To the editor:
RUNX1 (also known as AML1 or CBFA2) and CBFB encode the α and β subunits of a heterodimeric core binding transcription factor complex involved in the development of normal hematopoiesis (reviewed by de Bruijn and Speck1 ). Both genes are rearranged in acute myeloid leukemia (AML) with t(8;21)(q22;q22);RUNX1-RUNX1T1 and inv(16)(p13.1q22)/t(16;16)(p13.1;q22);CBFB-MYH11, which are collectively called core binding factor (CBF) AML. Targeted mutational studies have shown that both CBF AML subgroups are characterized by recurrent mutations in KIT, FLT3, NRAS, and KRAS.2-5 Additional mutations in ASXL2 and ASXL1 were also recently described in t(8;21) AML.6,7
Mutation analyses employing untargeted approaches in large CBF AML cohorts are still lacking. We previously described comparative transcriptomic approaches leading to the comprehensive description of the mutational and transcriptomic landscape of MLL,8 EVI1,9 and NUP98-NSD110 AML subgroups. Using this same methodology and cohort, we now report the results of the 48 CBF AML specimens included in our collection of 415 specimens. We identified novel mutations and differentially expressed genes and demonstrated that the RUNX1-CBFA2T3 AML sample is characterized by a gene expression profile highly similar to that of RUNX1-RUNX1T1 AML.
This study is part of the Leucegene project, an initiative approved by the research ethics boards of Université de Montréal and Maisonneuve-Rosemont Hospital. All AML samples were collected with an informed consent between 2001 and 2015 according to Quebec Leukemia Cell Bank (BCLQ) procedures. Workflow for sequencing, mutation analysis, and transcripts quantification have been described previously8-10 and are complemented in the supplemental Methods (available on the Blood Web site).
Forty-eight CBF AML specimens, including 28 samples with inv(16) and 20 with t(8;21), and 367 control AML specimens were part of this analysis. Patient characteristics are described in Figure 1A. The cytogenetic distribution of the entire cohort is shown in supplemental Figure 1.
Transcriptomic landscapes of CBF and RUNX1-CBFA2T3 AML. (A) Characteristics of CBF and non-CBF AML cohorts. (B-C) Comparative analyses of expressed genes in t(8;21) (B) and inv(16) (C) AML subgroups. Diamonds correspond to differentially expressed genes (difference ≥1 or ≤−1) listed in supplemental Tables 1 and 2 for panels B and C, respectively. Scales: mean{log10[(RPKM+0.0001)*10 000]}. (D) PCA performed in the Leucegene cohort (n = 415) using the 145-gene signature that characterizes AML with t(8;21)/RUNX1-RUNX1T1 (red dots). One sample with a t(16;21)/RUNX1-CBFA2T3 (lime green dot) clustered with t(8;21) AML. (E) PCA performed in the same cohort using the 127-gene signature that characterizes AML with inv(16)/CBFB-MYH11 (steel blue dots). A small constant of 0.0001 was added to all RPKM values prior to log10 transformation. FAB, French-American-British; RPKM, reads per kilobase per million mapped reads; T-AML, therapy-related AML; WBC, white blood cell.
Transcriptomic landscapes of CBF and RUNX1-CBFA2T3 AML. (A) Characteristics of CBF and non-CBF AML cohorts. (B-C) Comparative analyses of expressed genes in t(8;21) (B) and inv(16) (C) AML subgroups. Diamonds correspond to differentially expressed genes (difference ≥1 or ≤−1) listed in supplemental Tables 1 and 2 for panels B and C, respectively. Scales: mean{log10[(RPKM+0.0001)*10 000]}. (D) PCA performed in the Leucegene cohort (n = 415) using the 145-gene signature that characterizes AML with t(8;21)/RUNX1-RUNX1T1 (red dots). One sample with a t(16;21)/RUNX1-CBFA2T3 (lime green dot) clustered with t(8;21) AML. (E) PCA performed in the same cohort using the 127-gene signature that characterizes AML with inv(16)/CBFB-MYH11 (steel blue dots). A small constant of 0.0001 was added to all RPKM values prior to log10 transformation. FAB, French-American-British; RPKM, reads per kilobase per million mapped reads; T-AML, therapy-related AML; WBC, white blood cell.
Using the most minimally and differentially expressed genes, we identified signatures of 145 and 127 genes that best characterize t(8;21) and inv(16) subgroups, respectively (Figure 1B-C; supplemental Tables 1 and 2). Fusion partner genes, RUNX1T1 and MYH11, are among the single most differentially expressed genes in each corresponding group. Previously reported candidates such as POU4F1 [t(8;21)] and ST18 [inv(16)] were among the most discriminatory genes identified by our analysis. Other CBF microarray data sets were readily enriched in Gene Set Enrichment Analysis studies11-13 (supplemental Tables 1 and 2). Importantly, ∼80% of genes identified in our CBF AML signatures have not been previously described in those data sets. For example, ADARB2-AS1 and LINC00958 are typical for t(8;21) AML and MEGF10 and APLN for inv(16) specimens in our collection. Our signatures shared 50% and 25% of the most significantly overexpressed or underexpressed genes with pediatric t(8;21) and inv(16) AML cohorts, respectively (supplemental Tables 1 and 2).14 This suggests that similar networks are at play in pediatric and adult CBF AML.
Using the subgroup-specific gene signatures and performing principal component analyses (PCAs), each CBF subgroup homogeneously clustered together (Figure 1D-E). Most interestingly, 1 sample harboring a t(16;21)(q24;q22);RUNX1-CBFA2T3 unambiguously clustered with t(8;21) specimens, suggesting that the transcriptional network is shared between these 2 entities (Figure 1D; supplemental Figure 2). In agreement with this observation, a KIT D817V mutation was detected in this RUNX1-CBFA2T3 sample. The RUNX1-CBFA2T3 fusion is a rare but recurrent gene rearrangement in AML (recently reviewed by Athanasiadou et al15 ). In contrast to other RUNX1 fusion partners, CBFA2T3 (also known as MTG16) shows high sequence identity with RUNX1T1 leading to a RUNX1-CBFA2T3 chimeric protein that shares similar structural characteristics to RUNX1-RUNX1T1.16 Further analyses are needed to determine if these observations translate into a similar susceptibility to chemotherapy and clinical outcome.
Genes mutated in t(8;21) and inv(16) cohorts are shown in Figure 2A and detailed in supplemental Table 3. As previously reported, the most frequent mutations in both CBF subgroups were found in activated signaling genes (Figure 2A). Notably, 15 (31%) CBF samples contained 2 to 5 mutations in activated signaling genes. In the majority of cases, the sum of VAF did not exceed ∼50% (Figure 2B), hence suggesting that they occurred in different subclones. The loss of 2 signaling mutations and expansion of a third one in a relapse specimen further support this concept (Figure 2C).
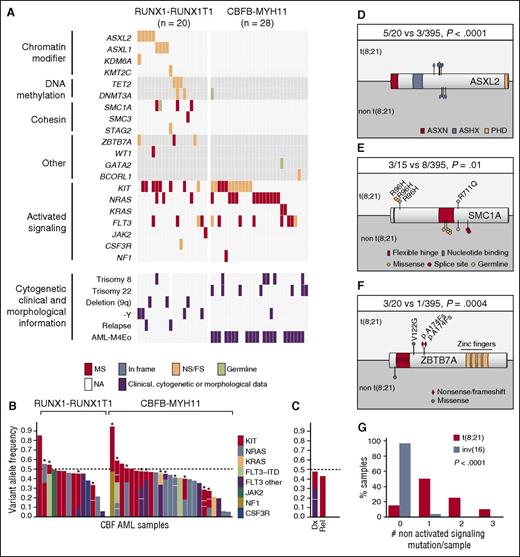
Mutational landscape of CBF AML. (A) Mutational, morphological, cytogenetic, and clinical information of CBF AML. Each column represents a patient sample. (B) Variant allele frequency (VAF) of mutations in activated signaling genes across t(8;21) and inv(16) genetic groups. Each bar represents a patient sample. Stars identify samples with 2 or more mutations. Note that for each mutation in a given specimen, VAFs are stacked with a no co-occurrence presumption to facilitate presentation of data. (C) Activated signaling mutations in a sample at diagnosis and relapse. (D-F) Primary structures of ASXL2, SMC1A, and ZBTB7A proteins, respectively, with corresponding positions of mutations. (G) Number of nonactivated signaling mutations in CBF AML subgroups. Statistics are based on Fisher’s exact test. ASX, additional sex combs; ASXH, ASX homology; ASXN, ASX N terminal; dx, diagnosis; FS, frameshift; MS, missense; NA, not available; NS, nonsense; PHD, plant homeodomain; rel, relapse.
Mutational landscape of CBF AML. (A) Mutational, morphological, cytogenetic, and clinical information of CBF AML. Each column represents a patient sample. (B) Variant allele frequency (VAF) of mutations in activated signaling genes across t(8;21) and inv(16) genetic groups. Each bar represents a patient sample. Stars identify samples with 2 or more mutations. Note that for each mutation in a given specimen, VAFs are stacked with a no co-occurrence presumption to facilitate presentation of data. (C) Activated signaling mutations in a sample at diagnosis and relapse. (D-F) Primary structures of ASXL2, SMC1A, and ZBTB7A proteins, respectively, with corresponding positions of mutations. (G) Number of nonactivated signaling mutations in CBF AML subgroups. Statistics are based on Fisher’s exact test. ASX, additional sex combs; ASXH, ASX homology; ASXN, ASX N terminal; dx, diagnosis; FS, frameshift; MS, missense; NA, not available; NS, nonsense; PHD, plant homeodomain; rel, relapse.
Sixteen different genes were mutated in t(8;21) AML cohort: KIT (8/20, 40%); ASXL2 (5/20, 25%); FLT3, ASXL1 (4/20, 20% each); ZBTB7A, NRAS, TET2, and SMC1A (3/20, 15%); DNMT3A (2/20, 10%); and KDM6A, KMT2C, SMC3, STAG2, WT1, JAK2, and CSF3R (1/20, 5% each) (Figure 2A). No association was found between mutations and additional cytogenetic aberrations and clinical or laboratory characteristics (Figure 2A). As previously described,17 an association was observed between t(8;21) and del(9q) or -Y. The activated signaling genes (14/20, 70%) were the most frequently mutated, followed by chromatin modifier (10/20, 50%), cohesin (5/20, 25% each), and DNA methylation (4/20, 20%) genes (Figure 2A). Mutations in the chromatin modifier ASXL2 were largely restricted to t(8;21) AML subgroup (5/20 vs 3/395, P < .0001, Fisher’s exact test; Figure 2D). Among the mutations identified in cohesin complex genes, 3 SMC1A acquired mutations occurred at position R96 (Figure 2E).
Three of 20 t(8;21) AML samples contained novel acquired mutations in ZBTB7A. Mutation in this gene occurred in only 1 other specimen, suggesting that it is specific to t(8;21) AML (3/20 vs 1/395, P = .0004, Fisher’s exact test; Figure 2F; supplemental Figure 3). Interestingly, ZBTB7A expression was the lowest in specimens with frameshift mutations suggesting that nonsense mediated decay could be at play (supplemental Figure 4). ZBTB7A (zinc finger and BTB domain containing 7A, also known as leukemia/lymphoma-related factor [LRF]) encodes a transcription factor of the POK (poxvirus and zinc finger and Krüppel)/ZBTB (zinc finger and broad complex, tramtrack, and bric-a-brac) family, which also includes BCL6 and PLZF (also known as ZBTB16) (reviewed in Lunardi et al18 ). ZBTB7A mutations were recently reported at a low frequency in various solid malignancies,19 in which they mostly consisted of missense mutations in the zinc finger domains. In contrast, 2/3 mutations identified in our analysis consisted of frameshift mutations, which are expected to lead to a truncated protein lacking the C-terminal zinc fingers. Analyses in other cohorts will determine whether ZBTB7A mutations in AML are specifically associated to t(8;21) subgroup.
As reported before, a strong association between inv(16) AML and trisomy 22 was noted (Figure 2A).17 Six genes were mutated in the inv(16) AML cohort: KIT and NRAS (12/28, 43% each), FLT3 (8/28, 29%), KRAS (2/28), and NF1 and BCORL1 (1/28 each). Strikingly, we found that the only recurrent mutations in this disease occurred in activated signaling genes, detected in 25/28 (89%) samples. This sharply contrasts with the high frequency of other nonactivated signaling gene mutations found in t(8;21) AML (Figure 2G).
In summary, this study represents the largest cohort of CBF AML analyzed by RNA sequencing to date, providing a comprehensive portrait of mutations and gene expression profiles in these AML subgroups. Our analysis complements the understanding of the mutational landscape of CBF AML, and has led to the identification of novel ZBTB7A mutations in t(8;21) AML subgroup. Finally, our transcriptomic subgroup-based approach unified the gene expression profiles of RUNX1-CBFA2T3 and RUNX1-RUNX1T1 AML. We hypothesize that additional AML genetic entities could be unified using a similar strategy.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Muriel Draoui for project coordination and Sophie Corneau for sample coordination, as well as Marianne Arteau and Raphaëlle Lambert at the Institute for Research in Immunology and Cancer genomics platform for RNA sequencing. The dedicated work of BCLQ staff, namely Giovanni d’Angelo, Claude Rondeau, and Sylvie Lavallée, is also acknowledged, as well as BCLQ coinvestigators for providing patient samples. G.S. and J.H. are recipients of research chairs from the Canada Research Chair program and Industrielle-Alliance (Université de Montréal), respectively. BCLQ is supported by grants from the Cancer Research Network of the Fonds de recherche du Québec–Santé. RNA-sequencing read mapping and transcript quantification were performed on the supercomputer Briaree from Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by grants from the Canada Foundation for Innovation, NanoQuébec, Réseau de médecine génétique appliquée, and the Fonds de recherche du Québec–Nature et technologies. V.-P.L. is the recipient of a Cole Foundation fellowship. This work was supported by the Government of Canada through Genome Canada and the Ministère de l’économie, de l’innovation et des exportations du Québec through Génome Québec, with supplementary funds from AmorChem.
Contribution: V.-P.L. contributed to project conception, analyzed transcriptomes of all samples, generated all figures, tables, and supplementary material, and was the main author of this manuscript; G.S. contributed to project conception and coordination and cowrote the manuscript; J.H. contributed to project conception, analyzed the cytogenetic and fluorescence in situ hybridization studies, provided all the AML samples, and cowrote the manuscript; R.N.A. contributed to sample sequencing and analysis; P.G. processed the raw next generation sequencing data and codeveloped the k-mer (km) approach (https://bitbucket.org/iric-soft/km); G.B. codeveloped the analytical pipeline; S.L. was responsible for supervision of the bioinformatics team and of statistical analyses and codeveloped the km approach; and I.B. performed data validation.
Conflict-of-interest: The authors declare no competing financial interests.
Correspondence: Josée Hébert, Banque de cellules leucémiques du Québec, 5415 L'Assomption Blvd, Montreal, QC H1T 2M4, Canada; e-mail: josee.hebert@umontreal.ca; and Guy Sauvageau, Institute for Research in Immunology and Cancer (IRIC), P.O. Box 6128, Station Centre-Ville, Montreal, QC H3C 3J7, Canada; e-mail: guy.sauvageau@umontreal.ca.

![Figure 1. Transcriptomic landscapes of CBF and RUNX1-CBFA2T3 AML. (A) Characteristics of CBF and non-CBF AML cohorts. (B-C) Comparative analyses of expressed genes in t(8;21) (B) and inv(16) (C) AML subgroups. Diamonds correspond to differentially expressed genes (difference ≥1 or ≤−1) listed in supplemental Tables 1 and 2 for panels B and C, respectively. Scales: mean{log10[(RPKM+0.0001)*10 000]}. (D) PCA performed in the Leucegene cohort (n = 415) using the 145-gene signature that characterizes AML with t(8;21)/RUNX1-RUNX1T1 (red dots). One sample with a t(16;21)/RUNX1-CBFA2T3 (lime green dot) clustered with t(8;21) AML. (E) PCA performed in the same cohort using the 127-gene signature that characterizes AML with inv(16)/CBFB-MYH11 (steel blue dots). A small constant of 0.0001 was added to all RPKM values prior to log10 transformation. FAB, French-American-British; RPKM, reads per kilobase per million mapped reads; T-AML, therapy-related AML; WBC, white blood cell.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/20/10.1182_blood-2016-03-703868/4/m_2498f1.jpeg?Expires=1769782092&Signature=EE~Lyh78g7O9fI6~b-xoow5NKiUa3zDOAI1EZvG9DJ-BLU5cOCBWzWt6mYd3GtETJGruro2I6atm9ewin~bsDV4adh-FMAsKvQnrx6Dmm5iboI5pgkLUZvVQYCA7MXAAavR0raW5zdoFdll1gdLErcjdR6j4L7s6gb7iieVFkXWWlGXo2KhBr7QUSABzUd5OITNIW8INFlhUnFIsBgziO08DDNPgH2OATPlJ0aY-~34QEFBWjm3unAX2WBdLUETbHmOSxzlVB-jZTsaKenO3edWAed1dsWPKB1aq2xD5brHBMSh30MgaOC~mcKj1Fs8PiM8w73T4tQwrvXkFvCxSzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)