Abstract
Background: The 2016 revision to the World Health Organization classification of myeloid neoplasms has recommended distinction between "proliferative" (WBC ≥ 13 x 10(9)/L) and "dysplastic" (WBC < 13 X 10(9)/L) subtypes of chronic myelomonocytic leukemia (CMML). In addition, CMML is characterized by a spectrum of cytogenetic and molecular abnormalities (Patnaik Leukemia 2014). In the current study, we looked for correlations between gene expression profiles (GEP) and morphologic or molecular categories of previously untreated patients with CMML.
Methods:35 patients with CMML were included in the study. Targeted capture assays were carried out on BM DNA specimens obtained at diagnosis for 28 myeloid-relevant genes (Patnaik Blood C. J.). RNA sequencing (Stranded Total RNA- Illumina, San Diego, CA), was performed on 9 normal BM controls, and 35 CMML samples (25 peripheral blood (PB) and 10 BM). The RNA-Seq data was analyzed using MAPRSeq v.2.0. Normalization and differential expression analysis was performed using edgeR 2.6.2. We selected genes that were differentially expressed with a False Discovery Rate (FDR) <0.1 and a fold change bigger than 2. Pathway enrichment analysis was performed using Ingenuity Pathway Analysis (IPA).
Results:Among the 35 study patients, 75% were males and median age was 71 years (range, 55-87). 21 (60%), 9 (25%) and 5 (15%) patients were classified as CMML-0, 1 and 2, respectively. 16 (45%) had a dysplastic while 19 (55%) had a proliferative phenotype. At a median follow-up of 16 months, 5 (14%) deaths and 4 (14%) leukemic transformations were documented. Mutational frequencies included; TET2 45%, ASXL1 45%, SRSF2 40%, NRAS 14%, SETBP1 13%, CBL 10%, JAK2 7%, RUNX1 6%, DNMT3A 6%, U2AF1 6%, SF3B1 5%, ZRSR2 4%, Tp53 4%, and IDH2 4%. Abnormal cytogenetics (n=9, 35%) included; -Y 8%, trisomy 8 6%, monosomy 7 3%, del(13q) 3% and monosomal karyotype 3%.
i) Differential GEP in PB samples:
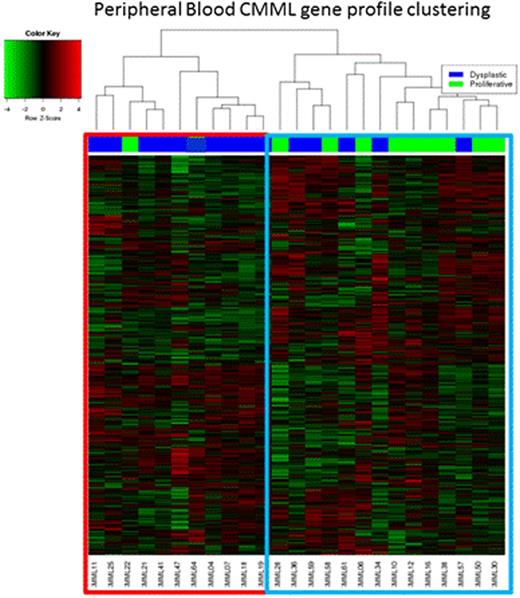
25 PB CMML samples were analyzed. Unsupervised GEP demonstrated two unique clusters (figure one); cluster one was enriched with dysplastic CMML (90%) and cluster two with proliferative CMML (64%). Consistent with the difference in the distribution of morphologic categories, in comparison to cluster one, patients in cluster two had a higher white blood count (p=0.003), higher monocyte count (p=0.02) and higher risk prognostication by the Mayo Model (p=0.016); there was no significant difference between the two clusters in terms of age and gender distribution, hemoglobin level, neutrophil and platelet counts, PB or BM blast content, cytogenetic risk stratification, ASXL1, TET2, RAS, CBL and SETBP1 mutational status. Genes significantly upregulated (FDR<1e-5 and fold change ≥ 16x); in cluster two (proliferative CMML) in comparison to cluster one included; PRG2, CTSG, BEX1, CD34, CRISP2, while those significantly down-regulated included (FDR<1e-15 and fold change ≥ 5x); IL2RB, PRF1, GNLY, PDGFRB. IPA analysis identified the following pathways preferentially expressed in cluster two (proliferative CMML) 1) mitotic roles of polo-like kinase (KIF23, WEE-1, PLK1, PLK4), 2) Cell cycle: G2/M checkpoint regulation (CDC25C, WEE-1, AURKA, CDK1), 3) Cell cycle: chromosomal replication (CDC45, RPA3, MCM2, CDC7), and 4) BRCA1 in DNA damage response (BARD1, RBBP8, PLK1, RAD51). Whereas in the cluster one (dysplastic CMML), preferentially expressed pathways included; 1) T-cell receptor signalling (CD247 PRKCQ, CD4, PLCG1, CD28) and 2) STAT3 signalling (SOCS3, MAP3K9).
ii) Differential GEP in BM samples:
10 BM CMML samples were analyzed along with 10 normal controls. Samples and controls clustered separately and within the CMML samples two clusters were identified (figure two). These clusters had equal representations of dysplastic (n=5) and proliferative (n=5) CMML sub-types. Additional samples are being processed.
Conclusions:Differential gene expression profiling in patients with CMML identifies two unique clusters, clinically demarcated, at least in PB samples, by "proliferative" and "dysplastic" CMML subtypes. The proliferative cluster is enriched in genes involved in cell cycle regulation and DNA damage response; whereas the dysplastic cluster is enriched in T-cell receptor and STAT3 signalling. The prognostic and therapeutic impact of these gene clusters remains to be assessed in a larger group of informative patients.
Al-Kali:Novartis: Research Funding; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal