Key Points
The transcriptional networks controlling breakthrough acute GVHD can be mapped, and correlate closely with clinical disease.
Breakthrough acute GVHD is transcriptionally controlled by T-cell persistence, inflammation, and Th/Tc17 skewing.
Abstract
One of the central challenges of transplantation is the development of alloreactivity despite the use of multiagent immunoprophylaxis. Effective control of this immune suppression–resistant T-cell activation represents one of the key unmet needs in the fields of both solid-organ and hematopoietic stem cell transplant (HCT). To address this unmet need, we have used a highly translational nonhuman primate (NHP) model to interrogate the transcriptional signature of T cells during breakthrough acute graft-versus-host disease (GVHD) that occurs in the setting of clinically relevant immune suppression and compared this to the hyperacute GVHD, which develops in unprophylaxed or suboptimally prophylaxed transplant recipients. Our results demonstrate the complex character of the alloreactivity that develops during ongoing immunoprophylaxis and identify 3 key transcriptional hallmarks of breakthrough acute GVHD that are not observed in hyperacute GVHD: (1) T-cell persistence rather than proliferation, (2) evidence for highly inflammatory transcriptional programming, and (3) skewing toward a T helper (Th)/T cytotoxic (Tc)17 transcriptional program. Importantly, the gene coexpression profiles from human HCT recipients who developed GVHD while on immunosuppressive prophylactic agents recapitulated the patterns observed in NHP, and demonstrated an evolution toward a more inflammatory signature as time posttransplant progressed. These results strongly implicate the evolution of both inflammatory and interleukin 17–based immune pathogenesis in GVHD, and provide the first map of this evolving process in primates in the setting of clinically relevant immunomodulation. This map represents a novel transcriptomic resource for further systems-based efforts to study the breakthrough alloresponse that occurs posttransplant despite immunoprophylaxis and to develop evidence-based strategies for effective treatment of this disease.
Introduction
Transplantation, encompassing both solid-organ transplantation and hematopoietic stem cell transplantation (HCT), is currently in a stage of short-term success but long-term failure for the majority of patients. This short-term success has relied on the use of broadly active, nontargeted immune suppression, which has succeeded in controlling very early immune activation.1 In solid-organ transplantation, this results in high 1-year survival times for many transplanted organs (eg, 90% 1-year survival for renal transplants) but with the ultimate occurrence of immune-mediated rejection in the vast majority of patients (with a half-life of ∼10 years for renal transplants2 ). In HCT, similar immunosuppressive strategies result in most patients engrafting, but with up to 70% of patients ultimately developing acute graft-versus-host disease (GVHD), with the most severe cases being untreatable and lethal.3 The field has thus far been unsuccessful in identifying the underlying mechanisms responsible for immune escape and alloreactivity that occur despite ongoing immunosuppression, resulting not only in high rates of immunosuppression failure, but also in a “one-size-fits-all” approach to the treatment of breakthrough alloimmunity, which still relies on global use of corticosteroids as first-line therapy.
To address the critical unmet need for a detailed molecular understanding of mechanisms driving clinically relevant alloreactivity, our group has developed a nonhuman primate (NHP) model of GVHD, which has been specifically designed to probe the mechanisms of immune escape that occur both in the absence and in the presence of clinical immunosuppression, and in which the potential targets of GVHD can be studied.4-6 To discover the transcriptional networks that drive GVHD during clinically relevant immunoprophylaxis, we have now mapped the T-cell dysregulation that occurs in the setting of a variety of immunoprophylactic settings. We find that 2 signatures predominate: (1) a highly proliferative, cytotoxic signature that occurs during hyperacute GVHD and (2) the much more complex immune signature of breakthrough acute GVHD, which retains some T helper (Th)/T cytotoxic (Tc)1 elements, but which is predominated by an inflammatory Th/Tc17-skewed and apoptosis-resistant T-cell profile. Importantly, we have also identified these breakthrough acute GVHD transcriptional signatures in transplanted patients. These results provide the first map of the transcriptional complexity of primate breakthrough acute GVHD and identify targeted, immediately clinically translatable strategies for treating this disease that promise to move the field of transplantation forward toward an evidence-based, risk-adapted approach to therapeutic decision-making.
Materials and methods
NHP study design
This was a prospective cohort study in NHP designed to compare the clinical and immunologic outcomes of transplantation, and to discern the transcriptome of autologous and allogeneic transplant recipients. Details of the experimental cohorts studied, the relatedness and major histocompatibility complex (MHC) disparity between transplant pairs, immunoprophylactic dosing, and study design are described in supplemental Materials and methods (available on the Blood Web site).
Rhesus HCT and GVHD clinical analysis
In this study, we used our previously described strategy for allogeneic HCT (allo-HCT) in rhesus macaques.4 Details of the HCT regimen and GVHD clinical analysis are described in detail in supplemental Materials and methods.
Human study design
This study was designed as a retrospective, case-control study. Available cryopreserved patient peripheral blood mononuclear cell (PBMC) samples were obtained from patients enrolled in HCT clinical trials performed at Emory University and the University of Minnesota, with and without GVHD. Clinical details pertaining to each of the samples included in the clinical gene array analysis are shown in supplemental Table 3. Details of sample identification and array preparation are described in supplemental Materials and methods.
Flow cytometry
Longitudinal multicolor flow cytometric analysis was performed on all NHP transplant recipients as described in detail in supplemental Materials and methods.
NHP and patient transcriptome analysis
Following T-cell purification, RNA was isolated and NHP samples were hybridized to a GeneChip Rhesus Macaque Genome Array (Affymetrix) and human samples to a Human Transcriptome 2.0 Array (Affymetrix). The resultant fluorescent signals were analyzed using both Expression Console software (Affymetrix) and the R statistical platform. Full details of T-cell sorting, RNA preparation, microarray completion, computational analysis of the transcriptome, and data code availability are provided in supplemental Materials and methods.
Results
Immune evidence for 2 distinct variants of acute GVHD: hyperacute and breakthrough acute
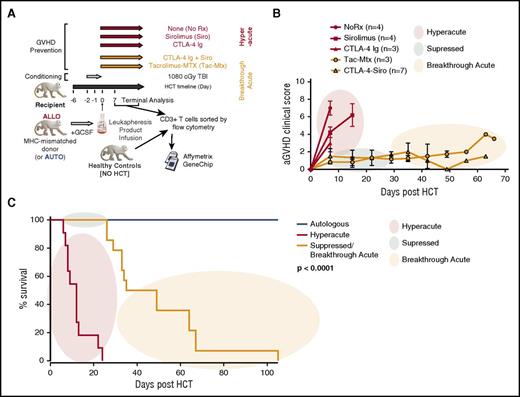
We have used a NHP GVHD model (schematized in Figure 1A) to perform a large series of autologous HCT and allo-HCT experiments interrogating multiple immunomodulating strategies. We have previously reported the flow cytometric4 and transcriptional attributes6 of T cells causing GVHD in the absence of immunosuppression (“No Rx”). These attributes include a highly proliferative T-cell program, characterized by multiple markers of Th/Tc1 skewing and expression of granzymes and interferon γ (IFNγ).4 In the current study, we compared the clinical and immunologic profile of the No Rx cohort with that of T cells purified from recipients who were only partially prophylaxed with either mammalian target of rapamycin inhibition (sirolimus [Siro]) as monotherapy or CD80/86-directed costimulation blockade (with CTLA4-immunoglobulin [CTLA4-Ig]) as monotherapy, or with clinically relevant combinatorial strategies (Figure 1B-C). Mirroring what we documented with the No Rx cohort, we observed flow cytometric evidence of an expanding T-cell proliferative and cytotoxic effector program when either sirolimus or CTLA4-Ig were used as monotherapies (Figure 2A), and a shift of the T-cell phenotype toward central memory (CM)/effector memory (EM) predominance (Figure 2B) with kinetics that approximated that of the No Rx cohort. Because of the commonalities in the pace and severity of the clinical syndromes, as well as the histopathologic severity and T-cell immune phenotype that developed in the No Rx, sirolimus monotherapy, and CTLA4-Ig monotherapy cohorts (Figures 1-2), we have characterized the GVHD in all of these recipients as “hyperacute GVHD.”
Immunoprophylactic strategies used in a NHP model of acute GVHD result in a clinical picture that can be categorized as hyperacute, suppressed, or breakthrough acute GVHD. (A) Experimental schema detailing the transplant protocol and immunoprophylaxis regimens used throughout this study. (B) Clinical score based on our previously described NHP GVHD clinical scoring system.4 Colored circles represent the clinical categories of hyperacute (red), suppressed (green) and breakthrough acute (orange) disease. (C) Comparison of survival curves between hyperacute and breakthrough acute cohorts. The Kaplan-Meier product-limit method was used to calculate survival. Significance was determined using log-rank statistics. aGVHD, acute GVHD; GCSF, granulocyte colony-stimulating factor; TBI, total body irradiation.
Immunoprophylactic strategies used in a NHP model of acute GVHD result in a clinical picture that can be categorized as hyperacute, suppressed, or breakthrough acute GVHD. (A) Experimental schema detailing the transplant protocol and immunoprophylaxis regimens used throughout this study. (B) Clinical score based on our previously described NHP GVHD clinical scoring system.4 Colored circles represent the clinical categories of hyperacute (red), suppressed (green) and breakthrough acute (orange) disease. (C) Comparison of survival curves between hyperacute and breakthrough acute cohorts. The Kaplan-Meier product-limit method was used to calculate survival. Significance was determined using log-rank statistics. aGVHD, acute GVHD; GCSF, granulocyte colony-stimulating factor; TBI, total body irradiation.
Animals with breakthrough acute GVHD develop clinical and pathological evidence of disease without the specific T-cell signatures that develop during hyperacute GVHD. (A) Expression of Ki67 and granzyme B (GrmB) in T cells from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test, and significant differences of the mean percentage of either CD4 (*) or CD8 (†) cell subtypes were determined using an unpaired Student t test. All other time-point comparisons were not significant. (B) Engraftment/expansion of CD4 and CD8 TCM and TEM from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days −7, 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test. *Significant differences of the absolute numbers of the CD4 and CD8 T-cell subpopulations. All other time-point comparisons were not significant. (C) The combined GVHD pathology score (summarizing liver, colon, skin, and lung scores [top panel]) and combined upper GI GVHD score (summarizing esophagus, stomach, and duodenum [Duod.]; [bottom panel]) of recipients from hyperacute (MST = 12 days) and breakthrough acute (MST = 34.5 days) cohorts. Histopathologic analysis was performed at the time of terminal analysis and analyzed by a pathologist (A.P.-M.) in a blinded manner. *P < .05 using the Mann-Whitney test on composite scores. NS, not significant. (D) Flow cytometric analysis of mononuclear cells isolated from peripheral blood, lymphoid, and nonlymphoid GVHD-target organs at the time of terminal analysis from animals from hyperacute and breakthrough acute cohorts and stained for Ki-67 (top and middle panel) and for granzyme B (bottom panel). Plots depict the percentages of CD4 and CD8 T cells expressing Ki-67 and the percentage of CD8 T cells expressing high levels of granzyme B (mean fluorescence intensity [MFI] ≥ 105). *P < .05 using the Mann-Whitney test. LN, lymph node.
Animals with breakthrough acute GVHD develop clinical and pathological evidence of disease without the specific T-cell signatures that develop during hyperacute GVHD. (A) Expression of Ki67 and granzyme B (GrmB) in T cells from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test, and significant differences of the mean percentage of either CD4 (*) or CD8 (†) cell subtypes were determined using an unpaired Student t test. All other time-point comparisons were not significant. (B) Engraftment/expansion of CD4 and CD8 TCM and TEM from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days −7, 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test. *Significant differences of the absolute numbers of the CD4 and CD8 T-cell subpopulations. All other time-point comparisons were not significant. (C) The combined GVHD pathology score (summarizing liver, colon, skin, and lung scores [top panel]) and combined upper GI GVHD score (summarizing esophagus, stomach, and duodenum [Duod.]; [bottom panel]) of recipients from hyperacute (MST = 12 days) and breakthrough acute (MST = 34.5 days) cohorts. Histopathologic analysis was performed at the time of terminal analysis and analyzed by a pathologist (A.P.-M.) in a blinded manner. *P < .05 using the Mann-Whitney test on composite scores. NS, not significant. (D) Flow cytometric analysis of mononuclear cells isolated from peripheral blood, lymphoid, and nonlymphoid GVHD-target organs at the time of terminal analysis from animals from hyperacute and breakthrough acute cohorts and stained for Ki-67 (top and middle panel) and for granzyme B (bottom panel). Plots depict the percentages of CD4 and CD8 T cells expressing Ki-67 and the percentage of CD8 T cells expressing high levels of granzyme B (mean fluorescence intensity [MFI] ≥ 105). *P < .05 using the Mann-Whitney test. LN, lymph node.
Unlike the hyperacute GVHD cohorts, recipients prophylaxed with combinatorial therapies (either with standard-of-care tacrolimus plus methotrexate [Tac/MTX], or Siro plus CTLA4-Ig [Siro/CTLA4-Ig])7,8 successfully suppressed hyperacute GVHD. Thus, in the first several weeks posttransplant, both the Tac/Mtx and Siro/CTLA4-Ig cohorts controlled the clinical symptoms of disease (Figure 1B-C). However, clinical acute GVHD eventually broke-through the ongoing immunoprophylaxis in both the Tac//MTX and Siro/CTLA4-Ig cohorts, which, in the absence of GVHD treatment, resulted in the recipient reaching protocol end point criteria due to clinical evidence of disease (median survival time [MST] = 34.5 days; Figure 1C). The clinical symptoms of breakthrough disease included vomiting and decreased appetite, which made it difficult for the animals to maintain nutritional status (although immunosuppression levels were maintained, as these agents were all delivered parenterally). Terminal analysis documented histopathologic signs of GVHD in canonical target organs in recipients with both hyperacute GVHD and breakthrough acute GVHD. As shown in Figure 2C, and in agreement with their clinical symptoms, there was less severe histopathologic GVHD in the skin, liver, lung, and colon in the breakthrough acute cohort compared with recipients with hyperacute disease. However, the amount of upper gastrointestinal (GI) disease (esophagus, stomach, duodenum) was equivalent in the 2 cohorts. Importantly, the clinical, histologic, flow cytometric, and transcriptomic characteristics of GVHD in both Tac/MTX and Siro/CTLA4-Ig cohort recipients were highly aligned, suggesting important commonalities in the mechanisms of immune escape with both prophylaxis regimens. Because of this, we characterized the disease that developed in each of these recipients as “breakthrough acute GVHD,” and performed subsequent analyses on the 2 prophylaxis regimens as 1 combined cohort. Importantly, clinical GVHD occurred in the breakthrough acute cohort despite continued flow cytometric evidence for control of T-cell proliferation and granzyme B expression, in both blood and GVHD target organs (Figure 2A,D). Taken together, these results suggest that unique mechanisms contributed to the alloreactivity in these 2 cohorts.
Transcriptomics distinguish hyperacute and breakthrough acute GVHD
To identify the T-cell programs that differentiate hyperacute vs breakthrough acute GVHD, we performed transcriptomic profiling on peripheral blood T cells from all NHP cohorts. We found that the broad transcriptional signature recapitulated the dichotomization of hyperacute vs breakthrough acute GVHD shown in Figures 1-2, with principal components of variance of terminal T-cell transcriptional data clustering by GVHD type (PC1 vs PC3, hyperacute vs breakthrough acute, P < .05; Figure 3A). Furthermore, PC1 specifically functioned as a significant independent predictor of survival (Figure 3B; P, .0038), underscoring the applicability of transcriptomic data to our clinical observations.
T-cell profiles from animals with hyperacute GVHD contain an abundance of transcripts associated with proliferation, and exhibit Th1 skewing. (A) First and third principal component projections reveal clustering of transplanted animals by immunoprophylactic strategy. Each dot represents an array sample. The center of inertia ellipses corresponds to the mean projections of the group (P < .05). (B) The first principal component shows a significant correlation with survival in a linear regression model (adjusted [Adj.], R2 = 0.3299; P < .004). (C) Heatmap of the top 25 gene sets enriched in the hyperacute cohort using ssGSEA. The constellation map to the right of the heatmap allows for the identification of clusters of these gene sets. Those gene sets plotted closer to the center have a greater degree of similarity to the phenotype of interest (measured by the normalized mutual information [NMI] score) whereas the angular distance corresponds to gene-set similarity with one another. A dark-green edge further indicates gene-set similarity with the thickness of the line proportional to the Jaccard index.10,11 (D) GSEA of transcripts previously shown to differentiate Th1 from Th17 cells.12 These molecules were found to be overrepresented in T cells from animals with hyperacute GVHD compared with healthy controls and the breakthrough acute cohort. *False discovery rate [FDR] < 0.01. (E) Flow cytometric analysis of PBMCs at the time of terminal analysis stimulated with phorbol myristate acetate (PMA)/ionomycin and measured for the production of IFNγ. *P < .05 using an unpaired t test.
T-cell profiles from animals with hyperacute GVHD contain an abundance of transcripts associated with proliferation, and exhibit Th1 skewing. (A) First and third principal component projections reveal clustering of transplanted animals by immunoprophylactic strategy. Each dot represents an array sample. The center of inertia ellipses corresponds to the mean projections of the group (P < .05). (B) The first principal component shows a significant correlation with survival in a linear regression model (adjusted [Adj.], R2 = 0.3299; P < .004). (C) Heatmap of the top 25 gene sets enriched in the hyperacute cohort using ssGSEA. The constellation map to the right of the heatmap allows for the identification of clusters of these gene sets. Those gene sets plotted closer to the center have a greater degree of similarity to the phenotype of interest (measured by the normalized mutual information [NMI] score) whereas the angular distance corresponds to gene-set similarity with one another. A dark-green edge further indicates gene-set similarity with the thickness of the line proportional to the Jaccard index.10,11 (D) GSEA of transcripts previously shown to differentiate Th1 from Th17 cells.12 These molecules were found to be overrepresented in T cells from animals with hyperacute GVHD compared with healthy controls and the breakthrough acute cohort. *False discovery rate [FDR] < 0.01. (E) Flow cytometric analysis of PBMCs at the time of terminal analysis stimulated with phorbol myristate acetate (PMA)/ionomycin and measured for the production of IFNγ. *P < .05 using an unpaired t test.
We then used differential expression (DE; supplemental Table 6) and single-sample gene-set enrichment analysis (ssGSEA)9 to dissect the T-cell immunopathology during hyperacute and breakthrough acute GVHD. In accordance with our previously published results,6 visualization of the top 25 gene sets in the hyperacute cohort using constellation mapping10,11 revealed that proliferative signals were a dominant theme in the T-cell transcriptional profile of hyperacute GVHD (Figure 3C), with 21 of 25 top correlating gene sets describing proliferative programing, and 15 of these demonstrating significant sharing of enriched transcripts (depicted as red circles in Figure 3C). Similar to our previous work, GSEA revealed that Th/Tc1 transcripts were overrepresented in animals with hyperacute GVHD compared with T cells purified from both healthy controls, and from recipients with breakthrough acute GVHD12 (Figure 3D). Flow cytometric assessment of IFNγ protein expression in T cells at the time of terminal analysis confirmed this observation on a single-cell level in both CD4 and CD8 T cells during hyperacute GVHD (Figure 3E).
In contrast to the uniform proliferation- and cytotoxicity-dominated T-cell activation program in hyperacute disease, transcriptomic analysis of breakthrough acute GVHD revealed a much more complex transcriptional landscape. This complexity was apparent in both DE analysis and ssGSEA/constellation mapping (Figure 4A-B; supplemental Table 6), which documented enrichment of a wide range of biological activities (Figure 4A-B). Some of these gene sets were similar to those found in hyperacute disease, and remained associated with Th/Tc1 skewing (eg, M2602 Biocarta RANKL pathway; Figure 4A). However, many pathways were unique to breakthrough acute GVHD. Moreover, and in agreement with the flow cytometric findings shown in Figure 2A, there was underrepresentation of the canonical proliferative and cytotoxicity transcripts, Ki67 and granzyme B (which were highly enriched in the hyperacute transcriptome; Figure 4C). Of note, the gene sets most similar to samples in the breakthrough GVHD cohort (gene sets 1-2; Figure 4B) were generated from a set of experiments that studied the expression of the Polycomb transcriptional regulators Bmi1 and Mel-18.13 These gene sets provided several important clues to the pathways active during breakthrough GVHD. Thus, the enrichment of Bmi-1 targets is notable, given the ability of Bmi-1 to promote survival14 and inhibit apoptosis in a variety of cell types, including T cells.15 The enrichment of Mel-18 targets was also important, given that Mel-18 has been observed to be a key component in Th17 cell programming.16
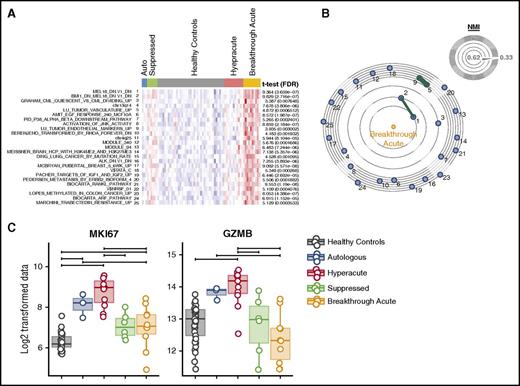
The T-cell transcriptome of breakthrough acute GVHD. (A) Heatmap of the top 25 gene sets enriched in the breakthrough acute cohort using ssGSEA and (B) constellation map visualization. (C) Box plots of expression data (Log2 normalized fluorescent intensity signal) for MKI67 and granzyme B (GZMB). Horizontal significance bars denote comparisons with a moderated t statistic < 0.05 corrected for multiple hypothesis testing.
The T-cell transcriptome of breakthrough acute GVHD. (A) Heatmap of the top 25 gene sets enriched in the breakthrough acute cohort using ssGSEA and (B) constellation map visualization. (C) Box plots of expression data (Log2 normalized fluorescent intensity signal) for MKI67 and granzyme B (GZMB). Horizontal significance bars denote comparisons with a moderated t statistic < 0.05 corrected for multiple hypothesis testing.
Antiapoptotic programing in T cells during breakthrough acute GVHD
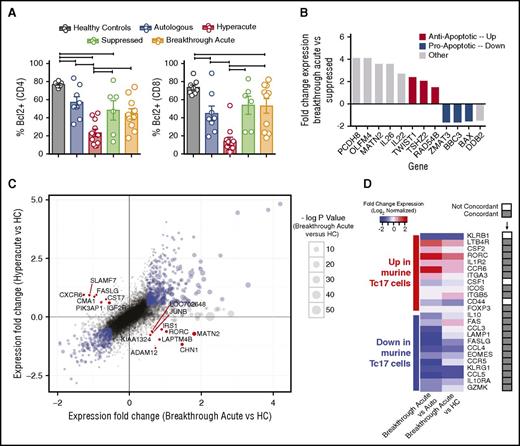
Given the survival of T cells during breakthrough acute GVHD despite the reduction in strong proliferative signaling, we hypothesized that the inhibition of apoptotic signals could be an important mechanism driving T-cell persistence during breakthrough acute GVHD. In agreement with this hypothesis, we observed normalization of expression of the antiapoptotic protein BCL-2 in breakthrough acute GVHD (Figure 5A) relative to hyperacute GVHD. Transcriptome analysis also revealed the emergence of a diverse antiapoptotic signature during breakthrough acute disease. This was perhaps best illustrated in the comparison of the transcriptome of T cells during breakthrough acute disease, vs the transcriptome of T cells from those same recipients earlier posttransplant, during which time clinical GVHD was suppressed. As shown in Figure 5B, of the 12 statistically significantly DE genes between the breakthrough acute and suppressed time points, 6 were involved in the control of apoptosis (TWIST1, TSHZ2, RAD54B, ZMAT3, BBC3 [PUMA], BAX), with the differential expression of each skewing the breakthrough acute transcriptome toward an antiapoptotic signature (Figure 5B; supplemental Table 4). Other genes identified in the breakthrough acute vs suppressed analysis were also notable, including the cytokines interleukin 22 (IL22) and IL26.17 Both IL22 and IL26 have been strongly linked to Th17 function,18 with IL22 increasingly appreciated to play a complex role in posttransplant outcome, with growing evidence for tissue-specific pathogenic as well as protective functions.19-21
T cells during breakthrough acute GVHD display a prosurvival and Th17-skewed phenotype. (A) Flow cytometric analysis of PBMCs measured for the expression of Bcl-2. Plots depict the percentages of CD4 and CD8 T cells expressing Bcl-2. *P < .05 using analysis of variance (ANOVA) with Tukey multiple comparison test. (B) Bar plot of fold-change expression values (log2 normalized) for transcripts in breakthrough acute vs suppressed samples that meet differential expression cutoffs (fold change > 1.3 and P < .05 using a moderated t statistic corrected using the Benjamini procedure.) Red bars, Antiapoptotic transcripts. Blue bars, Proapoptotic transcripts. Gray bars, Genes with other functional attributes. (C) Scatter plot of fold-change expression values (log2 normalized) of breakthrough acute vs healthy controls (HCs) (x-axis) and hyperacute vs healthy controls (y-axis). Blue points denote those transcripts whose expression is above threshold values (log2 fold change < 0.5, and P < .05 using a moderated t statistic corrected using the Benjamini procedure) and whose expression in both breakthrough acute and hyperacute samples is overrepresented or underrepresented relative to healthy controls. Red points denote pivot transcripts, whose expression meets cutoffs noted above, and whose differential expression in breakthrough acute and hyperacute samples compared with healthy controls are in opposite directions. (D) Heatmap showing the expression levels of inflammatory Tc17-related genes identified in a study of fate-mapped murine Tc17 cells35 in the NHP breakthrough acute GVHD arrays compared with either autologous controls (Auto, left column) or healthy controls (HC, right column). The rows in the heatmap contain 24 genes previously identified as overrepresented (n = 12, top half of heatmap) and underrepresented (n = 12, bottom half of heatmap) in murine Tc17 cells during GVHD relative to controls. One gene (Ly6c1) identified in the murine study was not included in this analysis, as it has no human/NHP ortholog.36 The columns reveal the expression of these genes in the breakthrough acute cohort vs either autologous (left) or healthy controls (right). The column to the right of the gene names designates whether the direction of expression in breakthrough acute vs autologous or healthy controls is concordant (gray boxes) or not (white boxes) with the expression in murine Tc17 cells.
T cells during breakthrough acute GVHD display a prosurvival and Th17-skewed phenotype. (A) Flow cytometric analysis of PBMCs measured for the expression of Bcl-2. Plots depict the percentages of CD4 and CD8 T cells expressing Bcl-2. *P < .05 using analysis of variance (ANOVA) with Tukey multiple comparison test. (B) Bar plot of fold-change expression values (log2 normalized) for transcripts in breakthrough acute vs suppressed samples that meet differential expression cutoffs (fold change > 1.3 and P < .05 using a moderated t statistic corrected using the Benjamini procedure.) Red bars, Antiapoptotic transcripts. Blue bars, Proapoptotic transcripts. Gray bars, Genes with other functional attributes. (C) Scatter plot of fold-change expression values (log2 normalized) of breakthrough acute vs healthy controls (HCs) (x-axis) and hyperacute vs healthy controls (y-axis). Blue points denote those transcripts whose expression is above threshold values (log2 fold change < 0.5, and P < .05 using a moderated t statistic corrected using the Benjamini procedure) and whose expression in both breakthrough acute and hyperacute samples is overrepresented or underrepresented relative to healthy controls. Red points denote pivot transcripts, whose expression meets cutoffs noted above, and whose differential expression in breakthrough acute and hyperacute samples compared with healthy controls are in opposite directions. (D) Heatmap showing the expression levels of inflammatory Tc17-related genes identified in a study of fate-mapped murine Tc17 cells35 in the NHP breakthrough acute GVHD arrays compared with either autologous controls (Auto, left column) or healthy controls (HC, right column). The rows in the heatmap contain 24 genes previously identified as overrepresented (n = 12, top half of heatmap) and underrepresented (n = 12, bottom half of heatmap) in murine Tc17 cells during GVHD relative to controls. One gene (Ly6c1) identified in the murine study was not included in this analysis, as it has no human/NHP ortholog.36 The columns reveal the expression of these genes in the breakthrough acute cohort vs either autologous (left) or healthy controls (right). The column to the right of the gene names designates whether the direction of expression in breakthrough acute vs autologous or healthy controls is concordant (gray boxes) or not (white boxes) with the expression in murine Tc17 cells.
The identification of “pivot transcripts” define dichotomies between hyperacute and breakthrough acute GVHD
In order to further probe the mechanistic underpinnings of breakthrough acute GVHD in more detail, we performed a comparative bioinformatic analysis of hyperacute and breakthrough acute GVHD compared with the healthy control cohort, with a specific focus on identifying what we term the “pivot transcripts.” We define pivot transcripts (Figure 5C; supplemental Table 5) as those genes found to be significantly DE in T cells during both hyperacute and breakthrough acute GVHD when compared with healthy controls but in opposite directions. Given the polar variability in the expression of these 16 transcripts, they serve as distinguishing marks for the 2 immune states defined by breakthrough acute vs hyperacute disease (Figure 5C; supplemental Table 5). These transcripts defined a dichotomy between hyperacute and breakthrough acute GVHD that included a pivot around (1) cytotoxicity (SLAMF7,22 CMA1,23,24 CST7,25 each underrepresented in breakthrough acute GVHD), (2) apoptosis (FASL26,27 and IGF2R,28,29 proapoptotic genes underrepresented in breakthrough acute GVHD and KIAA132430 and IRS1,31,32 antiapoptotic genes overrepresented in breakthrough acute GVHD), and (3) Th/Tc17 programming (ADAM1233 and RORC,34 overrepresented in breakthrough acute disease).
Significant overlap between T-cell programming in breakthrough acute disease and murine inflammatory Tc17 cells
Given the suggestion from multiple transcriptomic avenues (Figures 4A, 5B-C) for Th/Tc17-biased transcriptional programming during breakthrough acute GVHD, we sought to compare the NHP transcriptome with a recently annotated 25-member gene list from fate-mapped Tc17 cells identified in a murine GVHD model.35 As shown in Figure 5D, significant overlap existed between the genes identified in mice and the NHP breakthrough acute GVHD transcriptome. Thus, of 25 Tc17 genes identified in murine GVHD,35 21 were identified as concordantly differentially expressed in either the breakthrough acute vs autologous control or the breakthrough acute vs healthy control comparison. Three were not concordant and 1 gene (Ly6c1) has no human/NHP ortholog36 and was therefore not included in the analysis. This result suggests strong interspecies mechanistic ties between the 2 GVHD models.
An unsupervised, systems approach to breakthrough acute GVHD transcriptomics reveals further evidence for inflammatory Th/Tc17 signaling
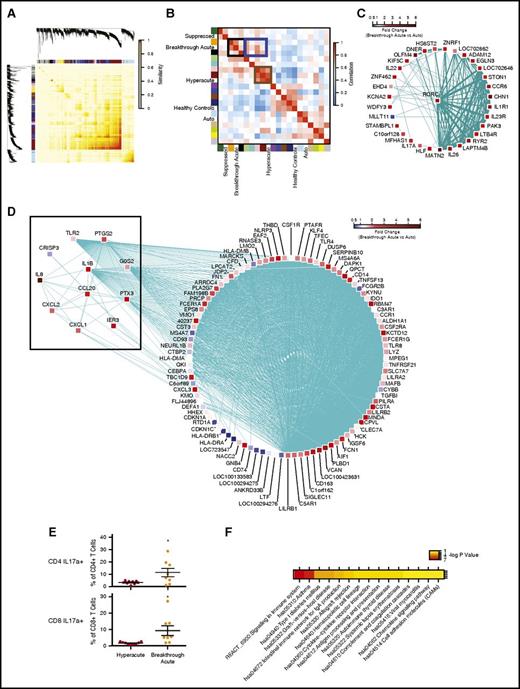
To deepen our understanding of the T-cell transcriptional program driving breakthrough acute GVHD, we applied weighted gene correlation network analysis (WGCNA)37 to construct a gene coexpression network from our data set. WGCNA is a computational method that has been used to perform large-scale organization of gene coexpression networks,38-40 and permits the construction of a gene correlation matrix (Figure 6A) from an entire data set. We were then able to perform hierarchical clustering of the matrix, identifying sets of genes that met a threshold of coexpression (representative examples of coexpression of RORC vs IL23R, IL1R1, CCR6, AHR, IL26, and IL22 are shown in supplemental Figure 3). This clustering analysis resulted in the identification of 16 discrete self-assembling modules (Figure 6B; supplemental Table 7). The module eigengenes41 (defined as a vector representation of the gene expression profile of a given module) were then clustered with clinical cohorts in order to identify metamodules (Figure 6B). We found that hyperacute GVHD clustered strongly in a metamodule with the dark-red and pink eigengenes (Figure 6B, brown box). The transcripts comprising these modules include canonical cytotoxic- and Th/Tc1-associated genes (supplemental Table 7). As predicted by the constellation mapping in Figure 4B, breakthrough acute GVHD clustered in a more complex metamodule, demonstrating the strongest statistical links to the orange and black eigengenes (Figure 6B, black box; supplemental Table 7) but with other links as well, including with the dark-red and pink eigengenes also associated hyperacute GVHD, as described in the previous section (Figure 6B blue box).
WGCNA reveals a Th/Tc17 transcriptional program mediating breakthrough acute GVHD in NHP. (A) Topological overlap matrix plot with hierarchical clustering tree and the resulting gene modules from a weighted network of T-cell transcripts. (B) Eigengene adjacency heatmap showing module eigengene similarity to NHP clinical cohorts. (C) Visualization of gene coexpression network connections between the most connected genes in the orange module using Cytoscape. Shown are nodes and network connections with topological overlap above a threshold of 0.05. Mean expression fold-change values of breakthrough acute vs autologous cohorts for each gene is visualized using a false-color scale. (D) Visualization of gene coexpression network connections between the most connected genes in the black module using Cytoscape. Shown are nodes with network connections whose topological overlap is above a threshold of 0.05. Edges with network connections above the threshold of 0.07 are shown. Mean expression fold change values of breakthrough acute vs autologous cohorts for each gene are visualized using a false-color scale. (E) Flow cytometric analysis of PBMCs at the time of terminal analysis stimulated with PMA/ionomycin and measured for the production of IL17a. *P < .05 using an unpaired t test. (F) Pathway enrichment for genes in the black WGCNA module performed using DAVID.59 Shown are those terms with a P < .05 (corrected for multiple hypothesis testing using the Benjamini procedure). Significance values are displayed using a false-color scale and are given in units of −log10 of the corrected P value.
WGCNA reveals a Th/Tc17 transcriptional program mediating breakthrough acute GVHD in NHP. (A) Topological overlap matrix plot with hierarchical clustering tree and the resulting gene modules from a weighted network of T-cell transcripts. (B) Eigengene adjacency heatmap showing module eigengene similarity to NHP clinical cohorts. (C) Visualization of gene coexpression network connections between the most connected genes in the orange module using Cytoscape. Shown are nodes and network connections with topological overlap above a threshold of 0.05. Mean expression fold-change values of breakthrough acute vs autologous cohorts for each gene is visualized using a false-color scale. (D) Visualization of gene coexpression network connections between the most connected genes in the black module using Cytoscape. Shown are nodes with network connections whose topological overlap is above a threshold of 0.05. Edges with network connections above the threshold of 0.07 are shown. Mean expression fold change values of breakthrough acute vs autologous cohorts for each gene are visualized using a false-color scale. (E) Flow cytometric analysis of PBMCs at the time of terminal analysis stimulated with PMA/ionomycin and measured for the production of IL17a. *P < .05 using an unpaired t test. (F) Pathway enrichment for genes in the black WGCNA module performed using DAVID.59 Shown are those terms with a P < .05 (corrected for multiple hypothesis testing using the Benjamini procedure). Significance values are displayed using a false-color scale and are given in units of −log10 of the corrected P value.
Having identified the orange and black eigengenes as strongly associated with breakthrough acute GVHD, we then used Cytoscape42 to visualize the significant coexpression patterns of the genes within these modules (Figure 6C-D). As shown in Figure 6C, expression of genes in the orange module was highly correlated with the expression of the canonical Th/Tc17 transcription factor, RORC (previously identified as a pivot transcript; Figure 5D), as well as with the RORC-related Th/Tc17 cytokines and their receptors IL17A, IL22, IL23R, IL26 among others.18,43 Given this strong Th/Tc17 transcriptional signal, we confirmed an increase in the proportion of IL17a-expressing CD4 and CD8 peripheral blood T cells in breakthrough acute relative to hyperacute GVHD using flow cytometry (Figure 6E).
In addition to the orange Th/Tc17-associated module, the black module was also strongly associated with breakthrough acute disease (Figure 6B,D). This module provided compelling evidence for inflammatory transcript enrichment in T cells during breakthrough acute disease, including the following inflammatory transcripts from the “Zhou_Inflammatory_response_FIMA_Up” gene set44 : IL8, CCL20, CXCL3, CXCL1, PTX3, IL1b, IER3, CRISP3, G0S2, TLR2, PTGS2, CXCL2 (highlighted in Figure 6D black box), the first 6 of which have been linked to Th/Tc17-mediated inflammation in both preclinical models and in human disease.45,46 The black module was also enriched for multiple other pathways of immune activation, including the KEGG and reactome “asthma,” “signaling in the immune system,” “GVHD” pathways as well as the KEGG “cytokine:cytokine receptor interaction” and “chemokine signaling” pathways (Figure 6F; supplemental Table 8).
Progressive enrichment of the orange and black modules after HCT in patients with GVHD
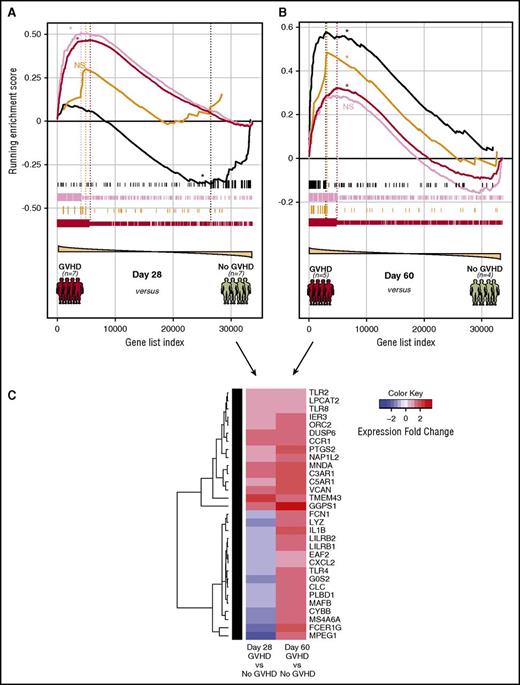
To interrogate the degree to which the NHP gene modules were represented in human patients with acute GVHD, we used GSEA to assay the enrichment of our NHP WGCNA modules in a pilot cohort of 23 posttransplant patients (12 with GVHD, 11 without GVHD). This cohort included patients diagnosed with GVHD either at day 28 ± 7 posttransplant or patients diagnosed at day 60 ± 7 posttransplant, along with time-matched controls (supplemental Table 3). As shown in Figure 7A, samples from patients who developed GVHD relatively early after transplant (by day 28 ± 7 days) exhibited statistically significant enrichment for the more Th/Tc1-dominated dark-red and pink modules (also associated with NHP breakthrough acute GVHD; Figure 6B blue box), without enrichment for the orange and black modules. In contrast, T cells purified from patients diagnosed with GVHD at day 60 ± 7 days had a distinct enrichment profile, with continued significant enrichment of the dark-red (but not the pink) module, and importantly, gain of enrichment for the orange and black modules (Figure 7B), with the highest degree of enrichment at this later time point in human samples for the inflammatory black module (heatmap demonstrating the evolution of enrichment of the black module in patients diagnosed at day 28 vs day 60 posttransplant shown in Figure 7C). The leading edge genes identified through GSEA (supplemental Table 9) suggest important commonalities between murine, NHP, and patient data. Thus, with respect to the orange module, patients with GVHD diagnosed at day 60 demonstrated enrichment for 3 key Tc17 transcripts (RORC, LTB4R, and CCR6), each of which was identified both in mice35 and in our NHP experiments (Figure 5E). Moreover, GSEA of the day 60 patient samples compared with the inflammatory black module also demonstrated important interspecies commonalities, with 6 of the Zhou_Inflammatory_response44 transcripts (IL1B, IER3, G0S2, TLR2, PTGS2, and CXCL2) enriched in both NHP and humans with GVHD. These data provide critical clinical corroboration of the NHP transcriptome data and suggest that an evolution of the mechanisms of GVHD pathogenesis may occur posttransplant, in both monkeys and humans.
Evolution of module enrichment in T cells from patients with GVHD. (A) Gene-set enrichment plot of selected NHP-WGCNA modules in patients who were diagnosed with GVHD at day 28 ± 7 posttransplant. Significant running enrichment scores were found in the dark-red and pink modules, but not for the orange and black modules for the day 28 samples. *FDR < 0.01. (B) Gene-set enrichment plot of selected NHP-WGCNA modules in patients who were diagnosed with GVHD day 60 ± 7 posttransplant. Significant running enrichment scores were found for both orange and black NHP gene modules at day 60 in patients with GVHD. Enrichment was also noted for the dark-red module, but not the pink module in the day 60 samples. *FDR < 0.01. (C) Heatmap showing the evolution of enrichment of the black module in patients diagnosed with GVHD at day 28 compared with day 60. Patients with GVHD were dichotomized into early GVHD (either at day 28 ± 7 posttransplant) or late GVHD (day 60 ± 7 posttransplant). Genes shown are those from the black NHP module exhibiting leading edge enrichment in patients at day 60 and their expression in either day 28 or day 60 GVHD patients relative to GVHD-negative controls.
Evolution of module enrichment in T cells from patients with GVHD. (A) Gene-set enrichment plot of selected NHP-WGCNA modules in patients who were diagnosed with GVHD at day 28 ± 7 posttransplant. Significant running enrichment scores were found in the dark-red and pink modules, but not for the orange and black modules for the day 28 samples. *FDR < 0.01. (B) Gene-set enrichment plot of selected NHP-WGCNA modules in patients who were diagnosed with GVHD day 60 ± 7 posttransplant. Significant running enrichment scores were found for both orange and black NHP gene modules at day 60 in patients with GVHD. Enrichment was also noted for the dark-red module, but not the pink module in the day 60 samples. *FDR < 0.01. (C) Heatmap showing the evolution of enrichment of the black module in patients diagnosed with GVHD at day 28 compared with day 60. Patients with GVHD were dichotomized into early GVHD (either at day 28 ± 7 posttransplant) or late GVHD (day 60 ± 7 posttransplant). Genes shown are those from the black NHP module exhibiting leading edge enrichment in patients at day 60 and their expression in either day 28 or day 60 GVHD patients relative to GVHD-negative controls.
Discussion
In this study, we present a T-cell transcriptional map of primate breakthrough acute GVHD developing in the setting of ongoing immunosuppression. This map has enabled us to identify the pathways that distinguish clinically relevant breakthrough acute GVHD from 3 key comparator groups: (1) healthy controls and autologous transplant controls (in which no GVHD occurs), (2) recipients who develop early, uncontrolled hyperacute GVHD, and (3) T cells from recipients that are purified at a time when they are suppressing GVHD, but who subsequently develop immune escape and breakthrough acute GVHD. Our results reveal striking transcriptional complexity in breakthrough acute GVHD that combines: (1) residual Th/Tc1 activation with (2) resistance to apoptosis, (3) a highly inflammatory transcriptional signature, and (4) skewing toward Th/Tc17 T-cell programing. Together, these results may provide a cogent explanation for the development of difficult-to-treat GVHD in immunosuppressed recipients, which is dominated not by rampant proliferation (as in hyperacute disease) but rather by T-cell persistence, inflammation, and Th/Tc17 programming. Our discovery that both an inflammatory signature and Th/Tc17 mechanisms evolve across divergent prophylaxis platforms also suggests that this may be an inherent attribute of T-cell immune escape during GVHD, rather than a regimen-specific mechanism.
Although many previous studies in mice have implicated Th/Tc1 cells as pathogenic mediators of disease (perhaps because murine studies are almost always designed to measure unprophylaxed hyperacute GVHD), there have been conflicting observations regarding the importance of Th/Tc17 cells to GVHD pathophysiology35,47-51 and few studies capable of identifying the more slow-to-evolve inflammatory nature of this disease. A recent murine study has shed important new light on the contribution that Tc17 cells, in particular the proinflammatory subset of this lineage, can make on GVHD. This study,35 which used sophisticated fate mapping, was able to track these cells in vivo even in unprophylaxed disease, despite their fleeting nature in mice. The amount of overlap of the transcriptional profile of these flow cytometrically purified murine T cells with T cells purified in the setting of clinically relevant breakthrough acute GVHD supports the mechanistic importance of these cells, and underscores the critical contribution of inflammatory T-cell pathophysiology during clinical disease. Lending further evidence for the diversity of Th/Tc17 immunopathology during acute GVHD is a recent description of a “Th17-prone” population of CD4+CD146+CCR5+ cells that can be detected prior to the onset of GI-GVHD.52
Given the time dependence of this shift in T-cell programing, these data also underscore the value of conducting in vivo GVHD experiments using a model that closely recapitulates the clinical milieu, where immune prophylaxis after transplantation is the rule and where detailed longitudinal analysis is feasible. Thus, likely due to the inclusion of serial sampling from recipients who ultimately developed breakthrough acute GVHD, as well as from pretransplant healthy controls and autologous transplant recipients, our NHP studies were sufficiently powered to complete a large series of statistical modeling and gene expression analyses. In contrast, the analysis that we could perform on the pilot patient cohort was restricted to GSEA, where statistical power is gained from the interrogation of gene sets rather than individual genes. This limitation resulted in the clinical data not being amenable to subset analysis of target organ involvement or immunoprophylaxis strategy used, which remain 2 key areas for future studies. Furthermore, differing immunoprophylactic strategies used in our NHP model (supplemental Table 2) and the human patients (supplemental Table 3) in this study likely limit strict gene-by-gene comparisons. Nevertheless, despite this limitation, the enrichment of the orange and black Th/Tc17 and inflammatory modules in day 60 patient arrays support the conclusions drawn from the NHP studies, which implicate the evolution of both signatures in GVHD. In interpreting the outcomes of this analysis, it is also important to note that the current study focused only on the peripheral blood T-cell transcriptome. The extent to which this is predictive of GVHD-target organ transcriptomics is currently unanswered. As such, these data suggest that a longitudinal transcriptional analysis of a large cohort of transplant patients (including both blood and target organ sampling) is an area of high priority for future investigations, in order to most accurately guide clinical investigation of novel GVHD prevention and treatment strategies.
In addition to the finding of inflammatory and Th/Tc17-driven mechanisms, the signature of antiapoptotic skewing in breakthrough acute GVHD T cells is of particular importance, and underscores the fact that classic models of proliferation-based T-cell pathogenicity derived from mouse models may not be predictive for clinical GVHD that occurs in the presence of ongoing immunosuppression. In this setting, and potentially more pronounced as time posttransplant lengthens, it may be the persistence rather than the proliferation of pathogenic T cells (through inhibition of apoptotic cell death) that is more responsible for pathology. This finding is consistent with the evidence of Th/Tc17 skewing during breakthrough acute GVHD, given that IL17-producing T cells are long-lived and have selected stem-cell like functional attributes.53-55 One of the salient questions that arises from this observation concerns the impact that GVHD treatment (with steroids as well as other modalities) makes on T-cell persistence and how this impacts effective vs ineffective GVHD control. It will be critical to determine the transcriptional networks associated with immune escape in the setting of corticosteroid treatment, which may shed new light on the mechanisms underlying life-threatening steroid-refractory GVHD, which occurs in some but not all patients. This is particularly relevant in the setting of a shift toward Th/Tc17 predominance, given that these cells have been shown to be more resistant to the proapoptotic effects of corticosteroid treatment.56-58
In addition to delineating the unique mechanisms driving both hyperacute and breakthrough acute GVHD, the data presented here provide a novel resource that takes a significant step forward toward enabling personalized, evidence-based decisions in GVHD diagnosis and treatment. These data provide critical support for the shifting landscape of T-cell immunopathology as time progresses posttransplant, and the first clues about how individual transcriptomes could, in the future, be compared with this transcriptional resource to drive patient-specific GVHD diagnosis and treatment decisions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge the technical assistance of Natalia Kozyr, the veterinary care provided by Elizabeth Strober, Joe Jenkins, Annie Torrence, Keith Vogel, and Jennifer Lane, in addition to the services provided by both the Vanderbilt Technologies for Advanced Genomics and the Oregon Health & Science University Gene Profiling Shared Resource. The authors are also grateful to Felix Wu and Jill Mesirov at the Broad Institute for their assistance with the constellation mapping software.
This work was supported by Yerkes National Primate Research Center base grant RR00165. The Washington National Primate Research Center at the University of Washington support was funded through the Office of Research Infrastructure Programs at the National Institutes of Health (NIH), Office of the Director (OD) grant P51 OD 010425. Funding was also provided through an Emory University Atlanta Clinical & Translational Science Institute pilot grant (E.K.W.), NIH National Heart, Lung, and Blood Institute (NHLBI) 2 R01 HL56067, National Institute of Allergy and Infectious Diseases (NIAID) R01 AI 34495 and P01 AI 056299 (B.R.B.), NHLBI 5 R01 HL095791, NIAID 5U19-AI051731 (L.S.K.), and Bristol-Myers Squibb (L.S.K.).
Authorship
Contribution: L.S.K. and B.R.B. conceived the study and designed experiments; S.N.F. developed computational methods; S.N.F., B.W., and V.T. analyzed the data; S.N.F., B.W., V.T., S.C., A.P.-M., K.B., M.B., D.J.H., J.B.S., K.Z., A.Y., C.R.G., E.K.W., J.S.M., B.R.B., and L.S.K. conducted the NHP and human experiments or provided clinical trial samples for analysis; and S.N.F., V.T., B.R.B., and L.S.K. wrote the paper with input from all of the authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leslie S. Kean, Ben Towne Center for Childhood Cancer Research, Seattle Children’s Research Institute, 1100 Olive Way, Suite 100, Seattle, WA 98101; e-mail: leslie.kean@seattlechildrens.org.
References
Author notes
S.N.F., B.W., and V.T. are joint first authors.

![Figure 2. Animals with breakthrough acute GVHD develop clinical and pathological evidence of disease without the specific T-cell signatures that develop during hyperacute GVHD. (A) Expression of Ki67 and granzyme B (GrmB) in T cells from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test, and significant differences of the mean percentage of either CD4 (*) or CD8 (†) cell subtypes were determined using an unpaired Student t test. All other time-point comparisons were not significant. (B) Engraftment/expansion of CD4 and CD8 TCM and TEM from peripheral blood, measured longitudinally posttransplant. Colored circles represent the clinical categories of hyperacute (red), suppressed (green), and breakthrough acute (orange) disease. Statistical differences between the hyperacute and suppressed cohorts were evaluated at days −7, 2, 5, 8, 12, 18, and 25 posttransplant using an unpaired Student t test. *Significant differences of the absolute numbers of the CD4 and CD8 T-cell subpopulations. All other time-point comparisons were not significant. (C) The combined GVHD pathology score (summarizing liver, colon, skin, and lung scores [top panel]) and combined upper GI GVHD score (summarizing esophagus, stomach, and duodenum [Duod.]; [bottom panel]) of recipients from hyperacute (MST = 12 days) and breakthrough acute (MST = 34.5 days) cohorts. Histopathologic analysis was performed at the time of terminal analysis and analyzed by a pathologist (A.P.-M.) in a blinded manner. *P < .05 using the Mann-Whitney test on composite scores. NS, not significant. (D) Flow cytometric analysis of mononuclear cells isolated from peripheral blood, lymphoid, and nonlymphoid GVHD-target organs at the time of terminal analysis from animals from hyperacute and breakthrough acute cohorts and stained for Ki-67 (top and middle panel) and for granzyme B (bottom panel). Plots depict the percentages of CD4 and CD8 T cells expressing Ki-67 and the percentage of CD8 T cells expressing high levels of granzyme B (mean fluorescence intensity [MFI] ≥ 105). *P < .05 using the Mann-Whitney test. LN, lymph node.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/21/10.1182_blood-2016-07-726547/4/m_blood726547f2.jpeg?Expires=1769777456&Signature=KUv7He72sNSDyJWTLsx0adGLnjyfLZk~RIzwjWNMgYACo9JBJSuKGTL-3JBSQOuBvhKsw0vPWx57pjnDZ-Mi4tp0YCqt9IC9PPpw1sdrZZkeusAKSJzh582cC~Q3WWjmDoc4A6OXwYHdGTayjNCO1fxa8DChpnBUthvpgpfYoTLZRk8qCEKZWKJnV2w7DyNVVyp8sf1r8E48g3ZuIVC49aV8MKf2wBwDtze5pfaiyorR2CSjVZTjGWD2HMy2AsWomuHgj8uvHuWeuA28FQRiZozKfq2H0d-2FhP-6l0X~fdcIQ3L4c4bhPEJ5~wJqh5YD~o91KZF0vq12TG64Zhl0w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. T-cell profiles from animals with hyperacute GVHD contain an abundance of transcripts associated with proliferation, and exhibit Th1 skewing. (A) First and third principal component projections reveal clustering of transplanted animals by immunoprophylactic strategy. Each dot represents an array sample. The center of inertia ellipses corresponds to the mean projections of the group (P < .05). (B) The first principal component shows a significant correlation with survival in a linear regression model (adjusted [Adj.], R2 = 0.3299; P < .004). (C) Heatmap of the top 25 gene sets enriched in the hyperacute cohort using ssGSEA. The constellation map to the right of the heatmap allows for the identification of clusters of these gene sets. Those gene sets plotted closer to the center have a greater degree of similarity to the phenotype of interest (measured by the normalized mutual information [NMI] score) whereas the angular distance corresponds to gene-set similarity with one another. A dark-green edge further indicates gene-set similarity with the thickness of the line proportional to the Jaccard index.10,11 (D) GSEA of transcripts previously shown to differentiate Th1 from Th17 cells.12 These molecules were found to be overrepresented in T cells from animals with hyperacute GVHD compared with healthy controls and the breakthrough acute cohort. *False discovery rate [FDR] < 0.01. (E) Flow cytometric analysis of PBMCs at the time of terminal analysis stimulated with phorbol myristate acetate (PMA)/ionomycin and measured for the production of IFNγ. *P < .05 using an unpaired t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/21/10.1182_blood-2016-07-726547/4/m_blood726547f3.jpeg?Expires=1769777456&Signature=coXDMRkVaW7JtnCHbh5tsAvEGxJ1DOzvN9WxPpY3IZNnAlyzpvuVFsOqx~sYOwqXPleENmh5ejwmJqE8tZBi3k0Gy2beBu9wUjF98y1DGy17HYoyETN3SgzXq9o4aUG5Tst0kApam342x~KhwiEZla-ZSJC9j7hNW-GQNuVpqwMOD-5-1XBezj65ONdz-FI9T0FVgW-ArDdd6bHQxYMczsMOyZVMtg9wTPJqlIbMkAUkk9hESHbVnz022zXrY5nKdJ39NOsjnQU0Dt539rnI7W4S7B3mzueCGIhob59V4EllzIb1pw5lWeON1ec4z3Lh2V~e6CS9MfDZ9Z99IN7l4g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)