To the editor:
Chronic lymphocytic leukemia (CLL) is one of the most common B-cell malignancies in individuals of European ancestry in the United States.1 Although advanced age, white ancestry, and family history of hematologic malignancies are risk factors, the etiology of CLL is unknown.2 No major susceptibility loci were identified for CLL from previous linkage studies,2 but genome-wide association studies conducted in CLL case-control samples have led to the identification of many single nucleotide polymorphisms with small effects.3,4 In this study, we identified a novel locus causing susceptibility to CLL using whole exome sequencing and follow-up targeted sequencing in a series of CLL high risk families.
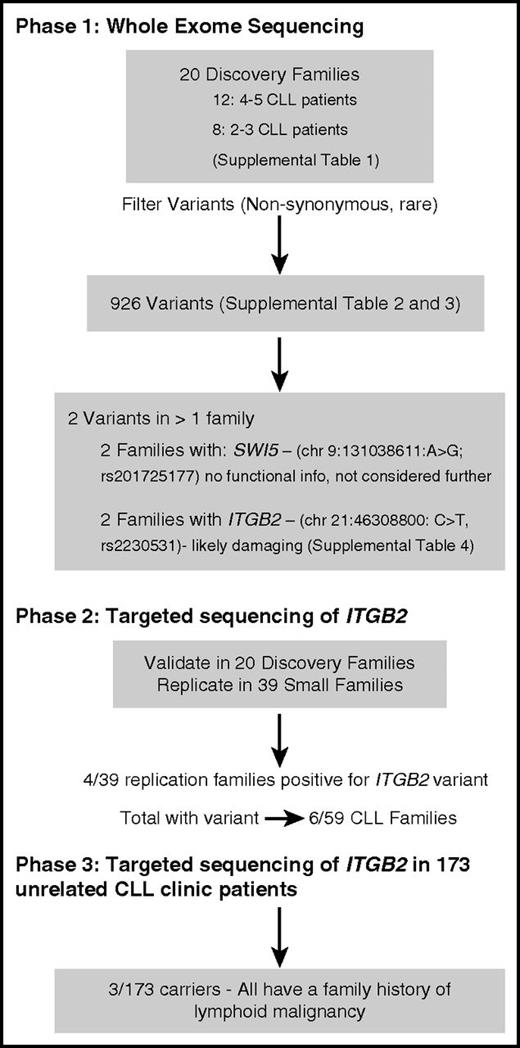
Figure 1 outlines the strategy that we used for screening families and patients in 3 phases. The study includes a total of 59 CLL-prone families (20 for discovery and 39 for replication; supplemental Table 1, available on the Blood Web site) and 173 unrelated CLL clinic patients unselected for family history. The clinical, laboratory, and bioinformatics methods were described previously5 and are summarized in the supplemental Methods. For each family, we filtered variants to consider only those segregating with illness with population allele frequency <1%. Our filtering pipeline resulted in 926 variants (supplemental Tables 2-3). Two were found in >1 family including missense variants in ITGB2 (chromosome 21:46308800: C>T, rs2230531) and SWI5. Due to lack of functional information and low sequencing coverage in some individuals, SWI5 was not pursued further. However, ITGB2 is involved in cell adhesion,6 and its expression in CLL cells predicts disease progression.7 In phase 2, we resequenced ITGB2 in the 20 discovery families and in 39 smaller replication families (Figure 1). The ITGB2 variant was technically validated in the 2 original families and was also found in 4 additional families in the follow-up sample for a total of 6/59 CLL families.
The frequency of the variant allele is 0.007 in Europeans from a large database (exome aggregation consortium8 ) and 0.009 among nonlymphoid cancer families sequenced in our laboratory. The variant results in a single amino acid residue change from glutamate to lysine (E630K). Bioinformatic analyses show the variant to be highly conserved and predicted to be “damaging” to the protein function, including a possible effect on splicing (supplemental Table 4). ITGB2 produces a protein, known as CD18, which is a cell surface marker expressed on lymphocytes. The integrin β chain forms heterodimers with α integrin protein and is involved in cell adhesion and cell-surface mediated signaling.
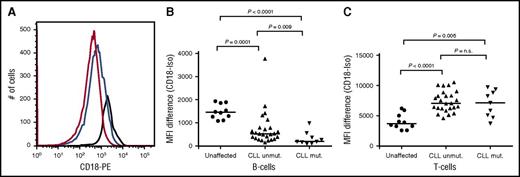
Among familial CLL patients carrying the ITGB2 variant, 9 individuals from 5 families had additional stored lymphocytes that could be examined for expression of CD18 by flow cytometry in both the B-cell and T-cell populations (Figure 2). These patients were matched by sex and age at diagnosis with 25 other CLL patients who were not variant carriers. As expected from previous studies in CLL patients, the CD18 expression in T cells is significantly higher than in B cells in all patients, and all patients have lower expression in B cells and higher expression in T cells than unaffected individuals (Figure 2C). The CLL variant carriers have significantly lower expression of CD18 on B cells than do the CLL patient noncarriers (Figure 2A-B) and no difference in expression on T cells (Figure 2C). We confirmed that these differences were not due to effects of age at diagnosis, age at blood sampling, or treatment history. The small sample size precluded additional correction for related individuals. In phase 3, we found that 3 of 173 unrelated CLL clinic patients carried the variant, and all had a family history of CLL or other lymphoid malignancy, as well as an early age at diagnosis (average = 55 years).
Analyses of CD18 expression on B cells and T cells of mutation carriers and noncarriers. Peripheral blood mononuclear cells were stained as previously described13 with the following antibodies: anti–CD19-allophycocyanin, anti–CD5–phycoerythrin cyanine dye (to identify the CLL cell population), anti–CD3-fluorescein isothiocyanate (to identify the T-cell population) and one of the following phycoerythrin-conjugated antibodies: anti–immunoglobulin G1-isotype control or anti-CD18 (BD Biosciences, Franklin Lakes, NJ). Each sample was run in duplicate. Cells were analyzed on a fluorescence-activated cell sorter (FACS) Canto II flow cytometer (BD Biosciences) using FACS-DIVA 6.1.1 and FlowJo software (Version 8.8.6; TreeStar, Ashland, OR). (A) Expression of CD18 in B cells of representative normal individual (black line), CLL patient without CD18 mutation (blue line), and CLL patient with CD18 mutation (red line). (B) Expression of CD18 in B cells of 9 CLL patients (with CD18 mutation), 25 CLL patients without CD18 mutation, and 10 unaffected individuals. (C) Expression of CD18 in T cells of 9 CLL patients (with CD18 mutation), 25 CLL patients without CD18 mutation, and 10 unaffected individuals.
Analyses of CD18 expression on B cells and T cells of mutation carriers and noncarriers. Peripheral blood mononuclear cells were stained as previously described13 with the following antibodies: anti–CD19-allophycocyanin, anti–CD5–phycoerythrin cyanine dye (to identify the CLL cell population), anti–CD3-fluorescein isothiocyanate (to identify the T-cell population) and one of the following phycoerythrin-conjugated antibodies: anti–immunoglobulin G1-isotype control or anti-CD18 (BD Biosciences, Franklin Lakes, NJ). Each sample was run in duplicate. Cells were analyzed on a fluorescence-activated cell sorter (FACS) Canto II flow cytometer (BD Biosciences) using FACS-DIVA 6.1.1 and FlowJo software (Version 8.8.6; TreeStar, Ashland, OR). (A) Expression of CD18 in B cells of representative normal individual (black line), CLL patient without CD18 mutation (blue line), and CLL patient with CD18 mutation (red line). (B) Expression of CD18 in B cells of 9 CLL patients (with CD18 mutation), 25 CLL patients without CD18 mutation, and 10 unaffected individuals. (C) Expression of CD18 in T cells of 9 CLL patients (with CD18 mutation), 25 CLL patients without CD18 mutation, and 10 unaffected individuals.
In our 6 of 59 high-risk CLL families, we found an uncommon, nonsynonymous variant, predicted to be damaging, in the ITGB2 gene shared by patients. In 1 family from the discovery set, 2 of 6 unaffected relatives also carried the variant. However, these 2 carrier individuals were in their early 30s when sampled and thus are unlikely to have manifested the disease. In addition, neither had evidence of monoclonal B-cell lymphocytosis (MBL) based on flow cytometry, although MBL is very rare at this age. We do not know if this variant is also associated with MBL as there were no MBL patients in the families carrying the variant. We do not have sufficient data to predict the likelihood of developing CLL among gene carriers. However, the variant may be associated with other lymphoid malignancies because we found the same variant segregating with Hodgkin lymphoma in 1 family.5 The finding that all 3 of the unrelated clinic patients who were carriers of the variant had a family history is striking because previous studies found 12% to 17% of patients from CLL clinic populations have such a family history.9,10
The cell surface marker, CD18, coded by the ITGB2 gene is expressed on lymphocytes and is involved in leukocyte adhesion. Its expression is reduced on CLL B cells compared with normal B cells.6,11 The low expression of CD18 on CLL cells impairs their migration to bone marrow and lymph nodes but not to the spleen, and thus they remain in circulation or migrate to the spleen.12 However, expression levels are increased on cells from patients with more advanced stage7 or higher prognostic risk groups.12 Although our numbers are small, we found that carriers of the germline ITGB2 variant had lower CD18 expression on B cells than a matched group of noncarriers, suggesting that decreased expression is an early event in the pathway to CLL. Future work should clarify how the mutation reduces CD18 expression and explore its biological consequences in cell lines or lymphocytes. The variant may cause differences in the ability of the lymphocytes to adhere, migrate, or respond to CD18 stimulation or it may affect protein stability or membrane transport. Given the long latency of CLL development, the effect of the mutation might also be studied in a genetic mouse model.
Our study has some limitations. Key susceptibility loci could be outside of exonic regions or not well covered by exome targets. Our filtering strategy may be too stringent, and we may have missed important variants. It is possible that variants identified in CLL patients derive from tumor. However, we required variants to be shared among all patients in a family making it most likely that they derive from germline. Although our findings could be a false positive given the large number of variants identified, our multistep filtering criteria minimizes this possibility. Other genetic mechanisms could be operating in familial CLL such as extensive genetic heterogeneity, leading to variants being “private” or susceptibility caused by multiple loci.
The strength of our study is the ability to comprehensively search for coding variants causing susceptibility to CLL in a large number of highly informative families. We identified a novel germline variant in the ITGB2 gene, which may account for familial CLL susceptibility in up to 10% of high-risk CLL families. The number of positive families from the replication sample, the finding of positive family history among all 3 carrier patients from the clinic population, and the expression differences found between carriers and noncarriers all indicate strong evidence in favor of this gene being important in CLL susceptibility. Further work is needed to confirm this association and understand its functional consequences.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors acknowledge the contribution of members of the National Cancer Institute (NCI) Division of Cancer Epidemiology and Genetics (DCEG) Cancer Sequencing Working Group: Bari Ballew, Sharon Savage, Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, and Guoqin Yu. The authors acknowledge the contribution of members of the NCI DCEG Cancer Genomics Research laboratory: Sarah Bass, Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. Hutchinson, Sally Larson, Kerry Lashley, Hyo Jung Lee, Wen Luo, Michael Malasky, Michelle Manning, Jason Mitchell, David Roberson, Aurelie Vogt, and Mingyi Wang.
This work was supported by the Intramural Research Program of the US National Institutes of Health (NIH), NCI, Division of Cancer Epidemiology and Genetics, and National Heart Lung and Blood Institute. This work was supported in part by funds from NCI/NIH contract HHSN261200800001E.
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Contribution: L.R.G., M.L.M., M.R., and A.M.G. performed and interpreted genetic analyses; M.L.M., M.A.T., L.F., G.M., and N.E.C. directed clinical work for the patients and families; S.E.M.H. and A.W. performed and interpreted flow cytometry expression analyses and provided DNA samples from CLL clinic patients; J.B., K.J., B.Z., L.B., B.H., M.Y., and S.J.C. conducted all whole-exome and targeting sequencing and associated bioinformatics; S.R. and B.T.L. performed functional prediction analyses; L.R.G., M.L.M., M.R., and N.E.C. drafted the manuscript; and all authors contributed to the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.F. is MedStar Georgetown Cancer Network, Washington, DC.
Correspondence: Lynn R. Goldin, Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 9609 Medical Center Dr, Rm 6E432, Bethesda, MD 20892-9769; e-mail: goldinl@mail.nih.gov.