Key Points
There is a high level of heterogeneity in cGVHD plasma biomarkers in a large cGVHD cohort, with CXCL10 being the most reproducible marker.
CXCR3+CD56bright natural killer regulatory cells have a strong inverse relationship with plasma CXCL10 in patients with or without cGVHD.
Abstract
Chronic graft-versus-host disease (cGVHD) remains one of the most significant long-term complications after allogeneic blood and marrow transplantation. Diagnostic biomarkers for cGVHD are needed for early diagnosis and may guide identification of prognostic markers. No cGVHD biomarker has yet been validated for use in clinical practice. We evaluated both previously known markers and performed discovery-based analysis for cGVHD biomarkers in a 2 independent test sets (total of 36 cases ≤1 month from diagnosis and 31 time-matched controls with no cGVHD). On the basis of these results, 11 markers were selected and evaluated in 2 independent replication cohorts (total of 134 cGVHD cases and 154 controls). cGVHD cases and controls were evaluated for several clinical covariates, and their impact on biomarkers was identified by univariate analysis. The 2 replications sets were relatively disparate in the biomarkers they replicated. Only sBAFF and, most consistently, CXCL10 were identified as significant in both replication sets. Other markers identified as significant in only 1 replication set included intercellular adhesion molecule 1 (ICAM-1), anti-LG3, aminopeptidase N, CXCL9, endothelin-1, and gelsolin. Multivariate analysis found that all covariates evaluated affected interpretation of the biomarkers. CXCL10 had an increased significance in combination with anti-LG3 and CXCL9, or inversely with CXCR3+CD56bright natural killer (NK) cells. There was significant heterogeneity of cGVHD biomarkers in a large comprehensive evaluation of cGVHD biomarkers impacted by several covariates. Only CXCL10 strongly correlated in both replication sets. Future analyses for plasma cGVHD biomarkers will need to be performed on very large patient groups with consideration of multiple covariates.
Introduction
Chronic graft-versus-host disease (cGVHD) remains one of the most significant long-term complications of allogeneic blood and marrow transplantation (BMT). Recent studies have shown that the quality of life of patients with cGVHD is much poorer, with a higher frequency of cardiovascular disease, metabolic syndrome, diabetes mellitus, obesity, cognitive issues, fatigue, sexual dysfunction, and endocrine abnormalities.1-4
Biomarkers that can act as diagnostic, prognostic, and predictive markers for cGVHD are needed to improve the ability to treat cGVHD effectively. Criteria for such biomarkers are well defined in the recent National Institutes of Health (NIH) Chronic GVHD consensus report.5 Many candidate plasma or serum cGVHD diagnostic biomarkers have been identified but none have been validated for use in clinical practice.6 Many recurring markers include soluble B-cell activating factor (sBAFF), aminopeptidase N (sCD13), interleukin-2 receptor alpha (IL-2Rα), and CXCL9.6-8
It has been 10 years since the first NIH consensus report was published, and the most recent report still concludes that there is a great need for biomarkers that can be rigorously replicated by several research groups. The conclusions of the most recent consensus report were that (1) larger replication studies are needed, (2) more work needs to focus on prognostic and predictive cGVHD biomarkers in addition to diagnostic markers, and (3) there is little consistency between the biomarkers currently identified to allow for clinical application. The report noted that collection methodology is highly variable, with little consideration of factors that may have an impact on the importance of the markers, and many potential covariates were identified.5 These covariates included method of sample collection, clinical data collected, clinical presentation, total body irradiation (TBI), previous acute GVHD (aGVHD), infection, type of preparative regimen, underlying disease, donor source, and level of HLA matching.
In this study, we performed a replication study of cGVHD diagnostic markers in adult cohorts after initial discovery analyses on independent test sets. We attempted to control our analyses for several covariates, including time of onset, presence of previous aGVHD, and TBI. We found that there was a high heterogeneity among plasma markers, but that CXCL10 was consistently elevated at diagnosis of cGVHD. We found that a high plasma CXCL10 level correlated with low numbers of peripheral blood CXCR3+CD56bright natural killer (NK) cells.
Methods
Sample collection and processing protocol
Samples in all test and replication sets were collected from patients age 18 years or older. Two test sets and 2 independent replication sets (Figure 1) were obtained, and laboratory analysis was performed after informed consent was provided in accordance with the Declaration of Helsinki and approval was given by the local ethics committee. Plasma samples for test set 1 were collected at the Fred Hutchinson Cancer Research Center from participants in a prospective longitudinal cohort study.9 Samples from late onset cases,10 diagnosed 9 or more months after transplantation, were collected within 1 month of the onset of cGVHD (n = 17). Control samples were obtained from patients with no cGVHD at the time of collection or in the subsequent 3 months. These samples were matched to cGVHD cases on the basis of time after BMT (n = 21). Any cases that had a previous diagnosis of cGVHD or a flare-up of old cGVHD were excluded from both groups.
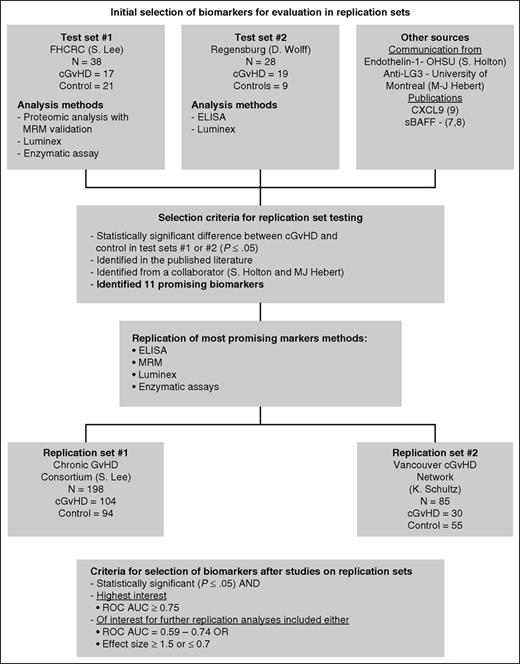
Biomarker selection algorithm. Biomarkers for testing in the replication sets were selected from 2 test sets with discovery-based evaluations using proteomic analysis followed by MRM-MS replication, Luminex, and enzymatic assays. Other markers were selected from communication with collaborators (M.-J.H. and S.G.H.) as well as from recent biomarker studies in the literature (Kitko et al8 ). Using the selection criteria as outlined, only 11 biomarkers were evaluated in the replication sets using ELISA, MRM-MS, Luminex, and enzymatic assays.
Biomarker selection algorithm. Biomarkers for testing in the replication sets were selected from 2 test sets with discovery-based evaluations using proteomic analysis followed by MRM-MS replication, Luminex, and enzymatic assays. Other markers were selected from communication with collaborators (M.-J.H. and S.G.H.) as well as from recent biomarker studies in the literature (Kitko et al8 ). Using the selection criteria as outlined, only 11 biomarkers were evaluated in the replication sets using ELISA, MRM-MS, Luminex, and enzymatic assays.
Twenty-three samples for test set 2 were collected from patients undergoing allogeneic BMT at the University Hospital Regensburg (19 cGVHD cases and 4 no GVHD controls). Serum samples were retrieved at diagnosis of cGVHD in the outpatient clinic and were prepared and stored at −80°C in aliquots within 4 hours. Clinical data were obtained within the prospective trial, including transplant characteristics, history of aGVHD, and NIH-based diagnosis, and organ grading of cGVHD, as well as intensity of immunosuppression at time of sampling.
Samples in replication set 1 (n = 198) were collected as part of the Chronic GVHD Consortium Protocol 6501 cohort study. In replication set 1, heparinized plasma was isolated from freshly processed blood without platelet depletion. Samples from cases were collected within 1 month of cGVHD diagnosis. Control samples were matched within 90 days of time since transplantation.
Samples in replication set 2 were collected as a part of a Canadian Institutes of Health Research (CIHR)–funded biomarker study based at the British Columbia Children’s Hospital Child and Family Research Institute from 9 Canadian BMT Group (CBMTG) centers, an additional 2 centers in the United States, and 1 BMT center in Saudi Arabia (n = 30 cGVHD cases; n = 53 no cGVHD controls). Onset samples were collected between 21 days before and 26 days after cGVHD diagnosis. In replication set 2, platelet-depleted plasmas were isolated and frozen within 24 hours of collection. Supplemental Table 1 (available on the Blood Web site) summarizes the comparison of the sample collection and processing differences between test sets and replication sets.
Proteomics for discovery of markers
Briefly, samples from cases and controls were sent to the University of Victoria Genome BC Proteomics Centre, where they were depleted of the most abundant proteins; albumin and immunoglobulins were precipitated, digested to form peptides, differentially labeled with iTRAQ, and separated by ion chromatography on a VISION workstation (Applied Biosystems). Resultant peptides were processed by using the 4800 MALDI TOF/TOF analyzer (Applied Biosystems). Spectra were searched against IPI Human (v3.2195) and MSDB (build 20063108) sequence databases.7
Validation with multiple reaction monitoring mass spectrometry
All experiments were performed on simple tryptic digests of human plasma without prior affinity depletion or enrichment. Stable isotope-labeled standard peptides were added immediately after tryptic digestion. Proteotypic tryptic peptides containing isotopically coded amino acids were synthesized for all 45 proteins. Peptide purity was assessed by capillary zone electrophoresis, and the peptide quantity was determined by amino acid analysis. For maximum sensitivity and specificity, instrumental parameters were empirically determined to generate the most abundant precursor ions and ion fragments. Concentrations of individual peptide standards in the mixture were optimized to approximate endogenous concentrations of analytes and to ensure the maximum linear dynamic range of the multiple reaction monitoring mass spectrometry (MRM-MS) assays.11
Anti-perlecan auto-antibodies (anti-LG-3)
Auto-antibody immunoglobulin G (IgG) titers against the bioactive C-terminal fragment of perlecan released by apoptotic endothelial cells is shown to be a biomarker of immune-mediated vascular injuries.12 Anti-LG3 titers were measured with previously described laboratory-made enzyme-linked immunosorbent assay (ELISA).13 Briefly, recombinant mouse LG-3 (10 μg/μL) was first coated on 96-well Nunc MaxiSorp plates (Thermo Scientific, Rochester, NY) for a total of 1 μg per well. The plasma was diluted (1:250), and 100 μL was added per well. After 1 hour of incubation at room temperature, the plates were washed, and bound IgG was detected by using horseradish peroxidase coupled with anti-human IgG antibody (Amersham, Little Chalfont, United Kingdom). Reactions were revealed with 100 μL of tetramethylbenzidine substrate (BD Biosciences) for 10 minutes and stopped with 50 μL of H2SO4. Spectrophotometric analysis was undertaken at 450 nm, and the results were expressed as optical density (OD) × 1000.
Cytokine measurements
ELISA and multiplex kits used to detect cytokines in replication sets were purchased from R&D Systems (Minneapolis, MN), except for CXCL9, which was purchased from RayBiotech (Norcross, GA). Manufacturer’s instructions were followed for every assay and the source of each assay is summarized in supplemental Table 2. The lowest standard concentration showed that OD ≥ 2.0 × OD of the blank was set as the detection limit. Samples evaluated for vascular markers at Oregon Health & Science University had levels of epidermal growth factor (EGF), fibroblast growth factor 1 (FGF-1,) FGF-2, heparin binding EGF-like growth factor, vascular endothelial growth factor A, (VEGFA), VEGFC, and VEGFD, angiopoietin-2, endothelin-1, endoglin, follistatin, leptin, and placental growth factor determined by Milliplex magnetic bead array (Millipore, Billerica, MA). For laboratory measurements that were detectable but below the lower limit of quantification, the values were imputed as the lower limit of quantification.
Immune cell phenotyping
Peripheral blood was collected, usually from a vein in the antecubital fossa, into blood collection tubes containing heparin (BD Vacutainer). Peripheral blood was processed, and immunophenotyping was performed on cryopreserved peripheral blood mononuclear cells with a panel consisting of FITC-CD3 (UCHT1; BioLegend), Brilliant Violet 510-CD8 (RPA-T8; BioLegend), PE-CD56 (MEM-188; BioLegend), Pacific Blue-CD4 (SK3; BioLegend), Brilliant Violet 785-CD45RA (HI100; BioLegend), APC/Cy7-CD183 (CXCR3) (G025H7; BioLegend), and 7-AAD viability staining solution (BioLegend). Cells were analyzed by using a Fortessa cell analyzer equipped with four lasers (yellow-green, blue, violet, and red) (BD Biosciences), and data were analyzed by using FlowJo, v9.8 software (Tree Star, Ashland, OR).
Criteria for selection of biomarkers
We used two separate criteria for biomarker selection (summarized in Figure 1). Markers identified in the test sets were selected for further evaluation in the replication sets if they were significantly different from the no cGVHD controls (P ≤ .05). They were considered of high interest if the difference was significant and the receiver operating characteristic area under the curve (ROC AUC) ≥0.75.
Statistical methods
Biomarker values were log-transformed before analysis; mean differences between groups were re-expressed as fold differences (case:control). Those recorded as below the lower limit of detection were assigned a value equal to one-half the lower limit. In test sets 1 and 2, differences between cGVHD case and control groups were assessed by Wilcoxon rank-sum test. In the replication sets, linear regression analysis was used to evaluate biomarker levels according to cGVHD case-control status, with or without inclusion of other covariates. In the replication data sets, logistic regression was used to compute ROC AUC by using single biomarkers or combinations of biomarkers to distinguish cGVHD cases and controls. Forward and backward selection was used with logistic regression to define optimal combinations of biomarkers.
Results
Discovery evaluations performed on test sets 1 and 2
Two separate test sets were evaluated for a broad number of plasma-based biomarkers to determine which panel would be tested in the larger replication sets. Test set 1 was obtained from adult patients at the Fred Hutchinson Cancer Research Center (Table 1) upon diagnosis of cGVHD (17 cGVHD cases, all collected within 1 month of diagnosis (supplemental Table 1) and from 21 patients who did not develop cGVHD (controls). Proteomic analysis on test set 1 plasma was performed with matrix-assisted laser desorption ionization time of flight/time of flight methodology by using iTRAQ labels. MRM-MS was used to validate 71 of the most promising candidates identified by the initial proteomic studies. Proteomic analysis followed by MRM-MS validation identified 2 proteins of highest statistical significance: aminopeptidase N (sCD13) (P = .004) and protease inhibitor 16 (PI-16) (P = .005). In addition, gelsolin and kallikrein, although less significant, were determined to be of sufficient interest for further evaluation (Table 2). Luminex assays identified 2 cytokine or chemokine markers. One of the markers was soluble IL-2Rα (sIL-2Rα [sCD25]), previously identified (by our group) in children7 (P = .005). The other was ICAM-1 (P = .04). Thus, test set 1 identified 6 candidate markers for replication, including gelsolin, kallikrein, aminopeptidase N, IL-2Rα, ICAM-1, and PI-16.
Patient characteristics
| Characteristic . | Test set 1 (Fred Hutchinson Cancer Research Center collection) (n = 38) . | Test set 2 (University Hospital Regensburg collection) (n = 23) . | Replication set 1 cGVHD Consortium (U54) (n = 198) . | Replication set 2 Vancouver cGvHD network collection (n = 83) . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cGVHD (n = 17) . | Control (n = 21) . | cGVHD (n = 19) . | Control (n = 4) . | cGVHD (n = 104) . | Control (n = 94) . | cGVHD (n = 30) . | Control (n = 53) . | |||||||||
| No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | |
| Time from HCT to sample, mo | ||||||||||||||||
| Early (<9) | 0 | 1 | 5 | 9 | 47 | 2 | 50 | 65 | 63 | 49 | 52 | 23 | 77 | 46 | 87 | |
| Late (>9) | 17 | 100 | 20 | 95 | 10 | 53 | 2 | 50 | 39 | 38 | 45 | 48 | 7 | 23 | 7 | 13 |
| Prior aGVHD | ||||||||||||||||
| No | 4 | 25 | 6 | 30 | 6 | 32 | 4 | 100 | 42 | 40 | 40 | 43 | 8 | 29 | 18 | 34 |
| Yes | 12 | 75 | 14 | 70 | 13 | 68 | 0 | 62 | 60 | 54 | 57 | 20 | 71 | 35 | 66 | |
| Age, y | ||||||||||||||||
| <50 | 9 | 53 | 8 | 38 | 12 | 63 | 3 | 75 | 41 | 39 | 37 | 39 | 17 | 57 | 30 | 57 |
| ≥50 | 8 | 47 | 13 | 62 | 7 | 37 | 1 | 25 | 63 | 61 | 57 | 61 | 13 | 43 | 23 | 43 |
| Donor | ||||||||||||||||
| Matched related | 6 | 35 | 11 | 52 | 6 | 32 | 1 | 25 | 39 | 38 | 27 | 29 | 11 | 38 | 14 | 27 |
| Matched unrelated | 10 | 59 | 5 | 24 | 10 | 53 | 2 | 50 | 45 | 43 | 33 | 35 | 14 | 48 | 30 | 58 |
| Mismatched | 1 | 6 | 5 | 24 | 3 | 16 | 1 | 25 | 20 | 19 | 34 | 36 | 4 | 14 | 8 | 15 |
| Stem cell source | ||||||||||||||||
| PBSCs | 16 | 94 | 15 | 71 | 19 | 100 | 3 | 75 | 93 | 89 | 57 | 61 | 28 | 97 | 46 | 92 |
| BM | 1 | 6 | 6 | 29 | 0 | 1 | 25 | 5 | 5 | 7 | 7 | 1 | 3 | 3 | 6 | |
| Umbilical cord blood | 0 | 0 | 0 | 0 | 6 | 6 | 30 | 32 | 0 | 1 | 2 | |||||
| Conditioning | ||||||||||||||||
| Myeloablative without TBI | 5 | 29 | 5 | 25 | 11 | 58 | 2 | 50 | 22 | 21 | 14 | 15 | 6 | 20 | 9 | 17 |
| Myeloablative with TBI | 3 | 18 | 6 | 30 | 8 | 42 | 2 | 50 | 22 | 21 | 25 | 27 | 16 | 53 | 13 | 25 |
| Nonmyeloablative | 9 | 53 | 9 | 45 | 0 | 0 | 60 | 58 | 55 | 59 | 8 | 27 | 31 | 58 | ||
| GVHD prophylaxis | ||||||||||||||||
| CNI + MTX/MMF with or without sirolimus | 16 | 94 | 20 | 95 | 6 | 32 | 1 | 25 | 94 | 90 | 71 | 76 | 24 | 89 | 25 | 47 |
| CNI with or without sirolimus | 0 | 0 | 0 | 0 | 8 | 8 | 16 | 17 | 0 | 0 | ||||||
| CNI + MTX/MMF + ATG | 0 | 0 | 13 | 68 | 3 | 75 | 0 | 0 | 3 | 11 | 28 | 53 | ||||
| Other | 1 | 6 | 1 | 5 | 0 | 0 | 2 | 2 | 7 | 7 | 0 | 0 | ||||
| Sex | ||||||||||||||||
| Male | 7 | 41 | 10 | 48 | 14 | 74 | 2 | 50 | 63 | 61 | 50 | 53 | 19 | 63 | 29 | 55 |
| Female | 10 | 59 | 11 | 52 | 5 | 26 | 2 | 50 | 41 | 39 | 44 | 47 | 11 | 37 | 24 | 45 |
| Female donor to male patient | ||||||||||||||||
| No | 13 | 76 | 17 | 81 | 17 | 89 | 4 | 100 | 77 | 74 | 77 | 82 | 26 | 90 | 45 | 88 |
| Yes | 4 | 24 | 4 | 19 | 2 | 11 | 0 | 27 | 26 | 17 | 18 | 3 | 10 | 6 | 12 | |
| CMV serostatus | ||||||||||||||||
| Negative | 5 | 29 | 12 | 57 | 15 | 79 | 4 | 100 | 48 | 47 | 41 | 44 | 13 | 43 | 29 | 57 |
| Positive | 12 | 71 | 9 | 43 | 4 | 21 | 0 | 55 | 53 | 53 | 56 | 17 | 57 | 22 | 43 | |
| Disease diagnosis | ||||||||||||||||
| AML | 4 | 24 | 9 | 43 | 11 | 58 | 2 | 50 | 30 | 29 | 34 | 36 | 12 | 41 | 17 | 43 |
| ALL | 3 | 18 | 3 | 14 | 2 | 11 | 0 | 14 | 13 | 13 | 14 | 6 | 21 | 4 | 10 | |
| CML | 0 | 1 | 5 | 0 | 0 | 8 | 8 | 5 | 5 | 2 | 7 | 2 | 5 | |||
| CLL | 1 | 6 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 2 | 7 | 5 | 13 | |||
| MDS | 1 | 6 | 3 | 14 | 1 | 5 | 0 | 23 | 22 | 14 | 15 | 1 | 3 | 8 | 20 | |
| HL/NHL | 4 | 24 | 4 | 19 | 4 | 21 | 1 | 25 | 14 | 13 | 15 | 16 | 2 | 7 | 4 | 10 |
| MM | 0 | 1 | 5 | 0 | 1 | 25 | 7 | 7 | 7 | 7 | 2 | 7 | 0 | |||
| Other | 4 | 24 | 0 | 1 | 5 | 0 | 5 | 5 | 3 | 3 | 2 | 7 | 0 | |||
| Receiving steroids at sample draw | ||||||||||||||||
| No | 14 | 82 | 21 | 100 | 10 | 53 | 4 | 100 | 55 | 53 | 83 | 88 | NA | |||
| Yes | 3 | 18 | 0 | 9 | 47 | 0 | 49 | 47 | 11 | 12 | ||||||
| cGVHD characteristics | ||||||||||||||||
| Median days from HCT to cGVHD (range) | 362 (257-818) | 281 (106-1953) | 203 (83-566) | 174 (74-383) | ||||||||||||
| Median days from cGVHD to sample (range) | 2 (0-24) | 0 (–1 to 0) | 6 (–12 to 29) | 2 (–21 to 27) | ||||||||||||
| Skin involvement | 59 | 79 | 63 | 65 | ||||||||||||
| Oral involvement | 65 | 53 | 61 | 83 | ||||||||||||
| GI involvement | 41 | 21 | 25 | 26 | ||||||||||||
| Eye involvement | 35 | 26 | 41 | 26 | ||||||||||||
| Joint involvement | 29 | 5 | 15 | 9 | ||||||||||||
| Lung involvement | 59 | 21 | 48 | 48 | ||||||||||||
| Liver involvement | 47 | 32 | 56 | 43 | ||||||||||||
| Genital involvement | 18 | 0 | 7 | 4 | ||||||||||||
| NIH overall severity | ||||||||||||||||
| Mild | 12 | 26 | 18 | 21 | ||||||||||||
| Moderate | 53 | 47 | 54 | 46 | ||||||||||||
| Severe | 35 | 26 | 28 | 32 | ||||||||||||
| Characteristic . | Test set 1 (Fred Hutchinson Cancer Research Center collection) (n = 38) . | Test set 2 (University Hospital Regensburg collection) (n = 23) . | Replication set 1 cGVHD Consortium (U54) (n = 198) . | Replication set 2 Vancouver cGvHD network collection (n = 83) . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cGVHD (n = 17) . | Control (n = 21) . | cGVHD (n = 19) . | Control (n = 4) . | cGVHD (n = 104) . | Control (n = 94) . | cGVHD (n = 30) . | Control (n = 53) . | |||||||||
| No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | No. . | % . | |
| Time from HCT to sample, mo | ||||||||||||||||
| Early (<9) | 0 | 1 | 5 | 9 | 47 | 2 | 50 | 65 | 63 | 49 | 52 | 23 | 77 | 46 | 87 | |
| Late (>9) | 17 | 100 | 20 | 95 | 10 | 53 | 2 | 50 | 39 | 38 | 45 | 48 | 7 | 23 | 7 | 13 |
| Prior aGVHD | ||||||||||||||||
| No | 4 | 25 | 6 | 30 | 6 | 32 | 4 | 100 | 42 | 40 | 40 | 43 | 8 | 29 | 18 | 34 |
| Yes | 12 | 75 | 14 | 70 | 13 | 68 | 0 | 62 | 60 | 54 | 57 | 20 | 71 | 35 | 66 | |
| Age, y | ||||||||||||||||
| <50 | 9 | 53 | 8 | 38 | 12 | 63 | 3 | 75 | 41 | 39 | 37 | 39 | 17 | 57 | 30 | 57 |
| ≥50 | 8 | 47 | 13 | 62 | 7 | 37 | 1 | 25 | 63 | 61 | 57 | 61 | 13 | 43 | 23 | 43 |
| Donor | ||||||||||||||||
| Matched related | 6 | 35 | 11 | 52 | 6 | 32 | 1 | 25 | 39 | 38 | 27 | 29 | 11 | 38 | 14 | 27 |
| Matched unrelated | 10 | 59 | 5 | 24 | 10 | 53 | 2 | 50 | 45 | 43 | 33 | 35 | 14 | 48 | 30 | 58 |
| Mismatched | 1 | 6 | 5 | 24 | 3 | 16 | 1 | 25 | 20 | 19 | 34 | 36 | 4 | 14 | 8 | 15 |
| Stem cell source | ||||||||||||||||
| PBSCs | 16 | 94 | 15 | 71 | 19 | 100 | 3 | 75 | 93 | 89 | 57 | 61 | 28 | 97 | 46 | 92 |
| BM | 1 | 6 | 6 | 29 | 0 | 1 | 25 | 5 | 5 | 7 | 7 | 1 | 3 | 3 | 6 | |
| Umbilical cord blood | 0 | 0 | 0 | 0 | 6 | 6 | 30 | 32 | 0 | 1 | 2 | |||||
| Conditioning | ||||||||||||||||
| Myeloablative without TBI | 5 | 29 | 5 | 25 | 11 | 58 | 2 | 50 | 22 | 21 | 14 | 15 | 6 | 20 | 9 | 17 |
| Myeloablative with TBI | 3 | 18 | 6 | 30 | 8 | 42 | 2 | 50 | 22 | 21 | 25 | 27 | 16 | 53 | 13 | 25 |
| Nonmyeloablative | 9 | 53 | 9 | 45 | 0 | 0 | 60 | 58 | 55 | 59 | 8 | 27 | 31 | 58 | ||
| GVHD prophylaxis | ||||||||||||||||
| CNI + MTX/MMF with or without sirolimus | 16 | 94 | 20 | 95 | 6 | 32 | 1 | 25 | 94 | 90 | 71 | 76 | 24 | 89 | 25 | 47 |
| CNI with or without sirolimus | 0 | 0 | 0 | 0 | 8 | 8 | 16 | 17 | 0 | 0 | ||||||
| CNI + MTX/MMF + ATG | 0 | 0 | 13 | 68 | 3 | 75 | 0 | 0 | 3 | 11 | 28 | 53 | ||||
| Other | 1 | 6 | 1 | 5 | 0 | 0 | 2 | 2 | 7 | 7 | 0 | 0 | ||||
| Sex | ||||||||||||||||
| Male | 7 | 41 | 10 | 48 | 14 | 74 | 2 | 50 | 63 | 61 | 50 | 53 | 19 | 63 | 29 | 55 |
| Female | 10 | 59 | 11 | 52 | 5 | 26 | 2 | 50 | 41 | 39 | 44 | 47 | 11 | 37 | 24 | 45 |
| Female donor to male patient | ||||||||||||||||
| No | 13 | 76 | 17 | 81 | 17 | 89 | 4 | 100 | 77 | 74 | 77 | 82 | 26 | 90 | 45 | 88 |
| Yes | 4 | 24 | 4 | 19 | 2 | 11 | 0 | 27 | 26 | 17 | 18 | 3 | 10 | 6 | 12 | |
| CMV serostatus | ||||||||||||||||
| Negative | 5 | 29 | 12 | 57 | 15 | 79 | 4 | 100 | 48 | 47 | 41 | 44 | 13 | 43 | 29 | 57 |
| Positive | 12 | 71 | 9 | 43 | 4 | 21 | 0 | 55 | 53 | 53 | 56 | 17 | 57 | 22 | 43 | |
| Disease diagnosis | ||||||||||||||||
| AML | 4 | 24 | 9 | 43 | 11 | 58 | 2 | 50 | 30 | 29 | 34 | 36 | 12 | 41 | 17 | 43 |
| ALL | 3 | 18 | 3 | 14 | 2 | 11 | 0 | 14 | 13 | 13 | 14 | 6 | 21 | 4 | 10 | |
| CML | 0 | 1 | 5 | 0 | 0 | 8 | 8 | 5 | 5 | 2 | 7 | 2 | 5 | |||
| CLL | 1 | 6 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 2 | 7 | 5 | 13 | |||
| MDS | 1 | 6 | 3 | 14 | 1 | 5 | 0 | 23 | 22 | 14 | 15 | 1 | 3 | 8 | 20 | |
| HL/NHL | 4 | 24 | 4 | 19 | 4 | 21 | 1 | 25 | 14 | 13 | 15 | 16 | 2 | 7 | 4 | 10 |
| MM | 0 | 1 | 5 | 0 | 1 | 25 | 7 | 7 | 7 | 7 | 2 | 7 | 0 | |||
| Other | 4 | 24 | 0 | 1 | 5 | 0 | 5 | 5 | 3 | 3 | 2 | 7 | 0 | |||
| Receiving steroids at sample draw | ||||||||||||||||
| No | 14 | 82 | 21 | 100 | 10 | 53 | 4 | 100 | 55 | 53 | 83 | 88 | NA | |||
| Yes | 3 | 18 | 0 | 9 | 47 | 0 | 49 | 47 | 11 | 12 | ||||||
| cGVHD characteristics | ||||||||||||||||
| Median days from HCT to cGVHD (range) | 362 (257-818) | 281 (106-1953) | 203 (83-566) | 174 (74-383) | ||||||||||||
| Median days from cGVHD to sample (range) | 2 (0-24) | 0 (–1 to 0) | 6 (–12 to 29) | 2 (–21 to 27) | ||||||||||||
| Skin involvement | 59 | 79 | 63 | 65 | ||||||||||||
| Oral involvement | 65 | 53 | 61 | 83 | ||||||||||||
| GI involvement | 41 | 21 | 25 | 26 | ||||||||||||
| Eye involvement | 35 | 26 | 41 | 26 | ||||||||||||
| Joint involvement | 29 | 5 | 15 | 9 | ||||||||||||
| Lung involvement | 59 | 21 | 48 | 48 | ||||||||||||
| Liver involvement | 47 | 32 | 56 | 43 | ||||||||||||
| Genital involvement | 18 | 0 | 7 | 4 | ||||||||||||
| NIH overall severity | ||||||||||||||||
| Mild | 12 | 26 | 18 | 21 | ||||||||||||
| Moderate | 53 | 47 | 54 | 46 | ||||||||||||
| Severe | 35 | 26 | 28 | 32 | ||||||||||||
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATG, antithymocyte globulin; BM, bone marrow; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CMV, cytomegalovirus; CNI, calcineurin inhibitor; HCT, hematopoietic stem cell transplantation; HL, Hodgkin lymphoma; MDS, myelodysplastic syndrome; MM, multiple myeloma; MMF, mycophenolate mofetil; MTX, methotrexate; NHL, non-Hodgkin lymphoma; PBSC, peripheral blood stem cell.
Test set univariate analysis results
| Candidate biomarkers . | Method of measurement . | cGVHD cases (n = 17) . | Controls (n = 21) . | Effect size* . | P . | ||
|---|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | ||||
| Test set 1 | |||||||
| Aminopeptidase N (sCD13), μU/mL | Enzymatic assay | 0.30 | 0.14 | 0.18 | 0.07 | 1.5× | .004 |
| Proteomic | 1.44 | 0.73 | 0.90 | 0.26 | 1.5× | .004 | |
| IL-2Rα (sCD25) (pg/mL) | Luminex | 3125 | 1825 | 1580 | 700 | 1.8× | .005 |
| PI-16, fmol | Proteomic | 0.81 | 0.27 | 1.12 | 0.35 | 0.7× | .005 |
| MRM-MS | 1.21 | 0.56 | 1.63 | 0.47 | 0.5× | .005 | |
| Gelsolin, fmol | Proteomic | 0.92 | 0.22 | 1.11 | 0.19 | 0.8× | .02 |
| MRM-MS | 0.063 | 0.040 | 0.133 | 0.090 | 0.7× | .02 | |
| Kallikrein, fmol | Proteomic | 0.80 | 0.32 | 1.00 | 0.32 | 0.8× | .06 |
| MRM-MS | 0.068 | 0.022 | 0.095 | 0.043 | 0.8× | .007 | |
| ICAM-1, pg/mL | Luminex | 140 | 110 | 80 | 40 | 1.4× | .04 |
| Test set 2 | (n = 19) | (n = 4) | |||||
| CXCL10, pg/mL | ELISA | 1423 | 765 | 349 | 157 | 3.8× | .005 |
| Candidate biomarkers . | Method of measurement . | cGVHD cases (n = 17) . | Controls (n = 21) . | Effect size* . | P . | ||
|---|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | ||||
| Test set 1 | |||||||
| Aminopeptidase N (sCD13), μU/mL | Enzymatic assay | 0.30 | 0.14 | 0.18 | 0.07 | 1.5× | .004 |
| Proteomic | 1.44 | 0.73 | 0.90 | 0.26 | 1.5× | .004 | |
| IL-2Rα (sCD25) (pg/mL) | Luminex | 3125 | 1825 | 1580 | 700 | 1.8× | .005 |
| PI-16, fmol | Proteomic | 0.81 | 0.27 | 1.12 | 0.35 | 0.7× | .005 |
| MRM-MS | 1.21 | 0.56 | 1.63 | 0.47 | 0.5× | .005 | |
| Gelsolin, fmol | Proteomic | 0.92 | 0.22 | 1.11 | 0.19 | 0.8× | .02 |
| MRM-MS | 0.063 | 0.040 | 0.133 | 0.090 | 0.7× | .02 | |
| Kallikrein, fmol | Proteomic | 0.80 | 0.32 | 1.00 | 0.32 | 0.8× | .06 |
| MRM-MS | 0.068 | 0.022 | 0.095 | 0.043 | 0.8× | .007 | |
| ICAM-1, pg/mL | Luminex | 140 | 110 | 80 | 40 | 1.4× | .04 |
| Test set 2 | (n = 19) | (n = 4) | |||||
| CXCL10, pg/mL | ELISA | 1423 | 765 | 349 | 157 | 3.8× | .005 |
SD, standard deviation; U, enzymatic unit.
Fold difference for cGVHD vs control.
Test set 2 (Table 2) was made up of 19 cGVHD onset samples and 4 time-matched controls obtained in collaboration with University Hospital Regensburg (D.W.). CXCL10 (P = .005) was identified by using a screening Luminex assay, ELISA, and enzymatic assays for markers previously identified in a pediatric cohort and evaluated in test set 1.7 Two collaborators (S.G.H. and M.-J.H.) identified 2 additional vascular inflammation markers. One marker was identified in collaboration with the Canadian National Transplantation Research Program (M.-J.H.)12,13 as anti-LG3, a potential validated kidney vascular rejection marker that may be elevated in GVHD.13 The second was identified in collaboration with Oregon Health & Science University (S.G.H.) by using a customized Luminex assay for vascular markers. Endothelin-114,15 was identified in 2 patients with samples drawn at the onset of cGVHD and 28 days later. Both had a significantly increased endothelin-1 concentration at diagnosis, with a 4.8-fold decrease in endothelin-1 after cGVHD treatment was initiated (19.7 to 4.1 pg/mL). We added two markers, CXCL98 and sBAFF,6,7 shown in previous publications as being significant for evaluation in the replication sets, even though they were not identified by the 2 test set evaluations or by our collaborators.
Replication of candidate diagnostic cGVHD biomarkers identified in test sets 1 and 2
On the basis of their significance in test sets 1 and 2 and in the literature, 11 biomarkers were selected for testing by using the algorithm in Figure 1. These included soluble aminopeptidase N (sCD13), gelsolin, kallikrein, PI-16, CXCL10, ICAM-1, endothelin-1, sBAFF, IL-2Rα (sCD25), anti-LG3, and CXCL9 (supplemental Table 2). Patient characteristics for replication sets 1 and 2 are summarized in Table 1. Replication set 1 included 198 patients (n = 104 [cGVHD]; n = 94 [controls after BMT]), and replication set 2 included 83 patients (n = 30 [cGVHD]; n = 53 [controls after BMT]).
In replication set 1 (Table 3), we found that in 4 significant biomarkers—sBAFF (P = .05), CXCL10 (P = .02), ICAM-1 (P = .01), and anti-LG3 (P = .04)—ROC AUCs were between 0.59 and 0.61. The highest ROC AUC was attained with CXCL10 in combination with anti-LG3 (0.65; Figure 2A).
Replication set univariate analysis for candidate biomarkers
| Candidate biomarkers . | No. of evaluable cases . | Effect size* . | P . | ROC AUC . |
|---|---|---|---|---|
| Replication set 1 | ||||
| Aminopeptidase N (sCD13) | 193 | 1.0× | .99 | |
| sIL-2Rα | 198 | 1.1× | .13 | 0.56 |
| sBAFF | 198 | 1.2× | .05 | 0.59 |
| CXCL10 | 198 | 1.5× | .02 | 0.61 |
| PI-16 | 173 | 0.9× | .30 | |
| ICAM-1 | 198 | 1.2× | .01 | 0.60 |
| CXCL9 | 187 | 1.4× | .06 | 0.60 |
| Anti-LG3† | 198 | 0.7× | .04 | 0.59 |
| Endothelin-1† | 160 | 1.3× | .20 | |
| Gelsolin† | 173 | 1.0× | .43 | |
| Kallikrein† | 173 | 1.0× | .48 | |
| Combination of markers | ||||
| CXCL10 + anti-LG3 | 198 | .003 | 0.65 | |
| CXCL10 + CXCL9 + sBAFF + anti-LG3 | 187 | .04 | 0.63 | |
| Replication set 2 | ||||
| Aminopeptidase N (sCD13) | 83 | 1.3× | .02 | 0.64 |
| sIL-2Rα | 83 | 1.1× | .28 | |
| sBAFF | 83 | 1.5× | .01 | 0.68 |
| CXCL10 | 83 | 3.9× | <.0001 | 0.78 |
| PI-16 | 83 | 0.8× | .11 | |
| ICAM-1 | 83 | 1.2× | .23 | |
| CXCL9 | 83 | 1.8× | .04 | 0.67 |
| Anti-LG3‡ | 83 | 0.9× | .55 | |
| Endothelin-1 | 80 | 0.7× | .02 | 0.65 |
| Gelsolin† | 83 | 0.8× | .0007 | 0.71 |
| Kallikrein† | 83 | 1.0× | .58 | |
| Combination of markers | ||||
| CXCL10 + CXCL9 | 83 | <.0001 | 0.83 | |
| CXCL10 + CXCL9 + sBAFF + anti-LG3 | 83 | <.0001 | 0.83 |
| Candidate biomarkers . | No. of evaluable cases . | Effect size* . | P . | ROC AUC . |
|---|---|---|---|---|
| Replication set 1 | ||||
| Aminopeptidase N (sCD13) | 193 | 1.0× | .99 | |
| sIL-2Rα | 198 | 1.1× | .13 | 0.56 |
| sBAFF | 198 | 1.2× | .05 | 0.59 |
| CXCL10 | 198 | 1.5× | .02 | 0.61 |
| PI-16 | 173 | 0.9× | .30 | |
| ICAM-1 | 198 | 1.2× | .01 | 0.60 |
| CXCL9 | 187 | 1.4× | .06 | 0.60 |
| Anti-LG3† | 198 | 0.7× | .04 | 0.59 |
| Endothelin-1† | 160 | 1.3× | .20 | |
| Gelsolin† | 173 | 1.0× | .43 | |
| Kallikrein† | 173 | 1.0× | .48 | |
| Combination of markers | ||||
| CXCL10 + anti-LG3 | 198 | .003 | 0.65 | |
| CXCL10 + CXCL9 + sBAFF + anti-LG3 | 187 | .04 | 0.63 | |
| Replication set 2 | ||||
| Aminopeptidase N (sCD13) | 83 | 1.3× | .02 | 0.64 |
| sIL-2Rα | 83 | 1.1× | .28 | |
| sBAFF | 83 | 1.5× | .01 | 0.68 |
| CXCL10 | 83 | 3.9× | <.0001 | 0.78 |
| PI-16 | 83 | 0.8× | .11 | |
| ICAM-1 | 83 | 1.2× | .23 | |
| CXCL9 | 83 | 1.8× | .04 | 0.67 |
| Anti-LG3‡ | 83 | 0.9× | .55 | |
| Endothelin-1 | 80 | 0.7× | .02 | 0.65 |
| Gelsolin† | 83 | 0.8× | .0007 | 0.71 |
| Kallikrein† | 83 | 1.0× | .58 | |
| Combination of markers | ||||
| CXCL10 + CXCL9 | 83 | <.0001 | 0.83 | |
| CXCL10 + CXCL9 + sBAFF + anti-LG3 | 83 | <.0001 | 0.83 |
Fold difference for cGVHD vs control.
Considered to be vascular markers.
Anti-LG3 has been associated with renal transplant rejection; reagents for anti-LG3 ELISA were provided by M.-J.H.
Receiver operator characteristic curves. (A) For replication set 1 with CXCL10 and anti-LG3 each alone and in combination; the combination gave the highest AUC and (B) for replication set 2 with CXCL10 and CXCL9 each alone and in combination; the combination gave the highest AUC.
Receiver operator characteristic curves. (A) For replication set 1 with CXCL10 and anti-LG3 each alone and in combination; the combination gave the highest AUC and (B) for replication set 2 with CXCL10 and CXCL9 each alone and in combination; the combination gave the highest AUC.
In replication set 2 (Table 3), an identical analysis was performed. Findings were different from those in replication set 1. Three biomarkers were identified as significant in replication set 2 that had not been identified in replication set 1, including aminopeptidase N (P = .02), endothelin-1 (P = .02), and gelsolin (P = .0007). Markers identified as significant in replication set 1, but not as significant in replication set 2, included ICAM-1 and anti-LG3. There were 2 markers identified as statistically significant in replication set 1 that had been previously identified in replication set 2. These were sBAFF (P = .01) and, most significantly, CXCL10 (P < .0001). Overall, the ROC AUCs were much higher in replication set 2 compared with replication set 1, with 4 markers—aminopeptidase N, sBAFF, CXCL9, and endothelin-1—having ROC AUCs between 0.60 and 0.69. Two markers, gelsolin (0.71) and CXCL10 (0.78), had ROC AUCs ≥0.70. The best model by forward or backward model selection was CXCL10 combined with CXCL9, with an AUC of 0.83 (Figure 2B). Analysis of the 4 markers, CXCL10, CXCL9, anti-LG3, and sBAFF, in combination in both replication sets 1 and 2 did not increase the AUC compared with the 2 combinations of CXCL10 plus anti-LG3 or CXCL10 and CXCL9 in the 2 replication sets (Table 3). The best cutoffs for CXCL10 (for highest sensitivity with high specificity) were 291 pg/mL (29% sensitivity and 90% specificity) in replication set 1 and 313 pg/mL (50% sensitivity and 91% specificity) in replication set 2.
Analysis of covariates that have an impact on cGVHD biomarkers
We performed a multivariate analysis evaluating major factors that may have an impact on the interpretation of biomarkers (Table 4): early vs late cGVHD, prior aGVHD, conditioning (myeloablative, myeloablative with TBI, or nonmyeloablative), female donor to male recipient, donor source (peripheral blood stem cells, bone marrow [BM], umbilical cord blood), donor matching (matched related, matched unrelated, mismatched), and age 50 years or older. For evaluation of time dependence, early cGVHD markers (cGVHD onset before 9 months) were matched with no GVHD controls (majority at 6 months after BMT) and late cGVHD (≥ 9 months cGVHD onset) with no GVHD controls (majority at 12 months after BMT). None of the control patients developed cGVHD within the first year after transplantation. Multiple factors were evaluated, including donor matching, donor source, TBI, previous aGVHD, time of onset of cGVHD (early vs late), and non-myeloablative preparatory regimen; all had an impact on the interpretation of at least 1 biomarker (Table 4). We found no concordance between the 2 replication sets for the impact of any of the factors on the marker results. Only 1 marker, PI-16, was not impacted by a covariate in either replication set. Some factors such as TBI, non-myeloablative transplantation, and donor mismatch were almost always associated with an increased effect size of the biomarker (CXCL10, endothelin-1, IL-2Rα). By contrast, the donor source of either BM or umbilical cord blood and late onset of cGVHD (≥ 9 months after BMT), were almost always (except for kallikrein) associated with a decreased biomarker effect size (sBAFF, endothelin-1, CXCL10) (Table 4). Evaluation for skin vs other organ involvement did not show any impact on the interpretation of the biomarker results (data not shown).
Multivariate analysis for association of biomarkers with cGVHD
| Biomarker . | Replication set 1 . | Replication set 2 . | ||||||
|---|---|---|---|---|---|---|---|---|
| Effect size* . | P . | Other effects† . | P . | Effect size* . | P . | Other effects† . | P . | |
| Aminopeptidase N | 1.1× | .41 | None | 1.2× | .13 | Age ≥ 50 y 1.3× | .03 | |
| ICAM-1 | 1.2× | .11 | Unrelated donor 0.8× | .02 | 1.2× | .45 | None | |
| CXCL10 | 1.5× | .04 | TBI 1.8× | .04 | 2.7× | .005 | None | |
| NMA 1.9× | .01 | |||||||
| Mismatch 2.1× | .01 | |||||||
| BM 0.5× | .04 | |||||||
| Endothelin-1 | 1.0× | .89 | aGVHD 1.6× | .02 | 0.7× | .04 | TBI 1.8× | .01 |
| Mismatch 2.0× | .04 | |||||||
| Umbilical cord blood 0.3× | .0005 | |||||||
| CXCL9 | 1.5× | .03 | aGVHD 0.6× | .003 | 1.5× | .29 | None | |
| NMA 2.3× | .001 | |||||||
| IL-2Rα | 1.2× | .02 | Late onset 0.8× | .003 | 1.0× | .99 | None | |
| TBI 1.3× | .05 | |||||||
| NMA 1.6× | .0001 | |||||||
| Anti-LG3 | 0.9× | .47 | aGVHD 0.6× | .0006 | 0.7× | .28 | Female donor to male recipient 0.4× | .01 |
| sBAFF | 1.2× | .05 | Late onset 0.8× | .04 | 1.2× | .38 | None | |
| BM 0.7× | .04 | |||||||
| Umbilical cord blood 0.7× | .02 | |||||||
| PI-16 | 0.9× | .46 | None | 0.6× | .008 | None | ||
| Gelsolin | 1.0× | .35 | TBI 0.9× | .03 | 0.8× | .03 | None | |
| Mismatch 0.9× | .04 | |||||||
| Kallikrein | 1.0 | .51 | None | 1.1× | .24 | BM 1.7× | .02 | |
| Biomarker . | Replication set 1 . | Replication set 2 . | ||||||
|---|---|---|---|---|---|---|---|---|
| Effect size* . | P . | Other effects† . | P . | Effect size* . | P . | Other effects† . | P . | |
| Aminopeptidase N | 1.1× | .41 | None | 1.2× | .13 | Age ≥ 50 y 1.3× | .03 | |
| ICAM-1 | 1.2× | .11 | Unrelated donor 0.8× | .02 | 1.2× | .45 | None | |
| CXCL10 | 1.5× | .04 | TBI 1.8× | .04 | 2.7× | .005 | None | |
| NMA 1.9× | .01 | |||||||
| Mismatch 2.1× | .01 | |||||||
| BM 0.5× | .04 | |||||||
| Endothelin-1 | 1.0× | .89 | aGVHD 1.6× | .02 | 0.7× | .04 | TBI 1.8× | .01 |
| Mismatch 2.0× | .04 | |||||||
| Umbilical cord blood 0.3× | .0005 | |||||||
| CXCL9 | 1.5× | .03 | aGVHD 0.6× | .003 | 1.5× | .29 | None | |
| NMA 2.3× | .001 | |||||||
| IL-2Rα | 1.2× | .02 | Late onset 0.8× | .003 | 1.0× | .99 | None | |
| TBI 1.3× | .05 | |||||||
| NMA 1.6× | .0001 | |||||||
| Anti-LG3 | 0.9× | .47 | aGVHD 0.6× | .0006 | 0.7× | .28 | Female donor to male recipient 0.4× | .01 |
| sBAFF | 1.2× | .05 | Late onset 0.8× | .04 | 1.2× | .38 | None | |
| BM 0.7× | .04 | |||||||
| Umbilical cord blood 0.7× | .02 | |||||||
| PI-16 | 0.9× | .46 | None | 0.6× | .008 | None | ||
| Gelsolin | 1.0× | .35 | TBI 0.9× | .03 | 0.8× | .03 | None | |
| Mismatch 0.9× | .04 | |||||||
| Kallikrein | 1.0 | .51 | None | 1.1× | .24 | BM 1.7× | .02 | |
Multivariate analysis was adjusted for early vs late onset, prior aGVHD, conditioning (myeloablative, myeloablative with total body irradiation [TBI], nonmyeloablative [NMA]), female donor to male recipient, source (peripheral blood stem cell, bone marrow [BM], umbilical cord blood), donor (matched related, matched unrelated, mismatched [HLA mismatch]), and age 50 years or older. Late onset is defined as cGVHD ≥9 months after BMT.
Fold difference for cGVHD vs control.
Other factors in model significant at 0.05 level.
Evaluation for a correlation between CXCL10 and CXCR3+ immune cell populations and cGVHD severity
It has previously been shown that CXCL9, CXCL10, and CXCL11 all influence trafficking of CXCR3+ T cells toward the peripheral tissues.16 In addition, a decrease in CD4+CXCR3+ T cells was seen in the blood of a small number of patients with active skin cGVHD and elevated serum CXCL10.17 In that study, CD4+CXCR3+ T cells were increased in skin biopsies from the same patients, suggesting a CXCL10-mediated recruitment of CXCR3+ cells to tissues. In addition to CD4+ and CD8+ T cells, another population, CD56bright NK cells, also express CXCR3.18
We evaluated whether there were any differences in the percentage of CXCR3+-expressing CD4+ T cells, CD8+ T cells, or CD56bright NK cells in the total lymphocyte count between patients with and those without cGVHD. Second, we evaluated whether there was a difference in the ratio of the plasma CXCL10 level to percentage of the 3 cell subsets in patients with or without cGVHD. These analyses were performed on a cohort of patients from replication sets 1 and 2 who had paired plasma and cell samples available. We found that the subset of replication sets 1 and 2 that was evaluated was representative of the larger replications sets with not more than a 20% difference between any of the clinical variables in the subset and the larger replication sets (supplemental Table 3). Supporting this conclusion, the CXCL10 ROC AUCs in the subsets of 0.61 and 0.73, respectively (Table 5), were similar to those in the larger replications sets. Evaluation of the 3 CXCR3+-expressing populations revealed that CXCR3+CD56bright NK cells were the only CXCR3+ population significantly associated with lower cell numbers in cGVHD for both replication set 1 (ROC AUC, 0.62; P = .03) and replication set 2 (ROC AUC, 0.70; P = .02). When a ratio of the 3 CXCR3+ cell populations was performed with CXCL10, all 3 populations had significant ROC AUCs in at least 1 replication set. CD56brightCXCR3+ NK cells remained the most significant in both replication sets 1 and 2 (Table 5). Correlation with CXCL9 and CXCR3+ populations was in fact slightly higher than that for CXCL10 in replication set 1 and the inverse in replication set 2 (Table 5). The most significant association of CXCL9 was with CXCR3+CD56bright NK cells, identical to that in CXCL10.
Association of CXCR3+ T-cell and NK-cell populations with plasma CXCL10 and CXCL9 in a subpopulation
| Chemokine . | Controls (n = 67/65)* . | cGVHD cases (n = 52/44)* . | P . | ROC AUC . | ||
|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | |||
| Replication set 1 | ||||||
| CXCL10, pg/mL | 129 | 105 | 607 | 2194 | .04 | 0.61 |
| CXCL9, pg/mL | 8969 | 9218 | 17715 | 18675 | .02 | 0.63 |
| Subset (% of parent population)† | ||||||
| CD3–CD56brightCXCR3+ | 12.6 | 10.8 | 10.3 | 11.5 | .03 | 0.62 |
| CD8+CXCR3+ | 23.4 | 12.9 | 22.1 | 12.8 | .50 | 0.54 |
| CD4+CXCR3+ | 13.2 | 8.5 | 11.0 | 8.2 | .10 | 0.59 |
| Ratio CXCL10/% | ||||||
| CD3–CD56brightCXCR3+ | 24.3 | 37.8 | 159 | 447 | .03 | 0.63 |
| CD8+CXCR3+ | 7.9 | 9.2 | 35.2 | 103.2 | .13 | 0.58 |
| CD4+CXCR3+ | 17.9 | 25.7 | 154 | 610 | .04 | 0.61 |
| Ratio CXCL9/% | ||||||
| CD3–CD56brightCXCR3+ | 1760 | 3090 | 7079 | 13023 | .008 | 0.65 |
| CD8+CXCR3+ | 574 | 808 | 1256 | 1570 | .04 | 0.62 |
| CD4+CXCR3+ | 1246 | 1883 | 3065 | 4870 | .01 | 0.64 |
| Replication set 2 | (n = 25) | (n = 24) | ||||
| CXCL10, pg/mL | 238.6 | 449.4 | 819.5 | 1370 | .005 | 0.73 |
| CXCL9, pg/mL | 16036 | 25116 | 25805 | 31451 | .12 | 0.63 |
| Subset (% of parent population)† | ||||||
| CD3–CD56brightCXCR3+ | 16.0 | 10.62 | 9.37 | 5.73 | .01 | 0.70 |
| CD8+CXCR3+ | 25.64 | 15.02 | 25.39 | 12.69 | .98 | 0.51 |
| CD4+CXCR3+ | 2.38 ± 2.43 | 1.90 | 2.32 | .38 | 0.57 | |
| Ratio CXCL10/% | ||||||
| CD3–CD56brightCXCR3+ | 19.08 | 31.48 | 143.0 | 201.9 | .0009 | 0.78 |
| CD8+CXCR3+ | 14.43 | 33.96 | 48.6 | 113.7 | .006 | 0.73 |
| CD4+CXCR3+ | 223.3 | 336.1 | 1087 | 2371 | .008 | 0.72 |
| Ratio CXCL9/% | ||||||
| CD3–CD56brightCXCR3+ | 1155 | 1350 | 5887 | 11457 | .05 | 0.67 |
| CD8+CXCR3+ | 848 | 1208 | 1105 | 1308 | .12 | 0.63 |
| CD4+CXCR3+ | 22075 | 53070 | 29705 | 35102 | .08 | 0.65 |
| Chemokine . | Controls (n = 67/65)* . | cGVHD cases (n = 52/44)* . | P . | ROC AUC . | ||
|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | |||
| Replication set 1 | ||||||
| CXCL10, pg/mL | 129 | 105 | 607 | 2194 | .04 | 0.61 |
| CXCL9, pg/mL | 8969 | 9218 | 17715 | 18675 | .02 | 0.63 |
| Subset (% of parent population)† | ||||||
| CD3–CD56brightCXCR3+ | 12.6 | 10.8 | 10.3 | 11.5 | .03 | 0.62 |
| CD8+CXCR3+ | 23.4 | 12.9 | 22.1 | 12.8 | .50 | 0.54 |
| CD4+CXCR3+ | 13.2 | 8.5 | 11.0 | 8.2 | .10 | 0.59 |
| Ratio CXCL10/% | ||||||
| CD3–CD56brightCXCR3+ | 24.3 | 37.8 | 159 | 447 | .03 | 0.63 |
| CD8+CXCR3+ | 7.9 | 9.2 | 35.2 | 103.2 | .13 | 0.58 |
| CD4+CXCR3+ | 17.9 | 25.7 | 154 | 610 | .04 | 0.61 |
| Ratio CXCL9/% | ||||||
| CD3–CD56brightCXCR3+ | 1760 | 3090 | 7079 | 13023 | .008 | 0.65 |
| CD8+CXCR3+ | 574 | 808 | 1256 | 1570 | .04 | 0.62 |
| CD4+CXCR3+ | 1246 | 1883 | 3065 | 4870 | .01 | 0.64 |
| Replication set 2 | (n = 25) | (n = 24) | ||||
| CXCL10, pg/mL | 238.6 | 449.4 | 819.5 | 1370 | .005 | 0.73 |
| CXCL9, pg/mL | 16036 | 25116 | 25805 | 31451 | .12 | 0.63 |
| Subset (% of parent population)† | ||||||
| CD3–CD56brightCXCR3+ | 16.0 | 10.62 | 9.37 | 5.73 | .01 | 0.70 |
| CD8+CXCR3+ | 25.64 | 15.02 | 25.39 | 12.69 | .98 | 0.51 |
| CD4+CXCR3+ | 2.38 ± 2.43 | 1.90 | 2.32 | .38 | 0.57 | |
| Ratio CXCL10/% | ||||||
| CD3–CD56brightCXCR3+ | 19.08 | 31.48 | 143.0 | 201.9 | .0009 | 0.78 |
| CD8+CXCR3+ | 14.43 | 33.96 | 48.6 | 113.7 | .006 | 0.73 |
| CD4+CXCR3+ | 223.3 | 336.1 | 1087 | 2371 | .008 | 0.72 |
| Ratio CXCL9/% | ||||||
| CD3–CD56brightCXCR3+ | 1155 | 1350 | 5887 | 11457 | .05 | 0.67 |
| CD8+CXCR3+ | 848 | 1208 | 1105 | 1308 | .12 | 0.63 |
| CD4+CXCR3+ | 22075 | 53070 | 29705 | 35102 | .08 | 0.65 |
Number for analysis of CXCL10/CXCL9.
Each CXCR3+ subpopulation was collected as a percentage of its parent population of NK, CD4+, and CD8+ T cells.
In replication set 1, cGVHD severity (overall NIH score) is positively correlated with the combined biomarker score derived from logistic regression of CXCL10 and anti-LG3 and cGVHD (Ptrend = .07 from linear regression). Among cases in replication set 2, there is no such evidence (Ptrend = .67), although the number of cases is smaller.
Discussion
The results summarized in this article show that, in 2 separate replication sets, CXCL10 was the most significant diagnostic biomarker that distinguished patients with cGVHD from those without. This represents one of the largest such replication studies (total of 350 patients, including 170 cGVHD cases and 180 controls) performed to date in cGVHD. Interestingly, CXCL10 was not identified in the initial test set 1 of patients with late onset (≥9 months after BMT). There was a high variability in its significance with an ROC of only 0.61 in replication set 1 and 0.77 in replication set 2. In addition, its highest predictive value was increased in combination with other biomarkers, but they were not identical in the 2 replication sets. Its association was highest in replication set 1 with anti-LG3, a vascular rejection biomarker in kidney transplantation,12,13 and in replication set 2, with CXCL9 and CXCR3+ regulatory NK (NKreg) cells.
CXCL10 is an inflammatory chemokine binding to CXCR3, which is involved in the activation and recruitment of T cells, eosinophils, monocytes, and NK cells.16 Previously, in a smaller group of patients (n = 8), all 3 CXCR3-binding chemokines (CXCL9, CXCL10, and CXCL11) were increased in patients with cGVHD of the skin17 and conjunctiva.19 Moreover, a significant decrease in CD4+CXCR3+ T cells was seen in the blood with concomitant increase in the number of central arterioles in the dermis. Another recent study found that both CXCL10 and CXCL11, along with sBAFF, were reproducible diagnostic biomarkers for cGVHD.20 CXCL10 is well associated with aGVHD in mice21 and humans22-24 and in solid organ rejection.25
Elevation of the CXCR3+ agonists—CXCL9, CXCL10, and CXCL11—will cause migration of CXCR3+ effector populations in the skin. We were not able to evaluate skin cell infiltration, but we did evaluate for an inverse relationship in peripheral blood. We evaluated 3 populations—CD4+ T cells, CD8+ T cells, and CD56bright NK cells—associated with NK regulatory function. Interestingly, we found the closest correlation of CXCL10 elevation with a decrease in CD56bright NK cells. There is now very solid evidence that the CD56bright NK cell population is a classic NKreg population. We found that the CXCR3+CD56bright populations demonstrated a significantly higher expression of CD335 (NKp46; not shown) compared with CD337 in both subpopulations, consistent with the NKreg phenotype. 1,2,26-28 CD56brightCD16– NK cells exert low cytotoxicity but produce high levels of cytokines upon stimulation, they primarily exhibit an immunoregulatory role, and they are usually called NKreg cells. There have been a number of NKreg populations with this phenotype associated with immune regulatory functions and immune tolerance.3,29
There was high heterogeneity in our results. Markers were markedly elevated in either test set 1 or 2, as in previous studies, including aminopeptidase N and IL-2Rα,7,8 sBAFF,6,7 and CXCL9.8 Only 1 of the markers, sBAFF, was significant in both replication sets (excluding CXCL10). Six other markers were significant in either replication set 1 (ICAM-1, anti-LG3, and IL-2Rα) or replication set 2 (gelsolin, endothelin-1, CXCL9, and aminopeptidase N). Interestingly, 3 of those markers—endothelin-1, gelsolin, and anti-LG3—are considered vascular inflammation markers, supporting that this mechanism is important in cGVHD. We consider that an ROC AUC of ≥0.75 represents a marker of highest interest, whereas an AUC between 0.59 and 0.74 represents a marker that is of interest and that warrants further investigation. CXCL10 was the only biomarker to meet our criteria for future validation with an AUC of ≥0.75. In our opinion, aminopeptidase N, CXCL9, sBAFF, gelsolin, endothlin-1, ICAM-1, anti-LG3, and IL-2Rα (with AUCs between 0.59 and 0.74) all warrant further investigation in future replication studies. CXCL10 has now been identified by 2 separate groups in addition to our own, 1 group in a small study17 and another group in a recent larger study with 2 centers.20 On the basis of the criteria set out by the NIH consensus report for biomarkers, CXCL10 has been validated, but we would argue that before it is used in the clinic as a validated diagnostic biomarker for cGVHD, it should be further validated by at least 1 additional separate large cohort of patients.5 Validation should focus on a CXCL10 cutoff for maximum sensitivity on plasma or serum concentrations between 291 and 313 pg/mL (approximately 90% specificity).
The high heterogeneity of biomarkers in this study points out a number of factors that may influence interpretation of plasma and serum biomarkers being evaluated and replicated in cGVHD. As summarized in supplemental Table 1, the fact that the time from collection to processing and whether the sample was plasma or serum were different for each of the sample sets, which may have influenced the heterogeneity of the results. The NIH cGVHD biomarker consensus report5 identified a number of covariates that may influence the interpretation of results. In light of these concerns, we evaluated multiple covariates. All covariates evaluated (Table 4) had an impact on the interpretation of at least 1 marker. These findings support the conclusion that each biomarker used to diagnose cGVHD may require correction for different covariates. Our results also support the lack of consistency between previous larger and smaller cGVHD biomarker trials (summarized in Paczesny et al5 ). Large trials that consider multiple covariates will be needed to evaluate these cGVHD markers before they can be used in clinical applications.
The complexity and heterogeneity of cGVHD continues to make replication and validation of candidate biomarkers difficult. In this large discovery and replication study, we found that only 1 marker—CXCL10—met strict criteria for replication as a clinical biomarker for the diagnosis of cGVHD. The high heterogeneity of our results within the study emphasizes the critical need for more clinically well-documented samples that have been collected in a standardized way. We hypothesize that the heterogeneity of results can be minimized by considering the impact of covariates on each individual marker in its interpretation to create a biomarker profile that can be used for clinical decisions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank centers that contributed to the Chronic GVHD Consortium, including Vanderbilt (Madan Jagasia), Roswell Park (George Chen), Stanford University (Sally Arai), Cleveland Clinic Taussig Cancer Institute (Betty Hamilton), Washington University (Iskra Pusic), and Cornell University (Sebastian Ma).
This work was supported by a Canadian Institutes of Health Operating Grant; by the CIHR/Wyeth Clinical Research Chair in Transplantation (K.R.S.); by the Canadian National Transplant Research Program (funded by the CIHR); by the Office of Research in Women’s Health and the National Institute of Child Health and Human Development, Oregon Building Interdisciplinary Research Careers in Women’s Health Award No. 2K12HD043488 (S.G.H.); by the Medical Foundation of Oregon (S.G.H.); and by the Deutsche José Carreras Leukämie-Stiftung (D.W.). The Chronic GVHD Consortium is funded via collaboration between the Center for Advancing Translational Sciences and the National Cancer Institute (NCI) (U54CA163438). Support for the discovery samples was provided by grant CA118953 from NCI. The Canadian Bone Marrow Transplant Group Clinical Trials Network provided samples from CBMTG 0601 (funded by National Institutes of Health/NCI 1R01CA108752-01A2) and samples from CBMTG 0801 (funded by CIHR).
Authorship
Contribution: A.K. designed and performed research, wrote the manuscript, and analyzed and interpreted data; S.G.H. performed research, wrote the manuscript, and analyzed and interpreted data; S.I. designed and performed research, performed statistical analysis, and analyzed and interpreted data; J.R. performed research, wrote the manuscript, performed statistical analysis, and analyzed and interpreted data; M.-J.H. contributed vital new reagents or analytical tools; P.J.M., S.J.L., D.W., and P.S. collected data and wrote the manuscript; S.A., S.S., J.S., M. Aljurf, M. Arora, C.C., G.G., J.K., J.L., T.J.N., L.F.N., J.P., G.P., D.S., J.T., C.L.T., I.W., and S.C. collected data; M.L. advised on experimental design and interpretation; T.P. analyzed data from the CBMTG 0801 and 0601 studies (Vancouver cohort, replication set 2); B.E.S. wrote the manuscript and performed statistical analysis; and K.R.S. designed the research, wrote the manuscript, and analyzed and interpreted data.
Conflict-of-interest disclosure: P.J.M. participated in a single scientific advisory board meeting for Pharmacyclics. The remaining authors declare no competing financial interests.
Correspondence: Kirk R. Schultz, British Columbia Children’s Hospital, University of British Columbia, 4480 Oak St, Room A119, Vancouver, BC V6H 3V4, Canada; e-mail: kschultz@mail.ubc.ca.