The clinical and prognostic relevance of many recently identified driver gene mutations in adult acute myeloid leukemia (AML) is poorly defined. We sequenced the coding regions or hotspot areas of 68 recurrently mutated genes in a cohort of 664 patients aged 18 to 86 years treated on 2 phase 3 trials of the German AML Cooperative Group (AMLCG). The median number of 4 mutations per patient varied according to cytogenetic subgroup, age, and history of previous hematologic disorder or antineoplastic therapy. We found patterns of significantly comutated driver genes suggesting functional synergism. Conversely, we identified 8 virtually nonoverlapping patient subgroups, jointly comprising 78% of AML patients, that are defined by mutually exclusive genetic alterations. These subgroups, likely representing distinct underlying pathways of leukemogenesis, show widely divergent outcomes. Furthermore, we provide detailed information on associations between gene mutations, clinical patient characteristics, and therapeutic outcomes in this large cohort of uniformly treated AML patients. In multivariate analyses including a comprehensive set of molecular and clinical variables, we identified DNMT3A and RUNX1 mutations as important predictors of shorter overall survival (OS) in AML patients <60 years, and particularly in those with intermediate-risk cytogenetics. NPM1 mutations in the absence of FLT3-ITD, mutated TP53, and biallelic CEBPA mutations were identified as important molecular prognosticators of OS irrespective of patient age. In summary, our study provides a comprehensive overview of the spectrum, clinical associations, and prognostic relevance of recurrent driver gene mutations in a large cohort representing a broad spectrum and age range of intensively treated AML patients.
We present comprehensive information on genetic driver events in a uniformly treated cohort of 664 adult AML patients aged 18 to 86 years.
Mutations in NPM1, FLT3, CEBPA, TP53, and, in patients <60 years, DNMT3A and RUNX1, are the most important molecular risk factors in AML.
Introduction
Next-generation sequencing (NGS) and other high-throughput techniques led to the discovery of numerous recurrently mutated genes in acute myeloid leukemia (AML).1-3 The Cancer Genome Atlas (TCGA) consortium studied 200 AML patients by whole-genome or whole-exome sequencing (WGS/WES), and identified 23 genes as “significantly mutated” at a higher-than-expected frequency.4 Although the functional consequences of many novel recurrent genetic alterations are not yet fully understood, these mutations may become clinically useful molecular markers for diagnosis, prognostication, prediction of treatment response, or disease monitoring. Furthermore, defining the spectrum and patterns of driver gene mutations in AML patients is an important step to identify relevant targets for new treatment approaches. Simultaneously, the increasing number of genes known to be mutated at intermediate or low frequencies leads to a complex interplay of genetic changes that can only be analyzed in sufficiently large cohorts. Therefore, we set out to comprehensively characterize the spectrum and clinical relevance of recurrent driver gene mutations in a large, well-characterized, and homogenously treated group of AML patients.
Subjects and methods
Patients
We studied 664 previously untreated AML patients who received intensive induction chemotherapy on 2 randomized multicenter phase 3 trials of the German AML Cooperative Group (AMLCG) between 1999 and 2012 (AMLCG-1999, ClinicalTrials.gov identifier NCT00266136, n = 390; and AMLCG-2008, NCT01382147, n = 274).5-8 Treatment regimens are described in the supplemental material, available on the Blood Web site. Patients were included in this study if a suitable bone marrow (BM) or peripheral blood (PB) specimen was available. The diagnosis of AML was based on World Health Organization (WHO) criteria. Cytogenetic risk groups were defined according to the Medical Research Council (MRC) classification.9 All study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards of the participating centers. All patients provided written informed consent for inclusion on the clinical trial and genetic analyses.
Targeted sequencing
We studied 68 genes recurrently mutated in myeloid malignancies (supplemental Tables 1 and 2). Mononuclear cells were enriched from pretreatment BM or PB by Ficoll density gradient centrifugation. We sequenced the entire coding sequences of 37 genes and recurrently mutated regions4,10 in 31 genes, using a custom amplicon-based targeted enrichment assay (Haloplex, Agilent, Boeblingen, Germany) and an Illumina MiSeq instrument (Illumina, San Diego, CA). Criteria used to differentiate leukemia-associated driver mutations from germline variants are detailed in the supplemental Methods and in supplemental Figure 1. Mutations in NPM1 and CEBPA, FLT3 internal tandem duplications (FLT3-ITD), and KMT2A (formerly MLL) partial tandem duplications (KMT2A-PTD) were also tested using standard methods.11,12
Statistical analyses
We studied associations between different gene mutations and between gene mutations and other patient characteristics, using Fisher's exact test for categorical and the Wilcoxon rank-sum test for continuous variables, with adjustment for multiple testing using the Benjamini-Hochberg method. Analyses of treatment outcomes used commonly accepted definitions of complete remission (CR), relapse-free survival (RFS), and overall survival (OS) (supplemental Methods).13 For survival end points, P values were calculated from the Cox proportional hazards model stratified for the trial arm, using the Wald test. Multivariate models were used to integrate known clinical and genetic risk factors and potential confounders (supplemental Methods). To account for baseline differences (supplemental Table 3) and different recruitment periods of the 2 clinical trial cohorts, all outcome analyses were adjusted for trial and treatment arm. Statistical analyses were performed using R version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria). Mutation data were deposited in the Catalogue of Somatic Mutations in Cancer, study reference COSP41536 (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
Results
Spectrum of driver mutations in adult AML
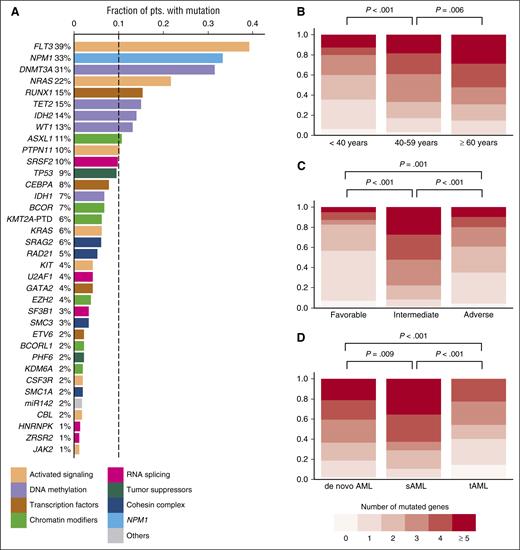
The median age of the 664 patients was 57 years (range, 18-86). Other baseline characteristics are listed in Table 1.14 The mean sequencing coverage of the target region across all samples was 430-fold, and, on average, 96% of the target region was covered at >30×. We identified a total of 2395 driver gene mutations in 59 different genes (supplemental Table 4A). Forty-eight genes were recurrently mutated, 35 genes were mutated in >1% of patients, and 10 genes were mutated in >10% (Figure 1A). Patients aged ≥60 years, representing 43% of the entire cohort, more commonly carried mutations in RUNX1, TET2, IDH2, SRSF2, TP53, BCOR, and SF3B1, and less often had mutations in WT1 compared with those <60 years (supplemental Figure 2).
Pretreatment patient characteristics
| Variable . | Total cohort, N = 664 . | Younger patients (<60 y), N = 376 . | Older patients (≥60 y), N = 288 . |
|---|---|---|---|
| Age (y) | |||
| Median | 57 | 45 | 67 |
| Range | 18-86 | 18-59 | 60-68 |
| Female gender, n (%) | 330 (50) | 203 (54) | 127 (44) |
| De novo AML | 570 (86) | 335 (89) | 235 (82) |
| Secondary AML | 59 (9) | 22 (6) | 37 (13) |
| Therapy-related AML | 35 (5) | 19 (5) | 16 (6) |
| ECOG performance status, n (%) | |||
| 0 | 172 (27) | 111 (30) | 61 (22) |
| 1 | 313 (48) | 175 (47) | 138 (49) |
| 2 | 125 (19) | 59 (16) | 66 (24) |
| 3 | 33 (5) | 20 (5) | 13 (5) |
| 4 | 6 (1) | 4 (1) | 2 (1) |
| WBC count, ×109/L | |||
| Median | 23.4 | 25.7 | 20.9 |
| Range | 0.1-666 | 0.1-486 | 0.5-666 |
| Hemoglobin, g/dL | |||
| Median | 9.0 | 9.0 | 9.0 |
| Range | 3.5-16.0 | 3.5-15.6 | 3.8-16 |
| Platelet count, ×109/L | |||
| Median | 53 | 52 | 55 |
| Range | 1-1760 | 1-1760 | 1-471 |
| Bone marrow blasts, % | |||
| Median | 80 | 80 | 80 |
| Range | 6-100 | 6-100 | 9-100 |
| FAB category, n | |||
| M0 | 35 | 16 | 19 |
| M1 | 157 | 80 | 77 |
| M2 | 178 | 93 | 85 |
| M4 | 163 | 107 | 56 |
| M5 | 83 | 51 | 32 |
| M6 | 19 | 11 | 8 |
| M7 | 3 | 3 | 0 |
| MRC cytogenetic risk category | |||
| Favorable | 65 (10) | 50 (14) | 15 (5) |
| Intermediate | 452 (70) | 254 (69) | 198 (71) |
| Adverse | 129 (20) | 65 (18) | 64 (23) |
| Modified ELN classification* | |||
| Favorable | 189 (29) | 128 (35) | 61 (22) |
| Intermediate-I | 205 (32) | 107 (29) | 98 (35) |
| Intermediate-II | 119 (18) | 62 (17) | 57 (21) |
| Adverse | 133 (21) | 72 (20) | 61 (22) |
| NPM1-mutated, n (%) | 221 (33) | 129 (34) | 92 (32) |
| FLT3-ITD present, n (%) | 197 (30) | 119 (32) | 78 (27) |
| CEBPA, n (%) | |||
| Single mutation | 25 (4) | 13 (3) | 12 (4) |
| Double mutation | 27 (4) | 22 (6) | 5 (2) |
| DNMT3A-mutated, n (%) | 209 (31) | 108 (29) | 101 (35) |
| RUNX1-mutated, no. (%) | 102 (15) | 35 (9) | 67 (23) |
| TP53 mutated, n (%) | 63 (9) | 23 (6) | 40 (14) |
| Variable . | Total cohort, N = 664 . | Younger patients (<60 y), N = 376 . | Older patients (≥60 y), N = 288 . |
|---|---|---|---|
| Age (y) | |||
| Median | 57 | 45 | 67 |
| Range | 18-86 | 18-59 | 60-68 |
| Female gender, n (%) | 330 (50) | 203 (54) | 127 (44) |
| De novo AML | 570 (86) | 335 (89) | 235 (82) |
| Secondary AML | 59 (9) | 22 (6) | 37 (13) |
| Therapy-related AML | 35 (5) | 19 (5) | 16 (6) |
| ECOG performance status, n (%) | |||
| 0 | 172 (27) | 111 (30) | 61 (22) |
| 1 | 313 (48) | 175 (47) | 138 (49) |
| 2 | 125 (19) | 59 (16) | 66 (24) |
| 3 | 33 (5) | 20 (5) | 13 (5) |
| 4 | 6 (1) | 4 (1) | 2 (1) |
| WBC count, ×109/L | |||
| Median | 23.4 | 25.7 | 20.9 |
| Range | 0.1-666 | 0.1-486 | 0.5-666 |
| Hemoglobin, g/dL | |||
| Median | 9.0 | 9.0 | 9.0 |
| Range | 3.5-16.0 | 3.5-15.6 | 3.8-16 |
| Platelet count, ×109/L | |||
| Median | 53 | 52 | 55 |
| Range | 1-1760 | 1-1760 | 1-471 |
| Bone marrow blasts, % | |||
| Median | 80 | 80 | 80 |
| Range | 6-100 | 6-100 | 9-100 |
| FAB category, n | |||
| M0 | 35 | 16 | 19 |
| M1 | 157 | 80 | 77 |
| M2 | 178 | 93 | 85 |
| M4 | 163 | 107 | 56 |
| M5 | 83 | 51 | 32 |
| M6 | 19 | 11 | 8 |
| M7 | 3 | 3 | 0 |
| MRC cytogenetic risk category | |||
| Favorable | 65 (10) | 50 (14) | 15 (5) |
| Intermediate | 452 (70) | 254 (69) | 198 (71) |
| Adverse | 129 (20) | 65 (18) | 64 (23) |
| Modified ELN classification* | |||
| Favorable | 189 (29) | 128 (35) | 61 (22) |
| Intermediate-I | 205 (32) | 107 (29) | 98 (35) |
| Intermediate-II | 119 (18) | 62 (17) | 57 (21) |
| Adverse | 133 (21) | 72 (20) | 61 (22) |
| NPM1-mutated, n (%) | 221 (33) | 129 (34) | 92 (32) |
| FLT3-ITD present, n (%) | 197 (30) | 119 (32) | 78 (27) |
| CEBPA, n (%) | |||
| Single mutation | 25 (4) | 13 (3) | 12 (4) |
| Double mutation | 27 (4) | 22 (6) | 5 (2) |
| DNMT3A-mutated, n (%) | 209 (31) | 108 (29) | 101 (35) |
| RUNX1-mutated, no. (%) | 102 (15) | 35 (9) | 67 (23) |
| TP53 mutated, n (%) | 63 (9) | 23 (6) | 40 (14) |
ECOG, Eastern Cooperative Oncology Group; ELN, European LeukemiaNet; FAB, French-American-British classification; ITD, internal tandem duplication; MRC, British Medical Research Council.
Modified ELN classification designates the ELN reporting system for genetic alterations14; however only patients with double, and not those with single, CEBPA mutations were classified as “favorable.”
Overview of driver gene mutations identified by targeted sequencing in 664 AML patients. (A) Histogram showing the frequency of driver gene mutations detected in >1% of patients. Bars are colored according to the functional category assigned to each driver gene. (B) Number of mutated driver genes per patient, according to age category (<40 years, n = 112; 40-59 years, n = 264; ≥60 years, n = 288). (C) Number of mutated driver genes per patient, according to MRC cytogenetic risk category (favorable, n = 65; intermediate, n = 452; adverse, n = 129). (D) Number of mutated driver genes per patient for patients with de novo AML (n = 570) compared with sAML (n = 59) or tAML (n = 35). P values in (B-D) were calculated by the Kruskal-Wallis test, without adjustment for multiple testing.
Overview of driver gene mutations identified by targeted sequencing in 664 AML patients. (A) Histogram showing the frequency of driver gene mutations detected in >1% of patients. Bars are colored according to the functional category assigned to each driver gene. (B) Number of mutated driver genes per patient, according to age category (<40 years, n = 112; 40-59 years, n = 264; ≥60 years, n = 288). (C) Number of mutated driver genes per patient, according to MRC cytogenetic risk category (favorable, n = 65; intermediate, n = 452; adverse, n = 129). (D) Number of mutated driver genes per patient for patients with de novo AML (n = 570) compared with sAML (n = 59) or tAML (n = 35). P values in (B-D) were calculated by the Kruskal-Wallis test, without adjustment for multiple testing.
At least 1 driver mutation was identified in 97% of patients (646/664). Of the remaining 18 patients, 15 had a recurrent balanced chromosomal translocation, resulting in a known fusion transcript. Thus, ≥1 molecularly defined driver lesion was detected in 99.5% of patients. The median number of mutations per patient was 4 (range, 0-10; supplemental Figure 3), and the median number of mutated genes was 3 (range, 0-9). The number of mutated driver genes increased with age (Figure 1B; P < .001 for association with age as a continuous variable). Patients with MRC intermediate-risk cytogenetics had a higher number of mutated driver genes (median, 4) than in those with favorable or adverse cytogenetics (median, 1 and 2 mutated genes, respectively) (Figure 1C). Finally, patients with secondary AML after an antecedent hematologic disorder (sAML) had more, and patients with treatment-related AML (tAML) had fewer mutations in genes covered by our panel, compared with those with de novo AML (Figure 1D).
Differences in variant allele frequencies between driver genes
The median variant allele frequency (VAF) of driver gene mutations was 0.36, significantly lower than the VAFs of heterozygous germline polymorphisms (median, 0.50; supplemental Figure 4). Figure 2 illustrates the spectrum of copy number–adjusted VAFs observed for genes mutated in ≥20 patients. Mutations in some genes including TP53, IDH2, DNMT3A, CEBPA, TET2, and NPM1 and balanced chromosomal rearrangements involving KMT2A or the core-binding-factor (CBF) genes had a median VAF slightly below 0.5, compatible with heterozygous mutations present in most cells in the specimen. In contrast, mutations in other genes showed considerably lower allele frequencies, indicating they were often present only in a subpopulation of the sequenced cells. Of note, 5 of the 6 genes with the lowest median VAFs (FLT3, KIT, NRAS, KRAS, and PTPN11) are involved in growth-factor signaling, suggesting that alterations in these pathways are often acquired relatively late during the evolution of the leukemic clone.
Analysis of variant allele frequencies (VAFs) of different driver gene mutations. The boxplot shows the median, 25th, and 75th percentiles, and minimum and maximum VAF observed across the entire cohort of 664 patients. VAFs were adjusted for local copy number. Boxes are colored according to the functional category assigned to each driver gene. Only mutations found in ≥20 patients were included. The black dashed line marks an allele frequency of 0.5, the expected VAF for a heterozygous variant present in all cells in the specimen. Insertions and deletions of ≥15 base pairs were excluded, because VAFs for larger variants were skewed, possibly because of amplification bias. VAFs for balanced translocations and inversions (ie, t(8;21), inv(16) and balanced translocations involving the KMT2A locus) were determined by interphase fluorescence in situ hybridization on ≥200 nuclei.
Analysis of variant allele frequencies (VAFs) of different driver gene mutations. The boxplot shows the median, 25th, and 75th percentiles, and minimum and maximum VAF observed across the entire cohort of 664 patients. VAFs were adjusted for local copy number. Boxes are colored according to the functional category assigned to each driver gene. Only mutations found in ≥20 patients were included. The black dashed line marks an allele frequency of 0.5, the expected VAF for a heterozygous variant present in all cells in the specimen. Insertions and deletions of ≥15 base pairs were excluded, because VAFs for larger variants were skewed, possibly because of amplification bias. VAFs for balanced translocations and inversions (ie, t(8;21), inv(16) and balanced translocations involving the KMT2A locus) were determined by interphase fluorescence in situ hybridization on ≥200 nuclei.
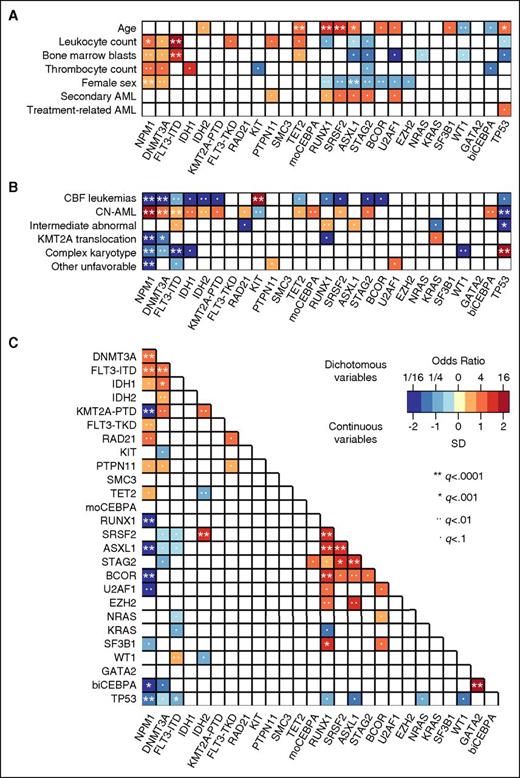
Association of driver gene mutations with clinical characteristics and cytogenetics
We studied associations of driver gene mutations occurring in ≥20 patients with baseline clinical characteristics, cytogenetic alterations, and other gene mutations (Figure 3 and supplemental Table 5A-C). NPM1 and DNMT3A mutations were more common, and mutations in RUNX1, SRSF2, ASXL1, STAG2, and BCOR less common in women compared with men (multiple testing–adjusted q < .01; Figure 3A). FLT3-ITD and NPM1 mutations were associated with higher leukocyte counts and higher BM blast percentages. Mutations in RUNX1, SRSF2, ASXL1, STAG2, U2AF1, and PTPN11 tended to be more frequent in sAML compared with de novo AML, whereas TP53 mutations were more common in patients with tAML. However, none of these associations between individual gene mutations and sAML/tAML was significant at q < .01, and no single mutation was highly specific for sAML or tAML in our cohort.
Associations between driver gene mutations, cytogenetics, and patient characteristics. Associations were studied for driver gene mutations found in ≥20 patients, and statistical significance was assessed using the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables, with adjustment for multiple testing using the Benjamini-Hochberg method. Only those pairings that were significant at an adjusted q < .1 are shown. The odds ratio of the association is color coded, and the significance level is indicated by the symbol in each field. Orange and red colors indicate a positive association (ie, 2 characteristics that frequently occurred together in the same patient, or for the association between a mutation and a continuous variable, a higher value in those carrying the mutation). Blue colors indicate a negative association (ie, 2 characteristics that rarely occurred together in the same patient, or for the association between a mutation and a continuous variable, a lower value in those carrying the mutation). (A) Pairwise associations between gene mutations and clinical patient characteristics. (B) Pairwise associations between gene mutations and cytogenetic findings. (C) Pairwise associations among gene mutations.
Associations between driver gene mutations, cytogenetics, and patient characteristics. Associations were studied for driver gene mutations found in ≥20 patients, and statistical significance was assessed using the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables, with adjustment for multiple testing using the Benjamini-Hochberg method. Only those pairings that were significant at an adjusted q < .1 are shown. The odds ratio of the association is color coded, and the significance level is indicated by the symbol in each field. Orange and red colors indicate a positive association (ie, 2 characteristics that frequently occurred together in the same patient, or for the association between a mutation and a continuous variable, a higher value in those carrying the mutation). Blue colors indicate a negative association (ie, 2 characteristics that rarely occurred together in the same patient, or for the association between a mutation and a continuous variable, a lower value in those carrying the mutation). (A) Pairwise associations between gene mutations and clinical patient characteristics. (B) Pairwise associations between gene mutations and cytogenetic findings. (C) Pairwise associations among gene mutations.
Mutations in NPM1, DNMT3A, FLT3-ITD, IDH1, IDH2, and single (monoallelic) or double (biallelic) CEBPA mutations mostly occurred in patients with normal cytogenetics (Figure 3B). KIT mutations strongly associated with CBF translocations, whereas many other gene mutations were rare or absent in this cytogenetic subset. TP53 mutations were closely linked to complex karyotypes, and KRAS mutations associated with KMT2A (MLL) translocations. Finally, RUNX1 mutations were enriched in patients with MRC intermediate-risk abnormal karyotypes, a finding partly driven by the known association between trisomy 13 and RUNX1 mutations.15
Patterns of co-occurrence and mutual exclusivity between driver gene mutations
When we investigated co-occurrence and mutual exclusivity of driver gene mutations, we found significant (q < .01) associations between mutated NPM1 and mutations in DNMT3A, FLT3, and RAD21, and between mutated DNMT3A and FLT3-ITD, IDH1, IDH2, and KMT2A-PTD mutations (Figure 3C and supplemental Table 5C). RUNX1, SRSF2, ASXL1, STAG2, and BCOR formed a separate cluster of genes that were frequently comutated. Strong pairwise associations were also observed between IDH2 and SRSF2, RUNX1 and SF3B1, and double CEBPA and GATA2 mutations.
Conversely, certain genetic alterations rarely coexisted in the same patient. We used mutually exclusive gene set analysis (MEGSA)16 to identify subsets of patients defined by mutually exclusive genetic alterations (supplemental Methods and Results). This analysis revealed 8 distinct and almost nonoverlapping subgroups, namely patients with CBF translocations (RUNX1-RUNX1T1 or CBFB-MYH11), GATA2-MECOM or DEK-NUP214 rearrangements, balanced rearrangements involving KMT2A, double CEBPA mutations, or mutations in TP53, NPM1, or RUNX1 (Figure 4A). Of the 664 patients, 503 (76%) had exactly one of these lesions. Only 16 patients (2.4%) carried more than one of these changes, and 145 patients (22%) did not harbor any of the aforementioned genetic alterations and thus could not be assigned to one of the clusters. The 8 genetically defined subgroups also showed widely divergent outcomes (Figure 4B-C)
Subgroups of AML patients defined by mutually exclusive genetic alterations. (A) Heatmap depicting gene mutations and balanced chromosomal translocations in 664 adult AML patients. Mutually exclusive gene signature analysis (MEGSA) identified 8 patient clusters (color coded at the top of the figure) based on almost mutually exclusive genetic alterations. The CBF rearrangements (RUNX1-RUNX1T1 and CBF-MYH11) were combined into 1 cluster in this analysis. The 9 resulting patient subgroups (8 mutually exclusive clusters and 1 subgroup comprising the remaining patients) differ with regard to co-occurring gene mutations and clinical characteristics. (B) RFS and (C) OS of the patient subgroups defined by mutually exclusive genetic alterations. Subgroups are color coded according to legend at the top of the figure. Patients with DEK-NUP214 were not included in both analyses, and patients with GATA2-MECOM were not included in the analysis of RFS, owing to small patient numbers.
Subgroups of AML patients defined by mutually exclusive genetic alterations. (A) Heatmap depicting gene mutations and balanced chromosomal translocations in 664 adult AML patients. Mutually exclusive gene signature analysis (MEGSA) identified 8 patient clusters (color coded at the top of the figure) based on almost mutually exclusive genetic alterations. The CBF rearrangements (RUNX1-RUNX1T1 and CBF-MYH11) were combined into 1 cluster in this analysis. The 9 resulting patient subgroups (8 mutually exclusive clusters and 1 subgroup comprising the remaining patients) differ with regard to co-occurring gene mutations and clinical characteristics. (B) RFS and (C) OS of the patient subgroups defined by mutually exclusive genetic alterations. Subgroups are color coded according to legend at the top of the figure. Patients with DEK-NUP214 were not included in both analyses, and patients with GATA2-MECOM were not included in the analysis of RFS, owing to small patient numbers.
Driver mutations and survival
We studied the associations of driver mutations found in ≥20 patients (≥3%) with treatment outcomes. The median OS of our patient cohort was 17.8 months, and the median follow-up for survivors was 58 months. Univariate analyses of gene mutations and OS are reported in supplemental Table 6. Patients with mutated NPM1, particularly in the absence of FLT3-ITD, and those with double CEBPA mutations had relatively favorable OS, whereas survival was significantly shorter for patients carrying mutations in DNMT3A, RUNX1, ASXL1, SRSF2, TP53, BCOR, U2AF1, or SF3B1 (supplemental Figure 5). However, these analyses may be biased because some gene mutations tended to co-occur, or showed associations with other prognostic markers including age and cytogenetics.
To account for the complex interrelations between gene mutations and other known risk factors, we constructed multivariate models. Because the frequency of several gene mutations differed between younger (<60 years) and older (≥60 years) patients, and treatment intensity was higher in younger patients, we also tested for interactions between each individual gene mutation and age. In a multivariate model for OS (Table 2), age <60 years, favorable cytogenetics, mutated NPM1 in the absence of FLT3-ITD, and, in trend, double CEBPA mutations associated with longer OS. The prognostic impact of DNMT3A and RUNX1 mutations differed significantly between younger and older patients. Both mutations were associated with inferior OS only among younger, but not among older patients. Therapy-related AML, decreased performance status, higher leukocyte counts, adverse cytogenetic changes, and TP53 mutations all were associated with shorter OS regardless of age group.
Multivariate analysis for overall survival
 |
 |
95% CI, 95% confidence interval; ECOG, Eastern Cooperative Oncology Group; MRC, Medical Research Council; ITD, internal tandem duplication.
629 patients were included in the multivariate model, whereas 35 patients (5%) were excluded because of missing data. Gene mutations were included in the multivariate model if they were detected in at least 20 patients and had a univariate P value for OS, not adjusted for multiple comparisons, of ≤.10. The model was stratified for trial arm. No variable selection or elimination technique was applied, and all variables were retained in the final model. Tests for interaction with age group (<60 y vs ≥60 y) were performed for each variable.
*The P value for the interaction between DNMT3A mutation status and age group was .018.
†The P value for the interaction between RUNX1 mutation status and age group was .015.
Univariate analyses of associations between gene mutations and achievement of CR and RFS are reported in supplemental Tables 7 and 8, and multivariate analyses of factors related to achievement of CR, RFS, and RFS censored at the time of allogeneic transplantation are shown in supplemental Tables 9-11. Similar to the OS model, the multivariate model for CR showed interactions between age group and DNMT3A and RUNX1 mutations, which were associated with lower chances of reaching a remission only among younger patients (supplemental Results). Mutations in both genes were also linked to shorter RFS, and no interactions with age group were observed for this end point.
Prognostic impact of gene mutations in defined age groups and cytogenetic subsets
Mutations in certain genes were closely associated with defined, prognostically distinct cytogenetic subgroups. Furthermore, multivariate analyses revealed that the impact of some mutations depends on patient age. To facilitate interpretation of these results and visualize the combined effects of co-occurring gene mutations on survival, we analyzed patient subsets stratified according to age and cytogenetics.
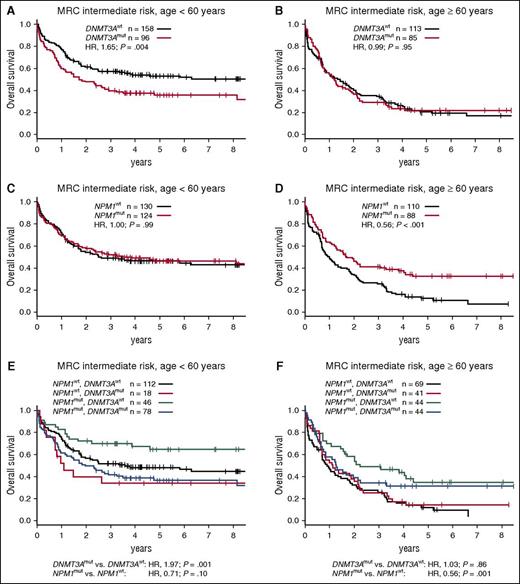
The 3 most common driver mutations in our cohort were strongly enriched in patients with intermediate-risk karyotypes. NPM1, DNMT3A, and FLT3-ITD mutations occurred in 47% (212/452), 40% (181/452), and 39% (175/452) of MRC intermediate-risk patients, respectively. Conversely, 97% of NPM1-mutated patients, 88% of DNMT3A-mutated patients, and 92% of FLT3-ITD–positive patients had intermediate-risk cytogenetics. Therefore, we examined the interplay of these mutations with regard to OS in the MRC intermediate-risk patient subset. DNMT3A and NPM1 mutations were closely associated in this subgroup (73% of DNMT3A-mutated patients also had mutated NPM1), yet their prognostic relevance was discordant. DNMT3A mutations associated with inferior OS among patients <60 years, but not among older patients (Figure 5A-B). Of note, no survival differences were observed between DNMT3A codon R882 and other DNMT3A mutations (data not shown).17 On the other hand, NPM1 mutations associated with favorable OS only among older patients (Figure 5C-D). When both mutations were analyzed jointly, DNMT3A mutations remained an unfavorable prognostic factor in younger patients, and NPM1 mutations associated with favorable OS among older patients (Figure 5E-F). Although there was no statistically significant interaction between the 2 mutations, patients with mutated NPM1 and wild-type DNMT3A had the most favorable outcomes in both age groups.
Survival of patients with MRC intermediate-risk cytogenetics according to age group and DNMT3A and NPM1 mutation status. (A) OS for MRC intermediate-risk patients aged <60 years or (B) aged ≥60 years, stratified according to DNMT3A mutation status. (C) OS for MRC intermediate-risk patients aged <60 years or (D) aged ≥60 years, stratified according to NPM1 mutation status. (E) OS for MRC intermediate-risk patients aged <60 years or (F) aged ≥60 years, stratified according to DNMT3A and NPM1 mutation status. P values in (A-D) were calculated from univariate Cox proportional hazards models, and in (E) and (F) from bivariate models, using the Wald test. All models were stratified for treatment arm. mut, mutated; wt, wild-type.
Survival of patients with MRC intermediate-risk cytogenetics according to age group and DNMT3A and NPM1 mutation status. (A) OS for MRC intermediate-risk patients aged <60 years or (B) aged ≥60 years, stratified according to DNMT3A mutation status. (C) OS for MRC intermediate-risk patients aged <60 years or (D) aged ≥60 years, stratified according to NPM1 mutation status. (E) OS for MRC intermediate-risk patients aged <60 years or (F) aged ≥60 years, stratified according to DNMT3A and NPM1 mutation status. P values in (A-D) were calculated from univariate Cox proportional hazards models, and in (E) and (F) from bivariate models, using the Wald test. All models were stratified for treatment arm. mut, mutated; wt, wild-type.
Patients with mutated NPM1 in the absence FLT3-ITD reportedly have favorable outcomes relative to all remaining patients (ie, those with wild-type NPM1 and/or FLT3-ITD), a finding that was confirmed in our cohort (supplemental Figure 6). When DNMT3A mutations were studied in conjunction with the combined NPM1/FLT3-ITD genotype, they again associated with significantly shorter OS among younger, but not among older, MRC intermediate-risk patients (Figure 6A-B).
Survival of patients with MRC intermediate-risk cytogenetics according to the combined NPM1 and FLT3-ITD mutations status and DNMT3A and RUNX1 mutations. (A) OS of MRC intermediate-risk patients aged <60 years or (B) aged ≥60 years, stratified according to the combined NPM1/FLT3-ITD genotype and DNMT3A mutations status. (C) OS of MRC intermediate-risk patients aged <60 years or (D) aged ≥60 years, comparing patients with mutated RUNX1 to RUNX1–wild-type patients with mutated NPM1 without FLT3-ITD (this prognostically favorable genotype was mutually exclusive with mutated RUNX1, and is thus shown separately), and all remaining patients (ie, wild-type NPM1 and/or FLT3-ITD and wild-type RUNX1). P values were calculated from bivariate Cox proportional hazards models stratified for trial arm, using the Wald test. mut, mutated; neg, negative; wt, wild-type.
Survival of patients with MRC intermediate-risk cytogenetics according to the combined NPM1 and FLT3-ITD mutations status and DNMT3A and RUNX1 mutations. (A) OS of MRC intermediate-risk patients aged <60 years or (B) aged ≥60 years, stratified according to the combined NPM1/FLT3-ITD genotype and DNMT3A mutations status. (C) OS of MRC intermediate-risk patients aged <60 years or (D) aged ≥60 years, comparing patients with mutated RUNX1 to RUNX1–wild-type patients with mutated NPM1 without FLT3-ITD (this prognostically favorable genotype was mutually exclusive with mutated RUNX1, and is thus shown separately), and all remaining patients (ie, wild-type NPM1 and/or FLT3-ITD and wild-type RUNX1). P values were calculated from bivariate Cox proportional hazards models stratified for trial arm, using the Wald test. mut, mutated; neg, negative; wt, wild-type.
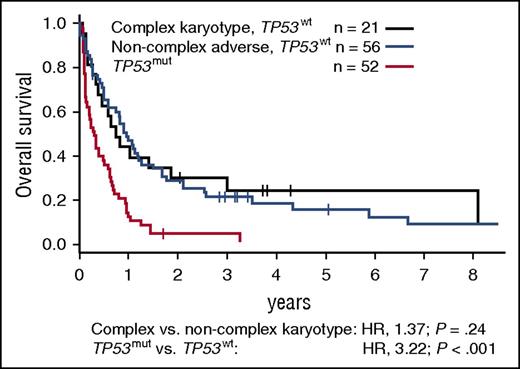
RUNX1 mutations most commonly occurred in patients with intermediate-risk or noncomplex adverse cytogenetics (prevalence, 17% and 22%, respectively). In MRC intermediate-risk patients, they were mutually exclusive with NPM1 mutations and, consequently, with the favorable NPM1-mutated, FLT3-ITD–negative genotype. RUNX1 mutations associated with shorter OS among younger, and in trend also among older, intermediate-risk patients (Figure 6C-D). supplemental Figure 7 illustrates the diversity of outcomes observed among patients with intermediate-risk cytogenetics, depending on age and NPM1, FLT3-ITD, DNMT3A, and RUNX1 mutations status. In patients with noncomplex adverse cytogenetics, RUNX1 mutation status did not affect outcomes (not shown). KIT mutations occurred in 26% (17/65) of patients with CBF rearrangements and did not associate with CR rate, RFS, or OS. Finally, TP53 mutations were present in 45% (52/116) of patients with adverse cytogenetics, including 70% (50/71) of those with complex karyotypes, but in only 1.5% (8/539) of patients with nonadverse karyotypes. TP53-mutated patients had significantly shorter OS compared with patients with complex karyotypes or noncomplex adverse cytogenetics without TP53 mutations (Figure 7).
Survival of patients with MRC adverse-risk cytogenetics according to TP53 mutation status. OS of MRC adverse-risk patients with mutated TP53, compared with patients with wild-type TP53 and complex or noncomplex adverse cytogenetics. A complex karyotype was defined as ≥4 unrelated abnormalities per the MRC classification. P values were calculated from a bivariate Cox proportional hazards model stratified for trial arm, using the Wald test. mut, mutated; wt, wild-type.
Survival of patients with MRC adverse-risk cytogenetics according to TP53 mutation status. OS of MRC adverse-risk patients with mutated TP53, compared with patients with wild-type TP53 and complex or noncomplex adverse cytogenetics. A complex karyotype was defined as ≥4 unrelated abnormalities per the MRC classification. P values were calculated from a bivariate Cox proportional hazards model stratified for trial arm, using the Wald test. mut, mutated; wt, wild-type.
Discussion
Our study presents comprehensive information on the incidence, patterns of comutation, clinical correlations, and prognostic relevance of mutations in 68 recurrently mutated genes in adult AML. To our knowledge, together with another recently published study,18 our work represents one of the largest patient cohorts characterized for an extensive set of known driver mutations, including the 23 “significantly mutated” genes identified by the TCGA consortium.4 Our study included intensively treated patients across a broad age range, including patients aged ≥60 years who are fit for intensive therapy, a clinically important population not included or underrepresented in other cohorts.18-20 Further strengths of our study include the fact that all patients received comparable treatment on 2 consecutive phase 3 trials, and the long duration of follow-up.
At least one genetic driver event was identified in >99% of patients when mutations and balanced translocations were combined. As reported in previous studies, patients with favorable or unfavorable abnormal cytogenetics, and particularly those with balanced translocations, had a lower number of additional gene mutations compared with those with cytogenetically normal AML or intermediate-risk abnormalities.19 Fusion genes seem to be strong oncogenic drivers requiring relatively few cooperating events. Patients with sAML had a larger number of mutated genes compared to de novo disease, possibly reflecting the stepwise gain of mutations during the evolution of myelodysplastic syndromes (MDS) or myeloproliferative neoplasms to AML.21,22 The number of driver gene mutations increased with age, even after adjustment for the higher proportion of sAML and the lower incidence of CBF translocations in older patients (data not shown). Older patients also were more likely to carry alterations in specific genes including TET2, RUNX1, and ASXL1, as previously reported,23-28 and SRSF2, genes that have recently been implicated in age-related clonal hematopoiesis.29 These data contribute to our understanding of differences in AML biology between younger and older patients. Coleman Lindsley and colleagues recently reported that mutations in 8 different genes are highly specific for sAML, based on a comparison of rigorously selected sAML and de novo AML patients.22 In our cohort, we only identified a modest association between clinically reported sAML and mutations in 4 of these genes, namely SRSF2, ASXL1, STAG2, and U2AF1. Of note, Coleman-Lindsley and coworkers also found secondary-type mutations in one third of patients in an unselected older cohort with clinically defined de novo AML. Emerging data suggest that age-related clonal hematopoiesis, clonal cytopenias of undetermined significance, and MDS form a spectrum of related conditions.30 Thus, some of the “de novo” AML patients in our report and in the unselected patient cohort studied by Coleman Lindsley may in fact have had preexisting clonal hematopoiesis or undiagnosed MDS. It remains to be determined whether a genetic definition of “secondary-type” AML is more relevant with regard to outcomes than an sAML diagnosis based on clinical history.
Our findings regarding the mutational landscape of adult AML are largely consistent with the TCGA consortium's WES and WGS data, with FLT3, NPM1, and DNMT3A identified as the most commonly mutated genes.4 We noted a higher prevalence of NRAS and KRAS mutations (26%) compared with the TCGA cohort (12%), a discrepancy potentially explained by the often low VAF of these mutations and the deeper sequencing coverage achieved by our targeted sequencing approach. Overall, 21% of driver mutations in our study had VAFs <10%, and such variants would likely be missed by conventional Sanger sequencing and standard WES/WGS approaches. A targeted NGS study from the Japan Acute Leukemia Study Group on 198 patients aged ≤65 years found only 6 genes (FLT3, NPM1, DNMT3A, CEBPA, KIT, and NRAS) mutated in >10%.19 The low frequency of RUNX1 and TET2 and the relatively high frequency of CEBPA and KIT mutations in the Japanese cohort may be a result of its younger age range. Mutations in NPM1 (19.2%) and DNMT3A (16%) also were found more rarely compared with our study and other cohorts from Europe and the US,4,20,31 suggesting that ethnicity may affect the mutation spectrum in AML.
NGS techniques allow estimating the proportion of cells in a specimen carrying a somatic mutation based on its allele frequency. We observed that mutations in some genes, including the epigenetic modifiers DNMT3A, TET2, and IDH2, frequently had VAFs close to 50%. This indicates they were usually present in a large majority of cells, consistent with the concept that these mutations constitute early, and potentially initiating, events during leukemogenesis.32,33 On the other hand, activating mutations in growth factor signaling pathways (eg, FLT3, KIT, NRAS, and KRAS mutations) commonly had lower VAFs. These changes apparently occur relatively late during the evolution of the malignant clone, a conclusion supported by analyses of paired diagnosis and relapse samples showing that FLT3 and RAS mutations are frequently lost during disease progression.34 We used VAFs corrected for copy number alterations to obtain a measure of the fraction of cells carrying each variant. Our assay did not allow us to detect possible copy-neutral loss of heterozygosity, yet this seems to be a relatively rare phenomenon in AML.35 Although the number of somatic variants identified through targeted sequencing is not sufficient to infer the precise clonal architecture in individual patients, our findings have implications for the development of targeted therapies aimed at eradicating the malignant “founder clone.”
Our study confirms previously reported associations of co-occurring driver mutations (eg, between mutations in DNMT3A, NPM1 and FLT3,4 between double CEBPA and GATA2 mutations,36 and between RUNX1 and ASXL1 mutations).24 Contrary to a recently published study suggesting a link between CSF3R p.T618I and biallelic CEBPA mutations, we found only one CSF3R mutation among 27 CEBPA double-mutated patients.37 We also identified associations that, to our knowledge, have not been described in AML before, including frequent comutation of SRSF2 with RUNX1 and IDH2, as previously reported in MDS.38,39 Frequent co-occurrence of mutations implies functional synergism, and our findings may inform further studies aiming to understand the underlying functional interactions.
Conversely, we also identified mutations that rarely coexist in the same patient, suggesting these alterations might have antagonistic or synthetic lethal effects. In particular, we identified a set of 8 genetic alterations that collectively occur in >75% of AML patients but show almost complete mutual exclusivity. The nonoverlapping, clinically distinct subgroups defined by these alterations likely correspond to different, and apparently incompatible, underlying pathways of leukemogenesis. Our data also support the inclusion of AML with RUNX1 or biallelic CEBPA mutations as separate entities in the upcoming revision of the WHO classification.40
Our large and uniformly treated cohort allowed us to comprehensively study the prognostic relevance of recurrent gene mutations. In multivariate analyses including molecular markers, cytogenetics and other potential risk factors and also testing for interactions with patient age, we found that NPM1 mutations in the absence of FLT3-ITD and, in trend, double CEBPA mutations associated with favorable OS, and patients with mutated TP53 had shorter OS, regardless of age. DNMT3A and RUNX1 mutations were linked to shorter OS only in patients <60 years, a finding that can at least partially be explained by the lower odds of reaching a CR for younger patients with these mutations.
Our results extend earlier studies that focused on individual markers or smaller gene panels, and were often limited to patients aged <60 years.19,20,24,25,28,31,41-44 We analyzed a comprehensive panel of known driver genes in patients across a broad age range and encompassing all cytogenetic subsets except acute promyelocytic leukemia. Therefore, our results provide strong evidence that TP53 and, in younger patients, DNMT3A and RUNX1, gene mutations are among the most relevant molecular risk factors in intensively treated adult AML. We confirm the importance of NPM1, FLT3-ITD, and CEBPA mutations that are already recognized in international guidelines.45 Despite the prognostic information provided by molecular markers, cytogenetics remained an important risk factor in multivariate analyses, and gene mutations were often linked to specific cytogenetic subsets. Subgroup analyses in patients stratified according to age group and cytogenetics illustrate that DNMT3A and RUNX1 mutations can refine risk stratification, particularly for younger patients with intermediate-risk karyotypes. Within the adverse cytogenetic risk group, TP53 mutations define a subset of patients with particularly dismal outcomes.
Recently, two studies suggested that a higher number of driver gene mutations may be an adverse prognostic factor in MDS and AML.39,45 Complex molecular genetic abnormalities, defined as ≥3 mutations in a 28-gene panel, were associated with shorter OS in a Japanese de novo AML cohort.45 We found that patients with >5 mutated genes had inferior survival in univariate analyses (supplemental Results), but not in multivariate models accounting for individual gene mutations and cytogenetics. Thus, the prognostic impact of specific driver gene mutations apparently outweighs the impact of total mutation number.
Integrating the prognostic information provided by gene mutations, cytogenetics, and other markers into clinically applicable risk stratification algorithms is an important yet challenging task. Several groups have proposed novel risk stratification schemes for AML based on gene mutations alone or in combination with other risk factors.19,20,44 Compared with our work, these studies focused on smaller sets of commonly mutated genes. Our cohort size was not sufficient to establish and independently validate another new molecular risk classification encompassing not only high- but also intermediate-frequency markers. Integration of the accumulating information on genetic risk factors in AML into clinically applicable algorithms should occur through collaboration of multiple study groups to increase sample size and validity.
One limitation of our study is that we focused on a predefined set of recurrently mutated genes, and not the entire coding sequence was analyzed for genes with well-defined “hotspots.” However, our assay covered 99% of the mutations in the 68 target genes that were found in the TCGA cohort. WES or WGS remain the preferred approaches for comprehensive mutation analyses. However, genome-wide somatic variant calling requires matched germline DNA, and suitable nonleukemic tissue was not available for our patients.
In summary, our study offers a detailed survey of recurrent driver gene mutations in one of the largest, well-characterized, and uniformly treated adult AML cohorts analyzed so far. We provide detailed insights into patterns of gene mutations, their clinical correlations, and their prognostic relevance. We hope our work will serve as an important reference for future research on AML pathogenesis, and as a starting point for a new, genetically-based disease classification that will contribute to improved risk stratification and, ultimately, better treatment results.
Acknowledgments
The authors thank Thorsten Haferlach, of the Munich Leukemia Laboratory (MLL) for providing patient samples. They are grateful to the patients who volunteered to participate in AMLCG clinical trials, and they acknowledge the contributions of physicians and other health care professionals at all participating centers.
This study was supported by a Clinical Research Fellowship from the European Hematology Association (EHA) (K.H.M.); and by grant support from Deutsche Forschungsgemeinschaft (DFG SFB 1243, TP A06 [K.H.M.]); Wilhelm Sander Stiftung (no. 2013.086.1 [T.H. and K.S.]); the German Cancer Consortium (Deutsches Konsortium für Translationale Krebsforschung, Heidelberg, Germany); and the Leukaemia & Blood Cancer New Zealand and the family of Marijanna Kumerich (S.K.B.). The AMLCG clinical trials were supported by Deutsche Krebshilfe (grants 70-2839-Bü4 and 108584).
Authorship
Contribution: K.H.M., T.H., and K.S. designed the study; K.H.M., T.H., and M.R.-T. analyzed sequencing data and performed statistical analyses; S.A., M.C.S., D.G., U.K., and A.F. were involved in data management and statistical analyses; K.H.M., M.R..-T., S.S., N.P.K., A.D., K.B., B.K., E.Z., L.H., P.A.G., M.S., S.K.B., and K.S. were involved in laboratory-based characterization of patient samples; K.H.M., T.H., M.F., M.S., S.K.B., U.K., W.E.B., B.W., T.B., W.H., J.B., and K.S. were involved in patient care and provided clinical and follow-up data; W.E.B., B.W., T.B., W.H., and J.B. are primary investigators of the AMLCG clinical trials, through which patients were recruited; all authors helped analyze and interpret the data; K.H.M. wrote the manuscript, and all authors revised and approved the final version of the manuscript.
A complete list of the members of the German Chronic Lymphocytic Leukemia Study Group appears in “Appendix.”
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Klaus H. Metzeler, Department of Internal Medicine III, University Hospital Grosshadern, Ludwig-Maximilans-Universität, Marchioninistrasse 15, 81377 München, Germany email: klaus.metzeler@med.uni-muenchen.de.
Appendix: study group members
The members of the German Acute Myeloid Leukemia Cooperative Group are: A. Heinecke, M. C. Sauerland, D. Görlich, T. H. Ittel, H. Fuß, S. Korsten, D. Hennesser, A. Mayr, A. Grüneisen, K. Possinger, J. Blau, R. Arnold, B. Dörken, G. Maschmeyer, C. Boewer, M Derwahl, H. J. Englisch, M. Notter, E. Thiel, W. D. Ludwig, D. Schöndube, E. Späth-Schwalbe, S. Hesse, J. Potenberg, A. J. Weh, A. Zumsprekel, C. Teschendorf, M. Stechstor, G. Trenn, B. Wörmann, B. Hertenstein, H. Thomssen, A. Peyn, M. Hallek, P. Staib, M. Heike, A. Niederste-Hollenberg, H. Pielken, H. Hindahl, A. Wehmeyer, A. Heyll, C. Aul, C. Giagounidis, W. Lange, S. E. Kuhlemann, M. Flaßhove, J. Karow, R. Fuchs, F. Schlegel, M. Kiehl, G. Meckenstock, D. Haase, L. Trümper, F. Griesinger, C. Gropp, R. Depenbusch, H. Eimermacher, W. Schütte, U. Haak, D. Braumann, L. Balleisen, J. G. Lange, U. Schmitz-Hübner, A. Fauser, M. Wolf, B. Ritter, H. Link, U. R. Fölsch, M. Kneba, A. Dormann, Th. Frieling, M. Planker, F. Hartmann, H. Middeke, C. Gründgens, C. Constantin, K.-A. Jost, Th. Wagner, S. Fetscher, J. Schmielau, M. Uppenkamp, M. Hoffmann, R. Hehlmann, E. Lengfelder, M. Schwonzen, H. Spangenberg, D. Graeven, D. Kohl, T. Heuer, B. Emmerich, R. Dengler, B. Schlag, W. Hiddemann, K. Nibler, D. Fleckenstein, W. E. Berdel, T. Büchner, J. Kienast, R. Mesters, C. Müller-Tidow, H. Serve, U. Krug, J. Hartlapp, T. Hegge, R. Peceny, O. Koch, G. Innig, Th. Südhoff, G. Maschmeyer, E. D. Kreuser, M. Schenk, A. Reichle, R. Andreesen, H. Huff, D. Schönberger, W. Gassmann, T. Gaske, N. Frickhofen, H.-G. Fuhr, W. Augener, W. Brugger, G. Papakonstantinou, U. Kreibich, G. Schott, S. Sommer, W. Zschille, J. Braess, S. Amler, S. Korsten, D. Hennesser, M. de Witt, B. Kallinich, B. Oldenkott, R. Ratei, D. Behringer, Y.-D. Ko, E.-B. Zinngrebe, K.-A. Kreuzer, M. Kiehl, G. Massenkeil, H.-W. Lindemann, C. M. Maciejewski, E. Roemer, H. Nowak, H. Biersack, E. Schleyer, K. Spiekermann, H. Braess, X. Schiel, M. Hentrich, R. Peceney, T. Gaska, J. Braees, and R. Schwerdtfeger.
References
Author notes
Presented in part at the 56th Annual Meeting of the American Society of Hematology, San Francisco, CA, December 6-9, 2014, and at the 57th Annual Meeting of the American Society of Hematology, Orlando, FL, December 5-8, 2015.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal