Abstract
Amplification of 1q is observed in 40% of Multiple Myeloma (MM) patients and is associated with a more aggressive clinical course of the disease. The frequency of 1q21 amplifications has been shown to increase significantly in the transition from monoclonal gammopathy of undetermined significance (MGUS) to overt myeloma and to relapse. Previous studies have reported genes on 1q such as ANP32E and CSK1B that have significant impact on survival. However, the biological mechanisms underlying disease aggressiveness associated to 1q amplification still remain unclear.
ADAR (Adenosine Deaminase Acting on RNA) is an enzyme responsible for A-to-I editing, a post-transcriptional modification of double stranded RNA consisting in the conversion of specific Adenosines (A) into Inosines (I) by deamination. As Inosine is structurally similar to Guanosine (G), editing events can result in functional consequences in RNA and protein structure, including non-synonymous changes in protein coding sequences and creation/disruption of miRNA binding sites on UTRs. Dysregulation of A-to-I editing by ADAR has been recently linked to cancer. Since the ADAR gene is located in 1q21.3 (the critical minimally amplified region in MM), we asked whether 1q amplification affected ADAR expression, RNA editing and overall prognosis in MM patients.
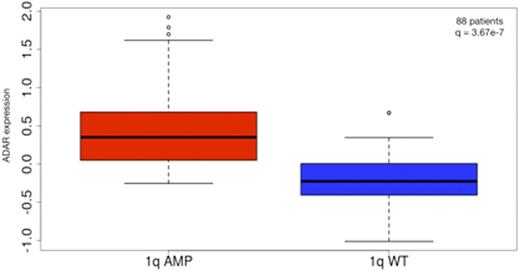
We identified 44 patients with 1q amplification from the IA6 release of the MMRF CoMMpass dataset. As a control group (wt), we selected an equal number of patients from CoMMpass without any 1q alteration. Gene expression analysis showed significantly higher expression of ADAR in 1q-amp patients compared to wt (q = 3.64e-7) (Fig. 1) and significant correlation between ADAR copy number and its expression (Spearman ρ=0.69, p = 4.52e-14).
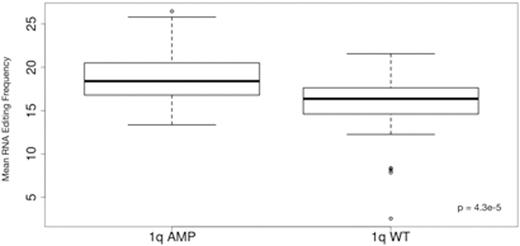
To evaluate the functional impact of ADAR up-regulation, we applied a computational pipeline based on the tool REDItools and our in-house scripts to detect A-to-I edited sites in RNA-Seq samples. The pipeline identified candidate A-to-G mutations in RNA sequences using corresponding Whole-Exome Sequencing data to filter out actual DNA mutations. We calculated sample-wise mean editing frequency across all edited sites and found significantly increased editing in 1q-amp patients compared to wt (p = 4.3e-5) (Fig. 2). Mean editing frequency was significantly correlated with ADAR expression (ρ = 0.62, p < 2e-16) and ADAR copy number (ρ= 0.5, p= 4.32e-7).
Our analysis identified 3,286 sites residing in Alu sequences and 1,303 in non-Alu regions. A-to-I editing has been shown to occur predominantly in Alu elements, repetitive sequences abundantly interspersed throughout the human genome, mostly within introns and untranslated regions (UTRs). As expected, most sites were reported within 3' UTRs (66%) and introns (12%).
Overall, at the site level, we observed increased editing in 1q-amp vs wt (p < 2e-16). We found that 2,173 sites (47%) had significant differential editing frequency between 1q-amp and wt patients (FDR < 20%).
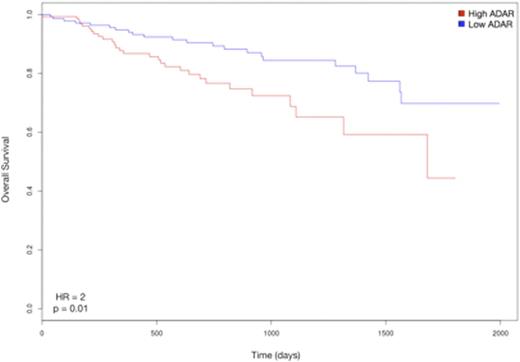
Next, we sought to assess the prognostic implications of ADAR activity. Cox regression analysis revealed a trend toward higher risk in terms of EFS (Event Free Survival) for 1q-amp vs wt (HR = 1.7, 95% CI = 0.83-3.59, p = 0.13), as well as for patients with higher expression of ADAR (HR = 2.4, 95% CI = 0.79-7.15, p = 0.11) and higher mean editing frequency (HR = 2, 95% CI = 0.72-5.59, p = 0.17). Since survival data in the CoMMpass dataset is not yet mature, we evaluated the effects of ADAR expression on survival on an independent dataset consisting of 559 samples from newly diagnosed patients pre-TT2 and -TT3 treatments (GSE2658, Shaughnessy et al, Blood 2007; 109:2276-84). Cox regression analysis showed a significant difference in terms of overall survival between patients with low and high ADAR expression, the latter being correlated with higher risk (HR = 2, 95% CI = 1.18-3.66, p = 0.01) (Fig. 3).
In conclusion, we found a significant increase in ADAR expression and aberrant A-to-I RNA editing in MM patients with amplification of 1q. These results demonstrate a novel mechanism by which 1q amplification can contribute to MM pathogenesis via induction of A-to-I RNA editing by ADAR.
Difference in mean RNA editing frequency between 1q-amp and wt patients.
Kaplan-Meier curves of overall survival in the Shaughnessy cohort stratified by ADAR expression (GSE2658)
Kaplan-Meier curves of overall survival in the Shaughnessy cohort stratified by ADAR expression (GSE2658)
Chari:Janssen: Consultancy, Research Funding; Pharmacyclics: Research Funding; Takeda: Consultancy, Research Funding; Celgene: Consultancy, Research Funding; Amgen Inc.: Honoraria, Research Funding; Array Biopharma: Consultancy, Research Funding; Novartis: Consultancy, Research Funding. Cho:Ludwig Institute for Cancer Research: Membership on an entity's Board of Directors or advisory committees; Agenus, Inc.: Research Funding; Genentech Roche: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Consultancy, Research Funding; Bristol-Myers Squibb: Membership on an entity's Board of Directors or advisory committees, Research Funding. Barlogie:Signal Genetics: Patents & Royalties. Jagannath:Janssen: Consultancy; Celgene: Consultancy; Merck: Consultancy; Bristol-Myers Squibb: Consultancy; Novartis: Consultancy. Dudley:NuMedii, Inc.: Patents & Royalties; AstraZeneca: Speakers Bureau; Ontomics, Inc.: Equity Ownership; NuMedii, Inc.: Equity Ownership; Ecoeos, Inc.: Equity Ownership; Ayasdi, Inc.: Equity Ownership; Janssen Pharmaceuticals, Inc.: Consultancy; GlaxoSmithKline: Consultancy; Personalis: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal