of BMPCs from responders (C) or nonresponders (D) with melphalan alone or in combination with an inhibitor. (E) Total amounts of monoadducts, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. Representative autoradiograms showing chromatin condensation of 3 untreated responders (R1, R2, R3) and 3 nonresponders (NR1, NR2, NR3) in the FN3 (F-G) and FN6 (H-I) fragments of the N-ras gene locus. Symbols M, D, and T represent the positions of nucleosome monomer, dimer, and trimer, respectively. (J) Kinetics of ICL formation and repair after treatment with melphalan alone. Formation and repair of ICLs 2 to 48 hours after treatment of BMPCs from responders (K) or nonresponders (L) with melphalan in combination or not with an inhibitor. (M) Total amounts of ICLs, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. Bar graph and error bars show mean ± SD.
Kinetics of monoadducts and ICLs repair and chromatin condensation in BMPCs. (A) The kinetics of monoadducts repair 0 to 2 hours (A) and 2 to 48 hours (B) after treatment with melphalan alone. R, responders; NR, nonresponders; NO INH, no inhibitor. The kinetics of monoadducts repair 2 to 48 hours after treatment of BMPCs from responders (C) or nonresponders (D) with melphalan alone or in combination with an inhibitor. (E) Total amounts of monoadducts, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. Representative autoradiograms showing chromatin condensation of 3 untreated responders (R1, R2, R3) and 3 nonresponders (NR1, NR2, NR3) in the FN3 (F-G) and FN6 (H-I) fragments of the N-ras gene locus. Symbols M, D, and T represent the positions of nucleosome monomer, dimer, and trimer, respectively. (J) Kinetics of ICL formation and repair after treatment with melphalan alone. Formation and repair of ICLs 2 to 48 hours after treatment of BMPCs from responders (K) or nonresponders (L) with melphalan in combination or not with an inhibitor. (M) Total amounts of ICLs, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. Bar graph and error bars show mean ± SD.
We next analyzed chromatin condensation in the N-ras gene locus (Figure 1I). In all patients with MM, the FN3 fragment showed greater looseness of chromatin structure than the FN6 fragment (Figure 3F-I). To note, responders to melphalan therapy (Figure 3F) are characterized by more condensed chromatin structure at the FN3 fragment than nonresponders (Figure 3G). Chromatin condensation of the FN6 fragment was similar in both groups of patients with MM (Figure 3H-I).
The formation and repair of ICLs were also evaluated. Peak ICL levels were observed 8 hours after melphalan treatment (Figure 3J). Thereafter, ICL levels were diminished, and differences in the rates of adduct loss between groups of patients were minor. On the other hand, ICL burden, expressed as AUC, was higher in responders’ BMPCs compared with nonresponders (Figure 3M; supplemental Table 2). The kinetics of ICLs showed no difference in the presence of DSB/R inhibitors (Figure 3K-M; supplemental Table 2).
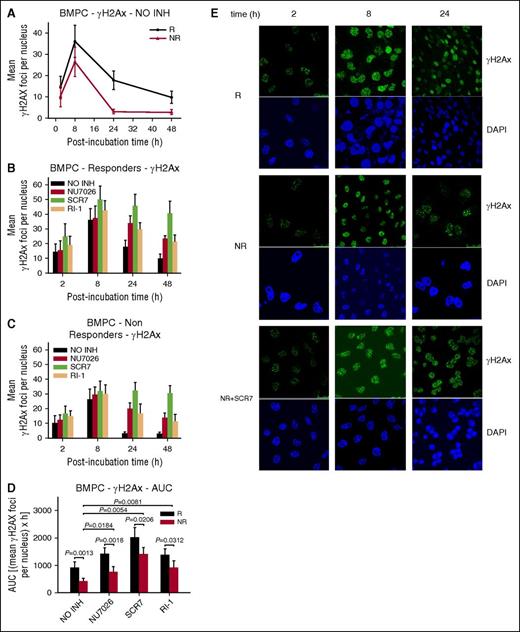
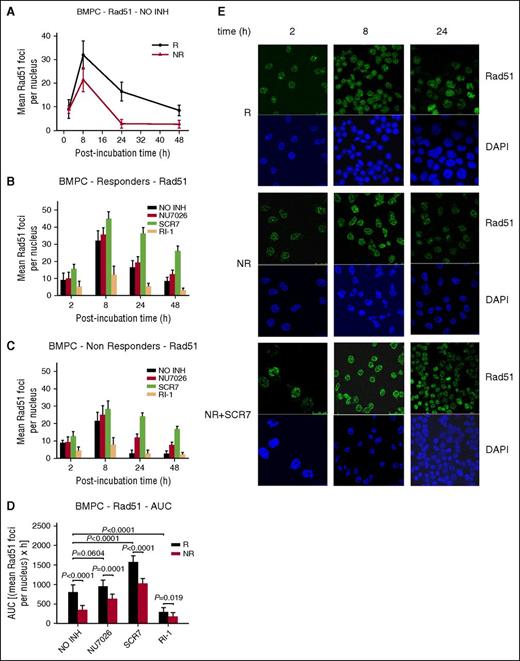
Furthermore, the formation and repair of DSBs were analyzed (Figure 4). Consistent with the results from the cell lines, peak γH2AX foci levels were observed within 8 hours of melphalan treatment and decreased thereafter (Figure 4A,E). Responders’ BMPCs showed lower γH2AX foci removal capacity (Figure 4A) and higher accumulation of γH2AX foci, expressed as AUC, compared with nonresponders (Figure 4D; supplemental Table 2). Interestingly, the combination of melphalan with an inhibitor resulted in inhibition of γH2AX foci removal and higher accumulation of γH2AX foci in all patients with MM (Figure 4B-D; supplemental Table 2), with SCR7 showing the strongest effect. The Rad51 response followed the same time course as the γH2AX response, peaking at 8 hours and declining thereafter (Figure 5A,E), with nonresponders showing lower Rad51 foci levels compared with responders (Figure 5A,D; supplemental Table 2). In line with the MM cell line results, homologous recombination was increased as a compensatory response to NHEJ inhibition, whereas IR-1 significantly reduced Rad51 foci levels (Figure 5B-D).
Formation of γH2AX foci in BMPCs. (A) γH2AX foci formation 2 to 48 hours after treatment with melphalan alone. Formation of γH2AX foci 2 to 48 hours after treatment of BMPCs from responders (B) or nonresponders (C) with melphalan in the presence or not of an inhibitor. NO INH, no inhibitor. R, responders; NR, nonresponders. (D) Total amounts of γH2AX foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. (E) Typical images showing the γH2AX staining at different time points after treatment of BMPCs from 1 representative responder (R) and 1 nonresponder (NR) with melphalan alone or in the presence of SCR7. Upper images, immunofluorescence antigen staining; bottom images, cell nuclei labeled with 4′,6-diamidino-2-phenylindole.
Formation of γH2AX foci in BMPCs. (A) γH2AX foci formation 2 to 48 hours after treatment with melphalan alone. Formation of γH2AX foci 2 to 48 hours after treatment of BMPCs from responders (B) or nonresponders (C) with melphalan in the presence or not of an inhibitor. NO INH, no inhibitor. R, responders; NR, nonresponders. (D) Total amounts of γH2AX foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. (E) Typical images showing the γH2AX staining at different time points after treatment of BMPCs from 1 representative responder (R) and 1 nonresponder (NR) with melphalan alone or in the presence of SCR7. Upper images, immunofluorescence antigen staining; bottom images, cell nuclei labeled with 4′,6-diamidino-2-phenylindole.
The role of homologous recombination in the repair of melphalan-induced DNA damage in BMPCs. (A) Rad51 foci formation 2 to 48 hours after treatment with melphalan alone. The formation of Rad51 foci 2 to 48 hours after treatment of BMPCs from responders (B) or nonresponders (C) with melphalan in the presence or not of an inhibitor. NO INH, no inhibitor. R, responders; NR, nonresponders. (D) Total amounts of Rad51 foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. (E) Typical images showing the Rad51 staining at different time points after treatment of BMPCs from 1 representative responder (R) and 1 nonresponder (NR) with melphalan alone or in the presence of SCR7. Upper images, immunofluorescence antigen staining; bottom images, cell nuclei labeled with 4′,6-diamidino-2-phenylindole.
The role of homologous recombination in the repair of melphalan-induced DNA damage in BMPCs. (A) Rad51 foci formation 2 to 48 hours after treatment with melphalan alone. The formation of Rad51 foci 2 to 48 hours after treatment of BMPCs from responders (B) or nonresponders (C) with melphalan in the presence or not of an inhibitor. NO INH, no inhibitor. R, responders; NR, nonresponders. (D) Total amounts of Rad51 foci, expressed as AUC, after treatment with melphalan in the presence or not of an inhibitor. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed. (E) Typical images showing the Rad51 staining at different time points after treatment of BMPCs from 1 representative responder (R) and 1 nonresponder (NR) with melphalan alone or in the presence of SCR7. Upper images, immunofluorescence antigen staining; bottom images, cell nuclei labeled with 4′,6-diamidino-2-phenylindole.
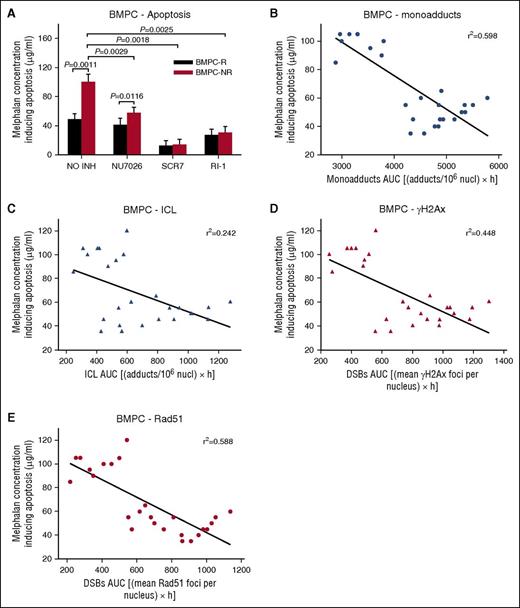
The induction of the apoptotic pathway in BMPCs was also evaluated. We observed that BMPCs from responders to melphalan therapy were characterized by higher rates of apoptosis compared with nonresponders. Using the Cell Death Detection ELISA-PLUS assay, we found that the melphalan concentrations for inducing apoptosis were much higher in nonresponders (96.7 ± 8.2 μg/mL) compared with responders (52.2 ± 7.5 μg/mL) (Figure 6A; supplemental Table 2). Moreover, the combination of melphalan with inhibitors significantly reduced the melphalan concentration that induced apoptosis in all patients with MM. Similar results were obtained by using ApoTox-Glo Triplex Assay 24 hours and 72 hours after treatment (supplemental Figures 7 and 8). Responders’ BMPCs showed increased apoptosis and cytotoxicity as well as decreased cell viability compared with nonresponders’ cells, especially 72 hours after melphalan treatment. The combination of melphalan with SCR7 led to significant enhancement of melphalan cytotoxicity on MM cells. In addition, an inverse correlation was observed between the melphalan concentration required for the induction of apoptosis and the levels of all types of DNA damage examined in the same individuals (linear regression analysis; monoadducts, r2 = 0.598, Figure 6B; ICLs, r2 = 0.242, Figure 6C; γH2AX, r2 = 0.448, Figure 6D; Rad51, r2 = 0.588, Figure 6E).
The induction of the apoptotic pathway in BMPCs. (A) The induction of apoptosis after treatment of BMPCs with melphalan alone or in combination with an inhibitor. Correlation between the melphalan doses required for the induction of apoptosis and the drug-induced monoadducts (B), ICLs (C), γH2AX (D), and Rad51 foci (E) in the same patients. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed.
The induction of the apoptotic pathway in BMPCs. (A) The induction of apoptosis after treatment of BMPCs with melphalan alone or in combination with an inhibitor. Correlation between the melphalan doses required for the induction of apoptosis and the drug-induced monoadducts (B), ICLs (C), γH2AX (D), and Rad51 foci (E) in the same patients. The experiments shown were based on a minimum of 3 independent repeats and the data reported are the mean ± SD of all the patients analyzed.
Discussion
The ability of MM cells to remove melphalan adducts through DNA repair pathways represents an important mechanism of resistance to melphalan therapy.46-48 Our previous studies12,35 have revealed a biphasic repair of melphalan monoadducts in the N-ras gene locus, reflecting rapid repair of adducts located immediately downstream of the transcription start site of the gene, together with slower repair of adducts located near the 3′-end of the gene. Moreover, the aforementioned observed variation in DNA repair efficiency has paralleled exactly with the variation in the degree of the local chromatin condensation, with more relaxed chromatin being associated with faster repair. Consistent with these results, we found that in all patients with MM, monoadducts were lost over 2 sharply demarcated phases, possibly because of differences in the chromatin structure along the transcribed genes. Indeed, in all individuals, a greater looseness of chromatin structure was found in regions close to the transcription start site of the N-ras gene compared with those at the 3′-end of the gene. Interestingly, responders to melphalan therapy are characterized by slower repair of monoadducts during the first phase of repair together with more condensed chromatin in regions in proximity to the transcription start site, which possibly impedes the access of DNA repair proteins to sites of DNA damage, thus reducing DNA repair capacity.49 No significant differences were observed between the 2 groups of patients regarding the efficiency of the second phase of repair and the chromatin condensation at the 3′-end of the N-ras gene. Furthermore, in all MM patients analyzed, similar formation of monoadducts was observed at the end of the 5-minute melphalan treatment, indicating that the local chromatin condensation did not affect their formation. To note, individual monoadducts’ repair efficiencies were inversely correlated with apoptosis rates in the same cells, suggesting that repair of monoadducts before crosslink formation may play an important role in protecting cells from melphalan cytotoxicity.
We next examined the ICLs “unhooking” step, which is the earliest DNA processing event in ICLs repair.44,50-53 Although “unhooking” rates were more or less similar in both groups of patients, ICLs burden was significantly higher in responders’ BMPCs because of the higher levels of monoadducts (precursors of ICL) that are left unrepaired in these cells. It is worth mentioning that individual ICL/R efficiencies were inversely correlated with apoptosis rates in the same cells, underlying the cytotoxic character of this lesion.
The formation and repair of melphalan-induced DSBs was also evaluated. We found that melphalan-induced DSB burden was significantly lower in nonresponder’s BMPCs first because of the lower accumulation of ICLs (precursors of DSBs), which are left unrepaired in these cells, and second, the significantly higher DSBs removal capacity of nonresponders’ BMPCs. Although we did not address the mechanistic basis for the increased DSBs’ repair efficiency of nonresponders’ BMPCs, it is possible that it could be related to the upregulation of several factors involved in DSBs repair, such as ligase IV, XRCC4, RPA2, Artemis, and PARP-1 found in the melphalan-resistant LR5 cells in previous studies.28 Moreover, individual DSB/R efficiencies were inversely correlated with apoptosis rates in the same cells, suggesting that the DSB/R capacity of BMPCs plays a crucial role in the successful outcome of chemotherapy. In addition, Sousa et al28 have shown that LR5 cells were significantly more sensitive to PARP-1 inhibition than RPMI 8226 cells, and that the DNA-PK inhibitor markedly increased the melphalan sensitivity of the resistant LR5 cell line. Herein, we found that DSB/R inhibitors significantly increased the accumulation of DSBs and the melphalan sensitivity of BMPCs in all patients with MM. Particularly, SCR7 showed the strongest effect, indicating that this compound might prove efficacious when used in combination with DNA-damaging chemotherapeutic drugs. Similar results were obtained using MM cell lines.
Collectively, responders to melphalan therapy are characterized by slower rates of NER and DSB/R mechanisms, resulting in higher accumulation of the extremely cytotoxic ICLs and DSBs lesions, which in turn triggers the induction of the apoptotic pathway, a priority for successful clinical outcome. Moreover, the enhancement of melphalan cytotoxicity by DSB/R inhibitors offers a promising strategy toward treatment of MM and improvement of existing regimens.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Enrique M Ocio (Hospital Universitario de Salamanca, Instituto de Investigacion Biomedica de Salamanca, Centro de Investigación del Cancer, Salamanca, Spain) and Jesus F. San Miguel (Clinica Universidad de Navarra, Centro de Investigacion Medica Aplicada, Instituto de Investigacion Sanitaria de Navarra, Pamplona, Spain) for the generous gift of MM cell lines.
Authorship
Contribution: M.G. performed the experiments, collected and analyzed the data, and wrote the manuscript; E.T. was responsible for the selection and clinical evaluation of patients, analyzed data, and reviewed the manuscript; C.B. performed the statistical analysis of data and reviewed the manuscript; N.C.M. evaluated results and reviewed the manuscript; M.A.D. was responsible for the selection and clinical evaluation of patients and reviewed the manuscript; and V.L.S. designed and supervised research, performed the experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vassilis L. Souliotis, Institute of Biology, Medicinal Chemistry and Biotechnology, National Hellenic Research Foundation, 48 Vassileos Constantinou Ave, 11635 Athens, Greece; e-mail: vls@eie.gr.
References
Author notes
M.A.D. and V.L.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal