Abstract
INTRODUCTION: Current therapy options for patients with myelodysplastic syndromes (MDS) are limited, and hypomethylating agents represent the standard of care for these patients. Next-generation sequencing (NGS) and the development of new targeted treatments are facilitating the arrival of personalized medicine as a feasible patient care paradigm. Although the concept of target therapy in MDS is new, a large number of targeted drugs are currently being tested in clinical trials. We report the experience of 812 patients with MDS who were tested using an NGS panel that included possible actionable alterations and its impact on treatment choice.
METHODS: We included all MDS patient for whom an NGS-based analysis for the detection of somatic mutations in the coding sequence of a total of 28 or 53 genes was performed at The University of Texas MD Anderson Cancer Center between October 2012 to October 2015. Clinical and demographic data were obtained from clinical records. 29 genes (ABL1, AKT1, ATM, BRAF, EGFR, FBXW7, FLT3, GNAQ, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MDM2, MLL, MPL, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RET, SMO, SRC, AND STK11) were considered potentially actionable due to the possibility to be targeted with established or investigational therapeutics, either directly or indirectly. Statistical analyses were performed with the IBM SPSS Statistics 23.0 software.
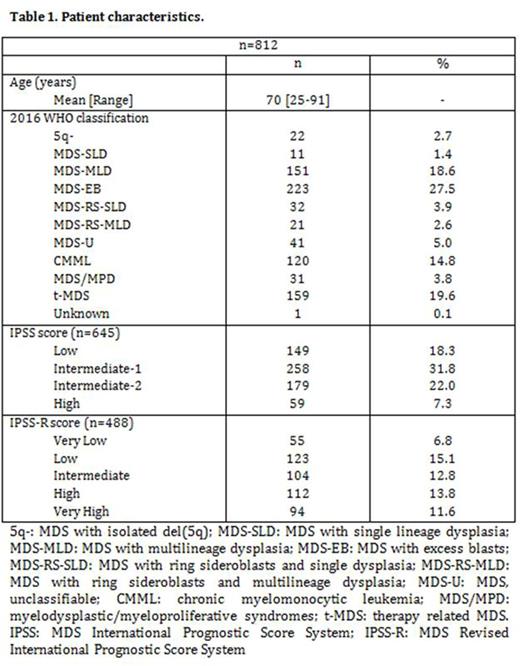
RESULTS:An NGS-based analysis for the detection of somatic mutations in the coding sequence of a total of 28 (n=603 patients) or 53 genes (n=209 patients) was performed in 812 patients with MDS. The median age was 70 years (range 25-91). Baseline characteristics are shown in Table 1.
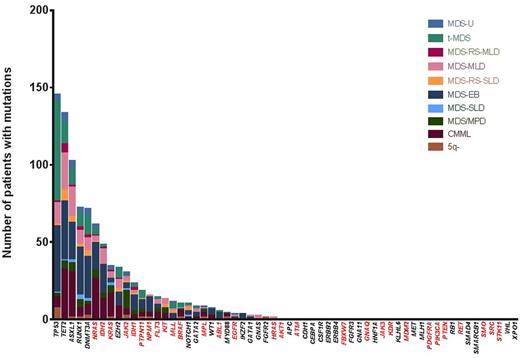
A total of 812 patients with MDS were evaluated. Patient characteristics are shown in Table 1. 522 patients (64%) had at least one detectable driver mutation: 234 (29%) on a potential actionable gene and 288 (35%) on non-actionable genes. The most frequently potentially actionable mutation was NRAS (n=61; 7.5%) followed by IDH2(N=48, 5.9%), and both were present on more than 5% of the patients. The frequency of all identified mutations is shown in Fig. 1.
A total of 497 patients were evaluated at the time of diagnosis, with 293 patients starting therapy for their MDS. 93 (32%) of these treated patients had potentially targetable alterations, and only 15 (5%) of these patients were treated with a targeted agent. Of the 315 remaining patients, 105 (33%) had a potentially actionable alteration, and 22 (7%) started a target therapy treatment (p=0.46).
197 of the patients (59%) who had a potentially actionable alteration never received a targeted therapy, with 14% of these patients (n=28) not returning to the institution after the mutation analysis and 18% (n=36) starting new treatment due to response to current therapy. From the remaining 133 patients, documentation of discussion of enrollment in a clinical trial was present on 74% (n=98), of which 14% (n=18) of these patients preferred a non-investigational treatment, 8% (n=11) were not eligible owing to poor status or other causes, and 8% (n=10) had difficulty in meeting the study visits. Patients with mutations in potentially actionable genes were more likely to be treated on a clinical trial (102/234 [44%] vs 201/578 [35%]) (p=0.02).
CONCLUSION:NGS technologies can be used to identify a significant proportion of patients (29%) with potentially actionable mutations. The presence of such mutations predicts for increased likelihood of treatment within a clinical trial. However, only a small proportion of patients (16%) with MDS and actionable alterations are treated with targeted therapy. Personalized medicine based on mutation profiles in MDS remains an area under development. The results of clinical trials currently in recruitment may change the way we treat these patients.
Jabbour:ARIAD: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Novartis: Research Funding; BMS: Consultancy. Cortes:ARIAD: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Teva: Research Funding. DiNardo:Abbvie: Research Funding; Agios: Research Funding; Celgene: Research Funding; Daiichi Sankyo: Research Funding; Novartis: Research Funding. Konopleva:Calithera: Research Funding; Cellectis: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal