Key Points
INF-α promotes engraftment of mid-gestation HSCs.
INF-α can restore the hematopoietic defect of Arid3a knockout embryos.
Abstract
In the developing mouse embryo, the first hematopoietic stem cells (HSCs) arise in the aorta-gonad-mesonephros (AGM) and mature as they transit through the fetal liver (FL). Compared with FL and adult HSCs, AGM HSCs have reduced repopulation potential in irradiated adult transplant recipients but mechanisms underlying this deficiency in AGM HSCs are poorly understood. By co-expression gene network analysis, we deduced that AGM HSCs show lower levels of interferon-α (IFN-α)/Jak-Stat1–associated gene expression than FL HSCs. Treatment of AGM HSCs with IFN-α enhanced long-term hematopoietic engraftment and donor chimerism. Conversely, IFN-α receptor–deficient AGMs (Ifnαr1−/−), had significantly reduced donor chimerism. We identify adenine-thymine–rich interactive domain-3a (Arid3a), a factor essential for FL and B lymphopoiesis, as a key transcriptional co-regulator of IFN-α/Stat1 signaling. Arid3a occupies the genomic loci of Stat1 as well as several IFN-α effector genes, acting to regulate their expression. Accordingly, Arid3a−/− AGM HSCs had significantly reduced transplant potential, which was rescued by IFN-α treatment. Our results implicate the inflammatory IFN-α/Jak-Stat pathway in the developmental maturation of embryonic HSCs, whose manipulation may lead to increased potency of reprogrammed HSCs for transplantation.
Introduction
In the developing mouse embryo, the first hematopoietic stem cells (HSCs), defined by their ability to repopulate lethally irradiated adult recipients, are detected in the aorta-gonad-mesonephros (AGM) at embryonic day 11.5 (E11.5).1,2 These HSCs later migrate to the fetal liver (FL) before reaching the bone marrow (BM) to sustain adult hematopoiesis. HSCs at various embryonic stages differ molecularly and functionally, reflecting a developmental maturation program. When their expression profiles are analyzed by a Bayesian classifier, AGM HSCs resemble macrophages and are enriched for gene-ontology terms such as “cell communication,” and “positive regulation of angiogenesis,” reflecting their migratory proclivity and endothelial origin.3 Such a molecular signature captures a known phenomenon termed the “endothelial-to-hematopoietic transition,” in which AGM-derived HSCs emerge through an endothelial intermediate.4-8 In contrast, FL HSCs resemble BM HSCs and their gene expression signature clusters away from AGM HSCs.3 When competing against BM cells, AGM HSCs demonstrate reduced repopulation potential in irradiated adult recipients compared with FL and BM HSCs.9 In contrast, transplantation of AGM HSCs is more permissive in neonatal than adult recipients.9 Hence, AGM HSCs are functionally less “mature” than FL and BM HSCs in their capacity to engraft in the adult BM niche.
One of several known regulators of HSC development is Sox17, a transcription factor that is required for the maintenance of embryonic and neonatal HSCs but not adult HSCs.10 However, the signaling pathways that promote the developmental maturation of AGM HSCs remain poorly characterized. Understanding the differences between AGM and adult HSCs is an important and challenging goal. Initial studies using HSCs derived from pluripotent stem cells via ectopic induction of the homeobox gene Hoxb4 demonstrated long-term primary and secondary transplant capability; however, skewed myeloid lineage potential and aberrant surface antigen phenotypes suggested developmental immaturity.11,12 Attempts to faithfully recapture HSCs via transgene induction from pluripotent cell-derived sources revealed similar defects in lymphoid reconstitution.13,14 Curiously, HSCs reprogrammed from committed adult blood cells seemed unaffected by these limitations.15 The correlation between the cell-of-origin of these induced HSCs and the nature of identified developmental defects highlights our imperfect understanding of the molecular programs governing the difference between AGM and adult HSCs.
Recent studies have suggested that inflammatory pathways mediated by interferon-γ (IFN-γ) and tumor necrosis factor (TNF) signaling are important for HSC emergence in the AGM.16-18 Here we focus on a distinct role of interferon-α (IFN-α) and its effects on the functional maturation of AGM HSCs.
Materials and methods
Mouse embryo culture
E11.5 staged mouse embryos were obtained using timed pregnancies of C57BL/6 females, unless indicated otherwise. The AGM region was isolated under a dissecting microscope, dissociated to single cells using DNase I, collagenase IV, and hyaluronidase for 15 minutes at 37°C and washed with Iscove modified Dulbecco medium (IMDM).19 Dissociated cells were cultured with IFN-α (PBL IFN source #12100-1 or Sigma #I8782) for 1.5 hours in 10% fetal calf serum/IMDM (v/v) in a 96-well V-bottom plate. The cells were then dissociated with enzyme-free dissociation buffer (Gibco) and washed with IMDM before further analysis.
Transplantation and peripheral blood analysis
B6.SJL-Ptprca Pepcb/Boy mice were used at 6 to 10 weeks of age. Mice were irradiated with a split dose of 10 Gy separated by 2.5 hours prior to transplantation. Each recipient was transplanted via tail vein injection along with red blood cell (RBC)-lysed 2 × 105 splenic helper cells per experiment group from B6.SJL-Ptprca Pepcb/Boy mice unless indicated otherwise. Peripheral blood was collected retro-orbitally at the indicated time points posttransplantation. RBCs were removed with 1% dextran sulfate/0.5% EDTA/phosphate-buffered saline (w/w/v) and lysed with RBC lysis buffer (Sigma) before analysis.
Statistical analysis
The number of biological replicates is represented by “n.” Two-tailed unpaired Student t tests were used, unless indicated. Error bars show standard deviation unless indicated otherwise. Statistical significance is indicated by *P < .05; **P < .01; ***P < .001.
Further experimental procedures are described in supplemental Materials and Methods, available on the Blood Web site.
Results
Jak-Stat correlates with HSC development in weighted gene co-expression network analysis (WGCNA)
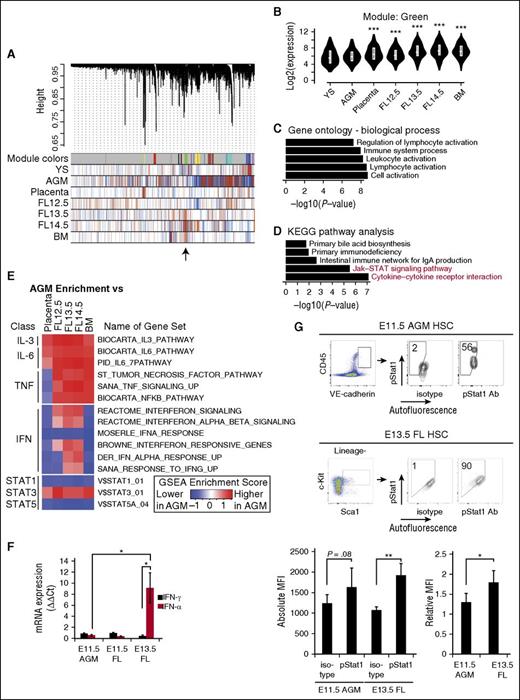
To identify co-regulated sets of genes (called modules) during hematopoietic development, we performed a WGCNA of our previously published microarray data set for HSCs at different developmental stages.3,20 In contrast to McKinney-Freeman et al3 in which 66 modules were reported, we merged similar modules to identify a single module that was expressed at low levels in AGM HSCs but higher levels in FL and adult BM HSCs, which yielded greater statistical power in further analyses (Figure 1A-B). Through gene ontology analysis,21 we identified enrichment for terms such as “immune system process,” “leukocyte activation,” and “lymphocyte activation” (Figure 1C). Similarly, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis identified the two highest enriched terms as “Jak-Stat signaling pathway” and “cytokine-cytokine receptor interaction” (Figure 1D).22 Thus, we determined that a co-expressed gene set is linked to the Jak-Stat signaling pathway, accounting for key differences between AGM and FL/adult BM HSCs.
Screen for pathways corresponding to developmental HSC maturation reveals Jak-Stat1. (A) WGCNA. The horizontal bar represents all genes in the sample. Identified co-expressed genes (modules) are assigned individual colors in the first row. In subsequent rows, red indicates upregulation and blue indicates downregulation in each sample. The module of interest, which shows low expression in the AGM and higher expression in the FL and BM is indicated by an arrow. (B) Expression levels of genes in this module identified in (A) during embryonic development. (C) Top 5 gene ontology biological process terms identified from genes in this module suggesting inflammatory pathway activation. (D) Top 5 KEGG pathway terms identified from genes in this module. Jak-Stat pathways are shown in red. (E) GSEA of samples displayed in columns compared against AGM HSCs. Red indicates that the gene set is more highly enriched in the AGM compared with tissues indicated, whereas blue indicates that the gene set is less enriched. The color scale is determined by the absolute enrichment score from the GSEA. (F) Quantitative RT-PCR for IFN-α and IFN-γ transcripts in whole E11.5 AGM, and E11.5 and E13.5 FL; n = 4. IFNs are detected in the AGM at Ct values <30, confirming their presence. (G) Percent of E11.5 AGM (VE-cadherin+CD45+) and E13.5 FL HSCs (Lineage−Sca1+c-Kit+), which are positive for intracellular staining of phospho-Stat1 (pacific orange). Absolute MFI for phospho-Stat1 is quantified. Relative MFI was determined by dividing MFI of sample by that of the IgG isotype control; n = 5-7. Statistical significance: *P < .05; **P < .01; ***P < .001. Ct, cycle threshold; IgA, immunoglobulin A; mRNA, messenger RNA; YS, yolk sac.
Screen for pathways corresponding to developmental HSC maturation reveals Jak-Stat1. (A) WGCNA. The horizontal bar represents all genes in the sample. Identified co-expressed genes (modules) are assigned individual colors in the first row. In subsequent rows, red indicates upregulation and blue indicates downregulation in each sample. The module of interest, which shows low expression in the AGM and higher expression in the FL and BM is indicated by an arrow. (B) Expression levels of genes in this module identified in (A) during embryonic development. (C) Top 5 gene ontology biological process terms identified from genes in this module suggesting inflammatory pathway activation. (D) Top 5 KEGG pathway terms identified from genes in this module. Jak-Stat pathways are shown in red. (E) GSEA of samples displayed in columns compared against AGM HSCs. Red indicates that the gene set is more highly enriched in the AGM compared with tissues indicated, whereas blue indicates that the gene set is less enriched. The color scale is determined by the absolute enrichment score from the GSEA. (F) Quantitative RT-PCR for IFN-α and IFN-γ transcripts in whole E11.5 AGM, and E11.5 and E13.5 FL; n = 4. IFNs are detected in the AGM at Ct values <30, confirming their presence. (G) Percent of E11.5 AGM (VE-cadherin+CD45+) and E13.5 FL HSCs (Lineage−Sca1+c-Kit+), which are positive for intracellular staining of phospho-Stat1 (pacific orange). Absolute MFI for phospho-Stat1 is quantified. Relative MFI was determined by dividing MFI of sample by that of the IgG isotype control; n = 5-7. Statistical significance: *P < .05; **P < .01; ***P < .001. Ct, cycle threshold; IgA, immunoglobulin A; mRNA, messenger RNA; YS, yolk sac.
Multiple cytokines, including interleukins (ILs), TNF, and IFNs, can activate the Jak-Stat signaling pathway, and many of these pathways have known roles in inflammation. Recent reports indicate that inflammatory pathways mediated by TNF and IFN-γ are required for the emergence of AGM HSCs.16-18 Therefore, we performed gene set enrichment analyses (GSEA) to analyze gene sets differentially expressed in AGM HSCs compared with FL and adult BM HSCs in response to ILs, TNF, and IFNs (Figure 1E).23 Because numerous Stat proteins exist, we examined Stat1, 3, and 5A responsive gene sets (Figure 1E). AGM HSCs are enriched for the activation of the IL-3 and IL-6 signaling pathways as well as their downstream target Stat3 as compared with FL and adult HSCs (Figure 1E), consistent with published reports implicating IL-3 and IL-6 in AGM HSC emergence.24,25 The IFN pathway, including the downstream Stat1-responsive genes, had lower enrichment scores in the AGM compared with that of the IL-3 and IL-6 signaling pathways (Figure 1E). Collectively, the informatics analyses predicted that the Jak-Stat pathway mediated by IFN/Stat1 signaling is low in AGM HSCs, albeit not absent.
Increasing IFN-α signaling during development
To validate this prediction, we examined IFN-α and IFN-γ expression levels in the E11.5 AGM and E13.5 FL via quantitative reverse-transcription polymerase chain reaction (RT-PCR) using whole tissues (Figure 1F). IFN-α and IFN-γ were expressed at low levels in the AGM but IFN-α was expressed at higher levels in the FL (Figure 1F; supplemental Figure 1A-B). To examine phospho-Stat1 expression in individual phenotypic HSCs, we performed intracellular flow cytometry on E11.5 vascular endothelial (VE)-cadherin+CD45+ AGM HSCs and E13.5 Lineage−Sca1+c-Kit+ (LSK) FL HSCs (Figure 1G; supplemental Figure 1C-D).3 In agreement with the informatics analysis (Figure 1A-E), AGM HSCs had less phospho-Stat1 as compared with FL HSCs via relative median fluorescence intensity (MFI) (Figure 1G).
Hematopoietic cells respond to IFN-α signaling
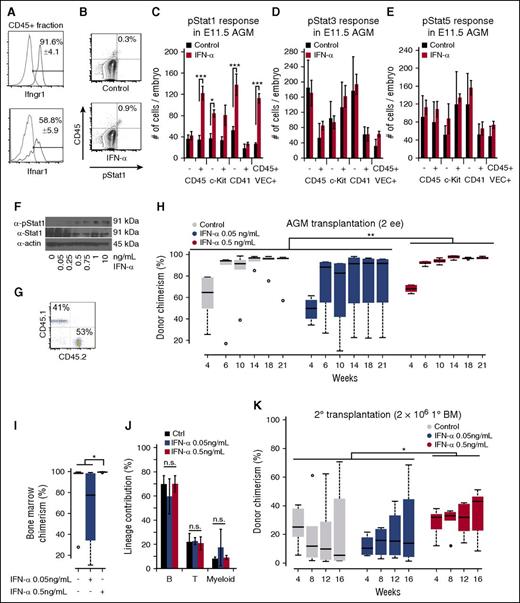
To determine which cells in the AGM are responsive to IFNs, we performed flow cytometry for Ifnαr1 and Ifnγr1, receptors for IFN-α and IFN-γ, respectively. Both receptors were expressed in the majority of hematopoietic cells (Figure 2A). To determine which Stat proteins become phosphorylated in response to IFN-α, we incubated AGM cells with IFN-α (0.5 ng/mL) for 90 minutes, an established duration for inflammatory molecules such as prostaglandin E2 (PGE2).26 Upon IFN-α treatment, hematopoietic CD45+ cells including VE-cadherin+CD45+ HSCs responded through phosphorylation of Stat1 (Figure 2B-C; supplemental Figure 1E-G). CD41− cells, but not CD41+ cells, responded significantly to IFN-α (Figure 2C).27 IFN-α also uniquely phosphorylated Stat1 as opposed to Stat3 or Stat5, indicating that IFN-α signaling is Stat1-mediated (Figure 2C-E). Taken together, our data indicate that AGM HSCs have low levels of Jak-Stat1 signaling but are poised to respond to IFN-α signaling.
IFN-α treatment promotes long-term engraftment of AGM HSCs. (A) Flow cytometry for IFN receptors Ifnγr1 (top) and Ifnαr1 (bottom), which are present in CD45+ cells of the AGM; n = 3. (B) Example of phospho-Stat1 activation in CD45+ E11.5 AGM cells treated with IFN-α (bottom) with isotype control (top). (C) Phospho-Stat1+ cells in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (D) Phospho-Stat3+ response in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (E) Phospho-Stat5+ response in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (F) Immunoblot for phospho-Stat1 in response to dose-titrations of IFN-α in adult splenocytes. (G) Example of donor chimerism analysis for multilineage engraftment. (H) Boxplots showing the effect of IFN-α on long-term hematopoietic engraftment of E11.5 AGM HSCs in the peripheral blood; 2 e.e. were transplanted with 2 × 105 helper splenocytes. Two-way analysis of variance (ANOVA) was performed. (I) Boxplots showing the effect of IFN-α on long-term hematopoietic engraftment of AGM HSCs in the BM at 21 weeks posttransplantation. Wilcoxon rank-sum test was performed; n = 4-6. (J) Quantification of lineage contributions of B, T, and myeloid cells at 21 weeks posttransplantation in the peripheral blood; n = 4-6. (K) Boxplots showing engraftment from secondary transplantation of 2 × 106 BM cells from IFN-α–treated and control AGMs in (H) with 3 × 105 competitor BM cells. Two-way ANOVA was performed. Statistical significance: *P < .05; **P < .01; ***P < .001. n.s., not significant.
IFN-α treatment promotes long-term engraftment of AGM HSCs. (A) Flow cytometry for IFN receptors Ifnγr1 (top) and Ifnαr1 (bottom), which are present in CD45+ cells of the AGM; n = 3. (B) Example of phospho-Stat1 activation in CD45+ E11.5 AGM cells treated with IFN-α (bottom) with isotype control (top). (C) Phospho-Stat1+ cells in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (D) Phospho-Stat3+ response in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (E) Phospho-Stat5+ response in the different cell compartments in the E11.5 AGM in response to IFN-α. The column indicates the absolute number of positive cells per embryo; n = 3. (F) Immunoblot for phospho-Stat1 in response to dose-titrations of IFN-α in adult splenocytes. (G) Example of donor chimerism analysis for multilineage engraftment. (H) Boxplots showing the effect of IFN-α on long-term hematopoietic engraftment of E11.5 AGM HSCs in the peripheral blood; 2 e.e. were transplanted with 2 × 105 helper splenocytes. Two-way analysis of variance (ANOVA) was performed. (I) Boxplots showing the effect of IFN-α on long-term hematopoietic engraftment of AGM HSCs in the BM at 21 weeks posttransplantation. Wilcoxon rank-sum test was performed; n = 4-6. (J) Quantification of lineage contributions of B, T, and myeloid cells at 21 weeks posttransplantation in the peripheral blood; n = 4-6. (K) Boxplots showing engraftment from secondary transplantation of 2 × 106 BM cells from IFN-α–treated and control AGMs in (H) with 3 × 105 competitor BM cells. Two-way ANOVA was performed. Statistical significance: *P < .05; **P < .01; ***P < .001. n.s., not significant.
IFN-α enhances AGM HSC transplantation
Previous work on IFN-γ signaling in mouse AGMs demonstrated that IFN-γ receptor deficiency reduces HSC emergence, although limiting dilution assays on IFN-α were lacking.16 IFN-α is expressed at low levels in the AGM and at higher levels in E13.5 FL (Figure 1F). We investigated its effect at E11.5, a stage at which HSCs that can engraft irradiated adults first, arise in the embryo.1,9 Because Stat1 signaling increases as AGM HSCs develop into FL HSCs (Figure 1E,G), we hypothesized that IFN-α signaling might promote the engraftment of AGM HSCs in adult recipients.
In adult HSCs, acute IFN-α treatment in vivo drives dormant HSCs into active cell-cycle progression, whereas chronic IFN-α treatment prompts HSC exhaustion.28 Because IFN-α exhibits dose sensitivity,29 we titrated the dose according to Stat1 phosphorylation levels in adult splenocytes and determined that 0.5 ng/mL resulted in an optimal phosphorylation (Figure 2F).
Next, we transplanted AGM HSCs from 2 embryo equivalents (e.e.) exposed for 90 minutes to 2 doses of IFN-α (0.05 ng/mL and 0.5 ng/mL) into irradiated adult mice without accompanying cells (Figure 2G-H). IFN-α treatment (0.5 ng/mL) promoted long-term engraftment at 21 weeks posttransplant as compared with control and subthreshold dose (0.05 ng/mL) (Figure 2H), although the difference was modest at this non-limiting dose of 2 e.e. in the absence of competitor cells due to BM saturation (Figure 2I). No significant lineage skewing occurred at 21 weeks posttransplant (Figure 2J). We also performed secondary transplantations using 2 × 106 BM cells derived from the primary transplants along with 3 × 105 recipient BM cells. At 12 weeks posttransplantation, recipients treated with the optimized dose of IFN-α had significantly higher donor chimerism than those treated with the suboptimal or control dose (Figure 2K). Collectively, our results show that IFN-α enhances long-term transplantation of AGM HSCs in irradiated adult recipients.
Increased competitiveness of IFN-α–treated AGM HSCs
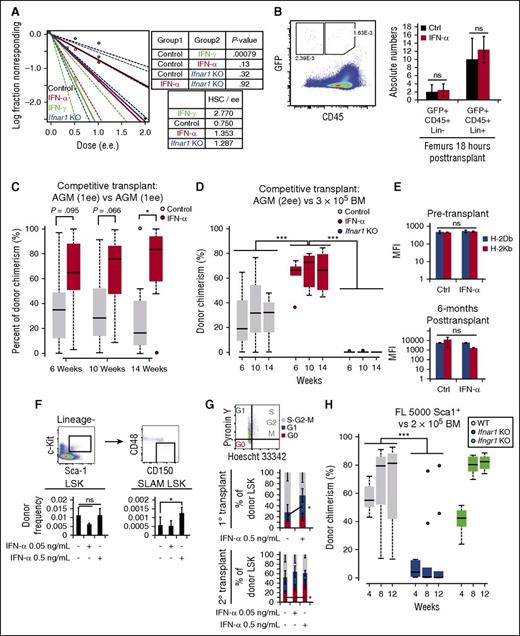
The above results prompted us to revisit whether IFN-α treatment had an effect on stem cell frequency, homing, or competitive engraftment potential. Thus, we performed limiting dilution assays with IFN-α–treated and untreated WT AGMs but observed no significant differences in the stem cell frequency in contrast to IFN-γ treatment, which increased stem cell frequency (Figure 3A).16 Similar results were obtained with Ifnαr1−/− AGMs, which lack the IFN-α receptor (Figure 3A).30
IFN-α enhances competitive transplant by enhancing HSC quiescence without affecting stem cell frequency or homing. (A) Limiting dilution assays with IFN-γ–treated, IFN-α–treated, untreated wild-type (WT), or Ifnαr1−/− E11.5 AGMs; n = 13 for a total of 89 mice. (B) Homing assay for detection of CD45+GFP+Lin+ and CD45+GFP+Lin− in the BM 18 hours posttransplantation; n = 5 (right). Representative flow cytometry plot showing gating for CD45 and GFP (left); n = 5. (C) Boxplots showing donor chimerism attributable to control or IFN-α treatment during AGM transplantation. (D) Boxplots showing engraftment from competitive transplant with IFN-α–treated, untreated, or Ifnαr1−/− AGMs against 3 × 105 competitor BM cells. Two-way ANOVA was performed. (E) MFI of MHC class I molecules detected on donor-derived hematopoietic cells 6 hours after IFN-α treatment (top) or in the periphery at 6 weeks posttransplantation (bottom); n = 3-6. (F) Cell-cycle analysis of the donor-derived (CD45.2+) LSK HSCs in the BM of transplantations involving IFN-α–treated or untreated AGMs. Samples were analyzed at 4 weeks posttransplant in the primary recipients (n = 3; middle) or 36 weeks posttransplant in the secondary recipients (n = 4-5; bottom) by flow cytometry with Pyronin Y and Hoescht 33342 (top); n = 4-5. (G) Cell-cycle analysis of the donor-derived (CD45.2+) LSK HSCs in the BM of transplantations involving IFN-α–treated or untreated AGMs. Samples were analyzed at 4 weeks posttransplant in the primary recipients (n = 3) or 36 weeks posttransplant in the secondary recipients (n = 4-5) by flow cytometry with Pyronin Y and Hoescht 33342. (H) Boxplots showing engraftment from competitive transplant with 5000 Sca1+ cells from E14.0 FL from WT, Ifnαr1−/−, or Ifnγr1−/− against 2 × 105 competitor BM cells. Two-way ANOVA was performed. Statistical significance: *P < .05; ***P < .001. ns, not significant; SLAM, signaling lymphocyte activation molecules.
IFN-α enhances competitive transplant by enhancing HSC quiescence without affecting stem cell frequency or homing. (A) Limiting dilution assays with IFN-γ–treated, IFN-α–treated, untreated wild-type (WT), or Ifnαr1−/− E11.5 AGMs; n = 13 for a total of 89 mice. (B) Homing assay for detection of CD45+GFP+Lin+ and CD45+GFP+Lin− in the BM 18 hours posttransplantation; n = 5 (right). Representative flow cytometry plot showing gating for CD45 and GFP (left); n = 5. (C) Boxplots showing donor chimerism attributable to control or IFN-α treatment during AGM transplantation. (D) Boxplots showing engraftment from competitive transplant with IFN-α–treated, untreated, or Ifnαr1−/− AGMs against 3 × 105 competitor BM cells. Two-way ANOVA was performed. (E) MFI of MHC class I molecules detected on donor-derived hematopoietic cells 6 hours after IFN-α treatment (top) or in the periphery at 6 weeks posttransplantation (bottom); n = 3-6. (F) Cell-cycle analysis of the donor-derived (CD45.2+) LSK HSCs in the BM of transplantations involving IFN-α–treated or untreated AGMs. Samples were analyzed at 4 weeks posttransplant in the primary recipients (n = 3; middle) or 36 weeks posttransplant in the secondary recipients (n = 4-5; bottom) by flow cytometry with Pyronin Y and Hoescht 33342 (top); n = 4-5. (G) Cell-cycle analysis of the donor-derived (CD45.2+) LSK HSCs in the BM of transplantations involving IFN-α–treated or untreated AGMs. Samples were analyzed at 4 weeks posttransplant in the primary recipients (n = 3) or 36 weeks posttransplant in the secondary recipients (n = 4-5) by flow cytometry with Pyronin Y and Hoescht 33342. (H) Boxplots showing engraftment from competitive transplant with 5000 Sca1+ cells from E14.0 FL from WT, Ifnαr1−/−, or Ifnγr1−/− against 2 × 105 competitor BM cells. Two-way ANOVA was performed. Statistical significance: *P < .05; ***P < .001. ns, not significant; SLAM, signaling lymphocyte activation molecules.
We next investigated whether IFN-α treatment, as with other inflammatory molecules such as PGE2, could promote homing of AGM HSCs to the BM.26 We transplanted green fluorescent protein (GFP+) AGMs (3 e.e.) from Ubiquitin-GFP transgenic mice, which allowed us to recover GFP+CD45+Lineage− cells from the BM 18 hours posttransplantation, a time at which the majority of stem and progenitor cells have not undergone cell divisions.31-33 No significant differences in homing were observed after IFN-α treatment (Figure 3B).
We then performed competitive transplants with AGMs derived from WT or Ubiquitin-GFP transgenic mice on the C57Bl/6 background using IFN-α–treated vs control AGMs (Figure 3C). AGM HSCs treated with 0.5 ng/mL IFN-α displayed increased donor chimerism over untreated, which was significant at 14 weeks posttransplant.
In our previous study, we noted a decrease in engraftment when AGMs were transplanted with 3 × 105 adult BM cells.9 We repeated these AGM transplantations with 2 e.e. in the presence of 3 × 105 adult BM competitor cells and observed that IFN-α treatment enhanced donor chimerism (Figure 3D). Conversely, transplantation of Ifnαr1−/− AGMs in the presence of adult BM competitor cells resulted in a decrease (Figure 3D).
A possible explanation for the increased donor chimerism observed for IFN-α–treated HSCs in irradiated adults is that major histocompatibility complex (MHC) class I antigens, which are normally expressed as early as E9 at low levels in midgestation embryonic tissues, are upregulated and prevent natural killer-mediated destruction.34,35 However, immediately before transplantation or at 6 weeks posttransplantation, we observed no significant changes in expression of strain-specific MHCb: H-2Kb and H-2Db (Figure 3E; supplemental Figure 2A).
Instead, the increased donor chimerism is likely explained by the increased frequency of donor-derived long-term CD150+CD48− HSCs in the BM (Figure 3F), and the corresponding increase in the percentage of quiescent HSCs (Figure 3G).36
Lastly, we hypothesized that a decrease in engraftment would be observed with transplantation of Ifnαr1−/− FL HSCs (Figure 1F-G). Because IFN signaling can affect Sca1 expression, which has a role in competitive transplantation,28,37 we normalized cell number by this marker and transplanted 5000 Sca1+ cells from the WT, Ifnαr1−/−, or Ifnγr1−/− E14 FLs with 2 × 105 adult BM cells. Hematopoietic engraftment from the Ifnαr1−/− but not Ifnγr1−/− FL was significantly lower than that of the WT (Figure 3H; supplemental Figure 2B).
Partial maturation of AGM HSCs by IFN-α treatment
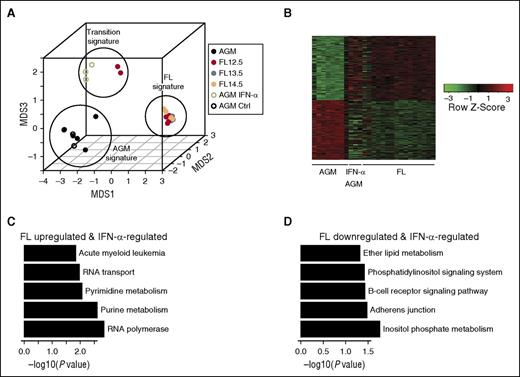
We previously observed that certain samples of E12.5 FL HSCs resemble E11.5 AGM HSCs, whereas others resemble adult-like E13-14.5 FL HSCs,3 suggesting a population in transition. By expression analysis, we observed that IFN-α–treated E11.5 AGM HSCs cluster with this transitional state of E12.5 FL HSCs (Figure 4A). Upon examining differentially expressed genes between E13-14.5 FL and AGM HSCs, we find that a modest 28% of these genes are stimulated by IFN-α treatment, suggesting partial maturation (Figure 4B). Among FL upregulated and IFN-α–responsive genes, nucleotide metabolism and RNA polymerase regulation were the top pathways identified (Figure 4C). Among FL downregulated and IFN-α–responsive genes, adherens junction and inositol phosphate pathways were the top pathways identified (Figure 4D).3,38,39
Microarray analysis shows partial maturation of IFN-α–treated AGM HSCs. (A) MDS shows 3 distinct groups of embryonic HSCs maturation, which correspond to AGM, “transition,” and FL signatures. IFN-α–treated AGM HSCs and subset of E12.5 FL HSCs are grouped as having this “transition” signature. Data from this study is depicted by open circles, whereas data from McKinney-Freeman et al (GSE37000) is depicted by closed circles. (B) Intersection of differentially expressed genes between E13.5-14.5 FL HSCs and AGM, and between AGM and IFN-α–treated AGM (nominal P value cutoff .05). (C) Top 5 KEGG pathway terms identified from FL upregulated and IFN-α–regulated gene set from (B). (D) Top 5 KEGG pathway terms identified from FL downregulated and IFN-α–regulated gene set from (B). MDS, multidimensional scaling.
Microarray analysis shows partial maturation of IFN-α–treated AGM HSCs. (A) MDS shows 3 distinct groups of embryonic HSCs maturation, which correspond to AGM, “transition,” and FL signatures. IFN-α–treated AGM HSCs and subset of E12.5 FL HSCs are grouped as having this “transition” signature. Data from this study is depicted by open circles, whereas data from McKinney-Freeman et al (GSE37000) is depicted by closed circles. (B) Intersection of differentially expressed genes between E13.5-14.5 FL HSCs and AGM, and between AGM and IFN-α–treated AGM (nominal P value cutoff .05). (C) Top 5 KEGG pathway terms identified from FL upregulated and IFN-α–regulated gene set from (B). (D) Top 5 KEGG pathway terms identified from FL downregulated and IFN-α–regulated gene set from (B). MDS, multidimensional scaling.
Adenine-thymine–rich interactive domain-3a (Arid3a) transcription factor is expressed in the AGM
To screen for co-regulators of IFN-α, we used a previously published microarray of HSCs collected after in vivo exposure to IFN-α.28 The transcription factors Arid3a, Irf7, and Id3 correlated with Stat1 activation after IFN-α (Figure 5A). We picked Arid3a as a candidate because of its known role in FL HSCs, which are exposed to higher levels of IFN-α (Figure 1F).40
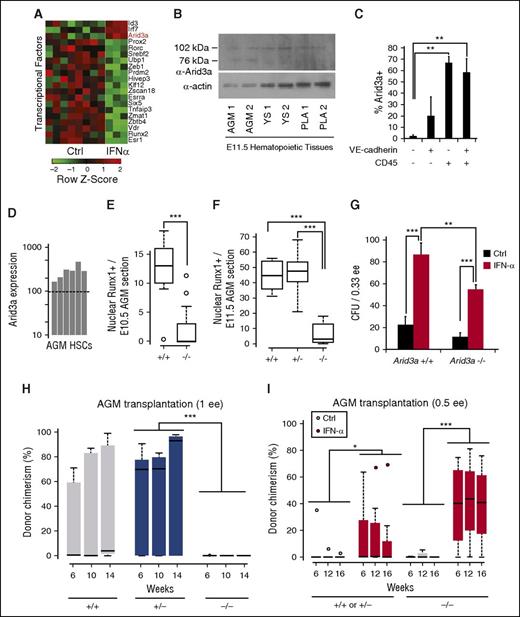
Hematopoietic defect in Arid3a KO embryos is rescued by IFN-α. (A) Microarray analysis of transcription factors upregulated by IFN-α in vivo in adult HSCs reveals Arid3a (GSE14361). (B) Immunoblot of Arid3a in the AGM, YS, and PLA. (C) Percentage of Arid3a+ cells in the various VE-cadherin/CD45 compartments showing abundance of Arid3a in hematopoietic cells. Sorted cells were cytospun and stained for Arid3a; n = 4-5. (D) Microarray analysis showing the presence of Arid3a expression in AGM HSCs (VE-cadherin+CD45+) (GSE37000) via presence/absence call. Dotted line indicates the presence/absence cutoff. (E) Boxplot quantification of nuclear Runx1+ surrounding the dorsal aorta in the E10.5 AGM of Arid3a+/+ and −/− sections; n = 13-19. (F) Boxplot quantification of nuclear Runx1+ surrounding the dorsal aorta in the E11.5 AGM of Arid3a+/+, +/−, and −/− sections; n = 6-9. (G) CFU assays from E11.5 Arid3a WT and KO AGMs; n = 3. Error bars indicate standard error of the mean (SEM). (H) Boxplots of donor chimerisms of Arid3a+/+, +/−, and −/− E11.5 to E12.5 AGMs transplanted at 1 e.e. and analyzed at 6, 10, and 14 weeks posttransplantation; 5 × 105 splenic helper cells were used. (I) Boxplots of donor chimerisms of E11.5 Arid3a+/+, +/−, and −/− AGMs transplanted at 0.5 e.e. and analyzed at 6, 12, and 16 weeks posttransplantation. Two-way ANOVA was performed. Statistical significance: *P < .05; **P < .01; ***P < .001. PLA, placenta; YS, yolk sac.
Hematopoietic defect in Arid3a KO embryos is rescued by IFN-α. (A) Microarray analysis of transcription factors upregulated by IFN-α in vivo in adult HSCs reveals Arid3a (GSE14361). (B) Immunoblot of Arid3a in the AGM, YS, and PLA. (C) Percentage of Arid3a+ cells in the various VE-cadherin/CD45 compartments showing abundance of Arid3a in hematopoietic cells. Sorted cells were cytospun and stained for Arid3a; n = 4-5. (D) Microarray analysis showing the presence of Arid3a expression in AGM HSCs (VE-cadherin+CD45+) (GSE37000) via presence/absence call. Dotted line indicates the presence/absence cutoff. (E) Boxplot quantification of nuclear Runx1+ surrounding the dorsal aorta in the E10.5 AGM of Arid3a+/+ and −/− sections; n = 13-19. (F) Boxplot quantification of nuclear Runx1+ surrounding the dorsal aorta in the E11.5 AGM of Arid3a+/+, +/−, and −/− sections; n = 6-9. (G) CFU assays from E11.5 Arid3a WT and KO AGMs; n = 3. Error bars indicate standard error of the mean (SEM). (H) Boxplots of donor chimerisms of Arid3a+/+, +/−, and −/− E11.5 to E12.5 AGMs transplanted at 1 e.e. and analyzed at 6, 10, and 14 weeks posttransplantation; 5 × 105 splenic helper cells were used. (I) Boxplots of donor chimerisms of E11.5 Arid3a+/+, +/−, and −/− AGMs transplanted at 0.5 e.e. and analyzed at 6, 12, and 16 weeks posttransplantation. Two-way ANOVA was performed. Statistical significance: *P < .05; **P < .01; ***P < .001. PLA, placenta; YS, yolk sac.
Examination of Arid3a protein levels in E11.5 embryonic tissues indicated that, in addition to the FL, Arid3a is expressed at sites of embryonic hematopoiesis, including the AGM, yolk sac, and placenta (Figure 5B).40 To determine whether Arid3a is expressed in endothelial or hematopoietic cells, we stained the AGM with CD45 and VE-cadherin, and sorted by flow cytometry into the four fractions (supplemental Figure 3A-E). Post-sort staining indicated that Arid3a+ cells fractionated within the CD45+ population, which includes the CD45+VE-cadherin+ HSC population, and less within the double-negative or single CD45−VE-cadherin+ endothelial population (Figure 5C). Analysis of previous microarray data confirmed that AGM HSCs express Arid3a (Figure 5D).3 Together, these data suggest a function for Arid3a within the same AGM HSCs that undergo IFN-α/Stat1 signaling.
IFN-α overcomes the hematopoietic defect in Arid3a knockout (KO) embryos
Arid3a KO mice die between E11.5 and E13.5 as a result of defects in erythroid lineage differentiation.40 They also have FL HSC defects,40 but whether they are impaired in AGM hematopoiesis is unknown. We first determined whether hematopoietic progenitors are present in the Arid3a KO AGM by immunostaining for Runx1 (supplemental Figure 3F-G). Nuclear Runx1+ cells were reduced in the dorsal aorta as compared with WT AGMs in both E10.5 and E11.5 embryos (Figure 5E-F). To determine if IFN-α treatment can compensate for the above hematopoietic defects, we treated WT and Arid3a KO AGMs with IFN-α and quantified the number of colony-forming units (CFUs). Arid3a KO embryos showed a trend toward lower numbers of CFUs, which were increased by IFN-α treatment (Figure 5G).
To determine whether Arid3a KO impairs AGM HSCs, we compared transplantation outcomes with those of WT and Arid3a+/− AGM controls. Arid3a−/− donor chimerism performed at 1 e.e. input was compromised (Figure 5H; P = .0004 by limiting dilution analysis), suggesting that the documented FL HSC hematopoietic defect in Arid3a KO mice originates within the AGM. Because in vivo treatment of mice with IFN-α has previously been shown to upregulate Arid3a,28 we initially presumed that IFN-α would fail to rescue the HSC defects in Arid3a KO mice. We split Arid3a KO AGMs into 0.5 e.e. each and treated one arm with IFN-α prior to transplantation, and were surprised to observe robust donor chimerism in mice transplanted with IFN-α–treated Arid3a KO AGMs (Figure 5I; supplemental Figure 4A-B). This unexpected observation prompted us to explore alternative mechanisms to account for the regulation of AGM HSCs by IFN-α and Arid3a.
Arid3a and Stat1 co-occupy promoters of IFN effector genes
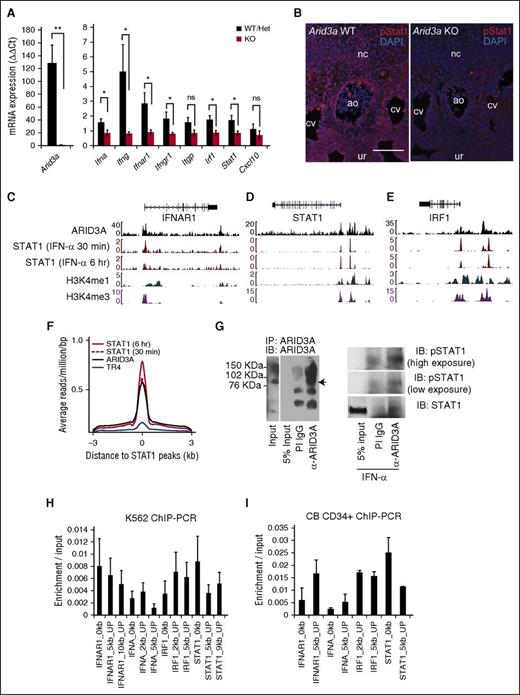
Because Arid3a KO HSCs engraft when treated with IFN-α, we hypothesized that IFN-α acts either downstream of Arid3a or in parallel. We found that the expression of IFNs (Ifnγ, Ifnα), receptors (Ifnγr1, Ifnαr1), and target genes (Irf1, Stat1) were downregulated in Arid3a KOs (Figure 6A). Similarly, phospho-Stat1 levels were decreased in Arid3a KO AGMs via immunofluorescence, indicating that Arid3a KO AGMs were deficient in IFN-α signaling (Figure 6B).
Genomic binding of ARID3A and STAT1. (A) Quantitative RT-PCR of Arid3a and IFN-related genes in Arid3a+/+, +/−, and −/− E11.5 AGM; n = 3-8. Error bars indicate SEM. (B) Immunostaining of E11.5 Arid3a WT and KO AGMs for phospho-Stat1. Scale bar = 100 μm. (C) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of IFN-αR1 in K562 cells. (D) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of IRF1 in K562 cells. (E) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of STAT1 in K562 cells. (F) ChIP-seq data showing global overlap of binding sites between ARID3A and STAT1 but not the hormone receptor TR4. (G) Immunoprecipitation of ARID3A (left). PI IgG was used as a control. Co-immunoprecipitation of STAT1 with ARID3A (right). K562 cells were exposed for 90 to 120 minutes of IFN-α (0.5 ng/mL). (H) Confirmation of ChIP-seq via quantitative ChIP-PCR in K562 cells normalized by input control; n = 4. Error bars indicate SEM. (I) Confirmation of ChIP-seq via quantitative ChIP-PCR in CB CD34+ cells normalized by input control; n = 2. Statistical significance: *P < .05; **P < .01. ao, dorsal aorta; CB, cord blood; cv, cardinal vein; DAPI, 4′,6-diamidino-2-phenylindole; het, heterozygous; IB, immunoblot; IgG, immunoglobulin G; nc, notochord; ns, not signicant; PI, pre-immune; ur, urogenital ridge.
Genomic binding of ARID3A and STAT1. (A) Quantitative RT-PCR of Arid3a and IFN-related genes in Arid3a+/+, +/−, and −/− E11.5 AGM; n = 3-8. Error bars indicate SEM. (B) Immunostaining of E11.5 Arid3a WT and KO AGMs for phospho-Stat1. Scale bar = 100 μm. (C) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of IFN-αR1 in K562 cells. (D) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of IRF1 in K562 cells. (E) ChIP-seq tracks ARID3A and STAT1 at the genomic loci of STAT1 in K562 cells. (F) ChIP-seq data showing global overlap of binding sites between ARID3A and STAT1 but not the hormone receptor TR4. (G) Immunoprecipitation of ARID3A (left). PI IgG was used as a control. Co-immunoprecipitation of STAT1 with ARID3A (right). K562 cells were exposed for 90 to 120 minutes of IFN-α (0.5 ng/mL). (H) Confirmation of ChIP-seq via quantitative ChIP-PCR in K562 cells normalized by input control; n = 4. Error bars indicate SEM. (I) Confirmation of ChIP-seq via quantitative ChIP-PCR in CB CD34+ cells normalized by input control; n = 2. Statistical significance: *P < .05; **P < .01. ao, dorsal aorta; CB, cord blood; cv, cardinal vein; DAPI, 4′,6-diamidino-2-phenylindole; het, heterozygous; IB, immunoblot; IgG, immunoglobulin G; nc, notochord; ns, not signicant; PI, pre-immune; ur, urogenital ridge.
To interrogate whether Arid3a plays a regulatory role in IFN-α signaling, we analyzed chromatin-immunoprecipitation sequencing (ChIP-seq) data from the human hematopoietic line K562.41 Assessment of ARID3A binding to select IFN/Stat1-related loci revealed scant binding to IFN-α cytokine genes (data not shown), suggesting that decreased IFN-α transcription observed in the Arid3a KO embryo is indirect (Figure 6A). However, ARID3A occupied the genomic loci of IFN-αR1, IRF1, and STAT1 at positions marked by euchromatic histone modifications (H3K4m3 and H3K4m1) (Figure 6C-E).42 Furthermore, STAT1-binding sites overlapped with ARID3A-binding sites but not with an unrelated protein TR4 (Figure 6F), implicating ARID3A as a transcriptional co-regulator of STAT1. Supporting this observation, STAT1 co-immunoprecipitated with ARID3A (Figure 6G). Lastly, ChIP-PCR confirmed positions of enrichment found in ChIP-seq data (Figure 6H). ChIP-PCR in CD34+ cord blood HSCs produced similar results (Figure 6I).
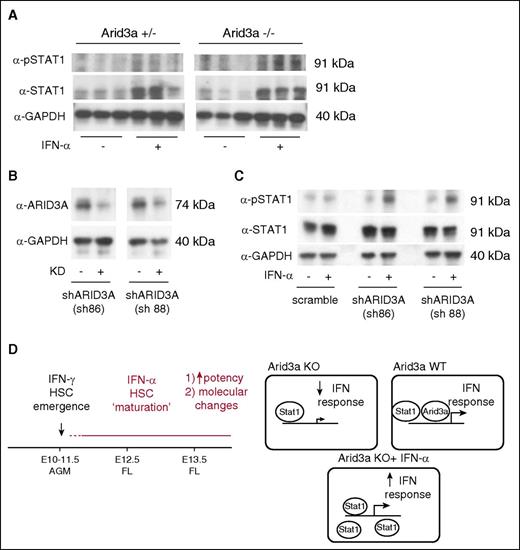
We finally investigated whether Arid3a was required for an IFN-α–mediated Stat1 response. In both Arid3a KO AGMs and short hairpin RNA-mediated knockdown of ARID3A in K562 cells, IFN-α treatment was able to increase phospho-Stat1, thus confirming that Arid3a and IFN-α–Stat1 are indeed parallel pathways that converge on Stat1 (Figure 7).
IFN-α response in Arid3a KO cells. (A) Immunoblot for phospho-Stat1 and Stat1 in Arid3a+/− and −/− AGMs treated with IFN-α. (B) Immunoblot confirmation of ARID3A KD in K562 cells via shRNAs. (C) Immunoblot for phospho-Stat1 and Stat1 in ARID3A KD cells showing response to IFN-α. (D) Representative model indicating the role of IFN-α during embryonic hematopoiesis. In contrast to IFN-γ, which promotes HSC emergence, IFN-α promotes partial maturation of AGM HSCs. Arid3a is a transcription co-regulator of IFN effector genes. When Arid3a is absent, inflammatory signaling via IFNs is dampened. Saturating the system with Stat1 via exogenous IFN-α treatment is able to overcome this defect. KD, knockdown; sh, short hairpin.
IFN-α response in Arid3a KO cells. (A) Immunoblot for phospho-Stat1 and Stat1 in Arid3a+/− and −/− AGMs treated with IFN-α. (B) Immunoblot confirmation of ARID3A KD in K562 cells via shRNAs. (C) Immunoblot for phospho-Stat1 and Stat1 in ARID3A KD cells showing response to IFN-α. (D) Representative model indicating the role of IFN-α during embryonic hematopoiesis. In contrast to IFN-γ, which promotes HSC emergence, IFN-α promotes partial maturation of AGM HSCs. Arid3a is a transcription co-regulator of IFN effector genes. When Arid3a is absent, inflammatory signaling via IFNs is dampened. Saturating the system with Stat1 via exogenous IFN-α treatment is able to overcome this defect. KD, knockdown; sh, short hairpin.
Discussion
Little is known about the signaling pathways that promote developmental maturation of AGM HSCs, which are the first definitive HSCs to arise in the mammalian embryo. Previously, we observed a reduced capacity for AGM HSCs to engraft irradiated adults relative to neonates, suggesting developmental immaturity.9 Our current study shows that AGM HSCs have lower levels of IFN-α–mediated Jak-Stat1 signaling than FL HSCs, and that treatment of AGM HSCs with IFN-α enhances long-term hematopoietic engraftment of AGM HSCs when transplanted into irradiated adults. We further show that Arid3a KO mice have defective AGM hematopoiesis that can be rescued by IFN-α treatment. It thus appears that Arid3a regulates the IFN pathway during normal embryogenesis via the modulation of IFN effector genes. These data explain how the embryo enlists an inflammatory gene regulatory network to promote the developmental maturation of nascent HSCs.
Our recent studies have been aimed at exploring the differences between AGM HSCs and FL or BM HSCs.9 Not surprisingly, nascent AGM HSCs exhibit molecular signatures reminiscent of their endothelial origin and need further maturation steps to reach the functional maturity of FL or adult HSCs.3,9 To achieve such a goal, the embryo must compartmentalize sites of HSC formation from maturation. Relative to the FL and BM, the AGM is enriched in IL-3 and IL-6 signaling pathways,3,24 which in concert with other pathways such as Notch signaling, participate in hematopoietic emergence.19,43 The AGM is enriched for shear-stress–mediated PGE2 and protein kinase A/cAMP-responsive element-binding protein signaling,44-46 which can activate IL-6.47-49 Subsequently, through exposure to IFN-α during maturation in the FL, AGM HSCs acquire expanded potential for engraftment in adults. This may explain the increased long-term repopulating activity observed when co-culturing AGM HSCs with nonhematopoietic cells from E14.5 liver.50 Indeed, the regulation of IFNs under normal developmental conditions is complex. The AGM and the FL sustain high levels of erythropoiesis to provide adequate oxygen carrying capacity in a hypoxic environment and to meet the metabolic demands of a rapidly developing embryo.51 One hypothesis is that this hypoxic environment and/or RBC enucleation are transiently pro-inflammatory, and the stabilization of hypoxia-inducible factor and activation of innate immunity may contribute to IFN-α production.52-54 A similar mechanism may exist in the placenta and serve as a niche for the maturation of HSCs.55
Distinct type I and II IFNs are used in different contexts for hematopoietic development.56,57 Previous work in zebrafish and mice suggest that IFN-γ promotes HSC emergence in midgestation embryos but zebrafish lack an IFN-α homolog, thus leaving the role of this type I IFN unexplored.16,18 Whereas IFN-γ promotes HSC emergence, our data argue that IFN-α promotes partial AGM HSC maturation. IFN-γ signaling results in the atypical activation of Stat3,18 which has known functions in HSC emergence.24,25 In our experiments involving IFN-α, we observed Stat1 activation but did not observe significant activation of Stat3 in the AGM. In postnatal development, the lack of maturation may explain the decreased number of LSK HSCs in Ifnαr1 KO mice but additional compensatory mechanisms must exist in adulthood to erase this difference.28,58
In contrast to the IFN-α receptor KO mice, which survive to adulthood although with immune compromise,30 the hematopoietic phenotype of Arid3a KO is more severe because Arid3a likely regulates a more complex expression network encompassing IFN-α and IFN-γ, their receptors, and downstream effector proteins. This implies that Arid3a has a broader role outside the context of IFN-α signaling. Indeed, in B lymphocytes, Arid3a is known to bind to specific adenosine thymine cytosine–rich matrix-associated regions within the immunoglobulin heavy chain enhancer (Eμ) and a subset of variable region (VH)-associated promoters.59-62 Matrix-associated regions compartmentalize specific loops of chromatin and in this case, juxtapose VH promoters with Eμ to mediate high level transcription of the locus during development.63-65 As with other members of the ARID family,66 Arid3a mediates chromatin remodeling.59-61
In agreement with this hypothesis, most Arid3a-expressing and IFN-α–responsive cells are CD45+, suggesting that the genetic interaction between Arid3a and IFN/Stat1 occurs within hematopoietic cells. Although we documented the downregulation of Ifnα and Ifnγ in Arid3a KO embryos, the regulation of IFN-α and IFN-γ transcription is unlikely to be directly mediated by Arid3a. In K562 cells, ARID3A failed to occupy the genomic loci of IFN-α cytokines; instead, ARID3A genomic occupancy overlapped with euchromatin histone marks within the STAT1 and IRF1 loci. This indicates that the relationship between Arid3a and IFN signaling is nonlinear and complex, involving direct ARID3A transactivation of IFN effectors. Perhaps, similar to the role played by Arid3a in regulating the accessibility of Eμ,61 Arid3a may function in the IFN-α/Stat1 pathway by catalyzing the chromatin accessibility of IFN effector genes (Figure 7D). Thus, by increasing Stat1 signals, IFN-α may overcome the hematopoietic defect in Arid3a KO embryos. Defining additional functions for Arid3a in CD45+ biology, and dissecting precise molecular relationships between Arid3a and IFN/Stat1 signaling will be subjects of future studies.
In conclusion, we have identified a novel role for IFN-α and a genetic interaction between Arid3a and IFN-α/Stat1 signaling in the developmental maturation of AGM HSCs. Understanding the differences between embryonic and adult HSCs may lead to novel ways of increasing the potency of HSCs for transplantation, especially when using HSCs from developmentally immature cell sources such as pluripotent stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
G.Q.D. is supported by grants from the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (R24-DK092760), the National Heart, Lung, and Blood Institute Progenitor Cell Biology Consortium (UO1-HL100001), and the Doris Duke Medical Foundation. G.Q.D. is an associate member of the Broad Institute, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. H.O.T. is supported by grants from the National Institutes of Health National Cancer Institute (R01CA31534), the Cancer Prevention and Research Institute of Texas (RP120459), and the Marie Betzner Morrow Centennial Endowment. S.H.O. is supported by the Center of Excellence in Molecular Hematology (P01HL032262 and P30DK049216).
Authorship
Contribution: P.G.K. and G.Q.D. conceived the study, analyzed the data, and wrote the manuscript; P.G.K., M.C.C., C.R., S.J.R., H.-C.T., and J.V.H. performed experiments; H.O.T. provided critical reagents and feedback; and S.H.O. provided feedback.
Conflict-of-interest disclosure: G.Q.D. is a member of the Scientific Advisory Boards of the following companies, and receives consulting fees and/or holds equity in True North Therapeutics, Inc., Solasia Pharma KK, and MPM Capital. H.O.T. is a member of the Advisory Boards and holds equity in Gene Screen, Inc. and Scarab, Ltd.
Correspondence: George Q. Daley, Howard Hughes Medical Institute/Boston Children’s Hospital, Karp Family Research Building, 7th Floor, 300 Longwood Ave, Boston, MA 02115; e-mail: george.daley@childrens.harvard.edu.