In this issue of Blood, Duployez et al present results from mutational profiling of >200 patients with core binding factor acute myeloid leukemia (CBF-AML). The identified mutational landscape is likely to have clinical relevance and hints at intriguing biological distinctions between these seemingly similar malignancies that will offer deep insights into the aberrant regulatory networks critical for leukemia development.1
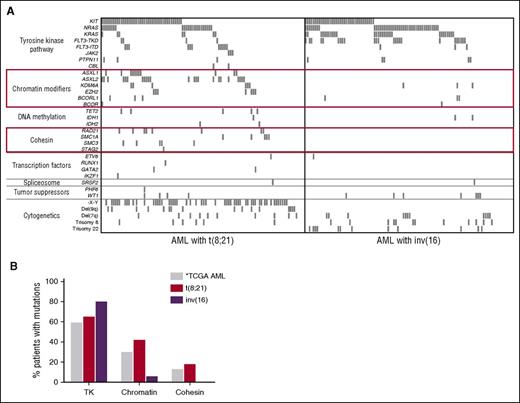
Mutational profile of CBF-AML. (A) Spectrum of recurrent mutations in 215 pediatric and adult patients with CBF-AML. ASXL1, ASXL2, and EZH2 mutations are mutually exclusive of each other as are mutations of members of the cohesin complex. These mutations were only detected in t(8;21) patients. (B) AML with inv(16) largely lack mutations of chromatin modifiers and cohesion complex members, which are common in t(8;21) AML and AML in general as reported by The Cancer Genome Atlas (TCGA) Research Network.9 *The TCGA data were derived by whole genome/exome sequencing and therefore are capable of detecting mutations of genes in each respective class not evaluated in this study. This figure has been adapted from Figure 1 in the article by Duployez et al that begins on page 2451.
Mutational profile of CBF-AML. (A) Spectrum of recurrent mutations in 215 pediatric and adult patients with CBF-AML. ASXL1, ASXL2, and EZH2 mutations are mutually exclusive of each other as are mutations of members of the cohesin complex. These mutations were only detected in t(8;21) patients. (B) AML with inv(16) largely lack mutations of chromatin modifiers and cohesion complex members, which are common in t(8;21) AML and AML in general as reported by The Cancer Genome Atlas (TCGA) Research Network.9 *The TCGA data were derived by whole genome/exome sequencing and therefore are capable of detecting mutations of genes in each respective class not evaluated in this study. This figure has been adapted from Figure 1 in the article by Duployez et al that begins on page 2451.
AMLs with t(8;21) and inv(16), corresponding to the RUNX1-RUNX1T1 and CBFB-MYH11 fusion genes, respectively, are collectively referred to as CBF-AML because each disrupts a member of the CBF complex.2 CBF-AML is considered a favorable risk disease; however, although most patients achieve remission, >40% will have a relapse, many of whom will ultimately die of their disease or complications of salvage therapy.3 It is likely that this clinical heterogeneity is largely driven by cooperating genetic or epigenetic events, which are undoubtedly present, as the CBF-AML fusions genes are necessary, yet insufficient, for leukemogenesis. Determining the full spectrum of co-occurring mutations may define subsets of patients at highest risk for relapse and identify therapeutically targetable pathways.
It is well established that CBF-AMLs frequently harbor tyrosine kinase (TK) pathway mutations including KIT, FLT3, and NRAS/KRAS mutations, and recent evaluations by these investigators and others have identified recurrent mutations in ASXL1/2 in patients with t(8;21).4,5 Here the authors expand on these results by probing for mutations in 40 genes commonly mutated in myeloid disease. Using high-throughput sequencing capable of detecting mutations even at very low variant allele frequencies (VAFs), they identified additional genetic aberrations in >90% of the 215 CBF-AML patients examined.
As expected, TK mutations were the most frequent events in the cohort. Interestingly, more than one third of all patients had >1 TK mutation, indicating the importance of these mutations in CBF-AML and the presence of remarkable intratumoral heterogeneity, as the VAFs of the co-occurring TK mutations suggest they arose in distinct clones. Many studies have examined the impact of TK pathway mutations on outcome, most consistently finding a poorer prognosis in patients with KIT mutations. Here, the authors found that the mutational burden impacted the prognostic significance with KIT only being prognostic at a VAF ≥35%. Surprisingly, FLT3–tyrosine kinase domain (TKD) mutations with a VAF down to 10% were associated with a high risk of relapse in this cohort. This finding suggests that in CBF-AML, FLT3-TKD might render cells particularly resistant to chemotherapy, allowing for survival of even a low-frequency subordinate clone after treatment, with subsequent emergence as the predominant clone at relapse. Although interesting, this finding requires validation in larger studies including paired diagnostic and relapse samples.
Although the mutational landscape of t(8;21) and inv(16) AMLs share mutations of TKs, this report demonstrates that these 2 distinct disease entities otherwise strongly diverge. In addition to the ASXL1/2 mutations, the authors identified mutations of other chromatin modifiers including EZH2, KDM6A, BCOR, and BCORL1. Strikingly, mutations of chromatin modifiers occurred in 42% of patients with t(8;21), yet were rare in those with inv(16). Additionally, the authors identified recurrent mutations of members of the cohesin complex in 18% of t(8;21) patients and none of the inv(16) patients, consistent with other small recent studies6,7 (see figure panel A). These findings suggest there may be very specific cooperative relationships between the RUNX1-RUNX1T1 fusion and mutations of chromatin modifiers and cohesin complex members.
Cohesin is a multiprotein ring-like complex made up of RAD21, STAG2, SMC1A, and SMC3 critically involved in cohesion of sister chromatids, DNA damage repair, and transcriptional regulation through recruitment of transcription factors and interaction with CCCTC-binding factor.8 Mutually exclusive mutations of members of this complex are present in ∼13% of adults with AML.9 Relevant here is the recent finding that cohesin mutants increase chromatin accessibility and binding of RUNX1,8 the DNA binding portion of which is retained by the RUNX1-RUNX1T1 fusion protein. Therefore, it is possible that mutations of the cohesin complex could contribute to leukemogenesis by altering the DNA binding of the fusion protein. ASXL1, ASXL2, and EZH2 mutations, which are exclusive to t(8;21), would be expected to result in deregulation of the polycomb repressive complex PRC2, leading to alteration of the repressive histone mark of methylated H3K27, possibly contributing to disease development by impacting accessibility of RUNX1 binding sites to both remaining wild-type RUNX1 and the mutant fusion protein.
The negative correlation between inv(16) and cohesin and chromatin modifier mutations is equally intriguing, begging the question of why such mutations, recurrent in t(8;21) and non–CBF-AML, are essentially absent in inv(16) AML (see figure panel B)? It is possible CBFB-MYH11 functionally overlaps with mutant chromatin modifier/cohesins, or perhaps intact PRC2 and cohesin complex function is required for CBFB-MYH11–induced leukemogenesis. Evaluation of the functional basis for these interactions will be important avenues of future research.
The authors went on to determine that of the t(8;21) patients, those harboring both a TK mutation and either a chromatin modifier or cohesin mutation had the highest risk of relapse, indicating a possible contribution to therapy failure. Interestingly, mutational profiling of paired diagnostic and relapse samples from a small cohort of CBF-AML patients, found a high frequency and remarkable stability of chromatin modifier and cohesin mutations, suggesting importance of these mutations in relapse,7 although additional studies are necessary.
This is an important study, providing many potential new clinical and biological insights into CBF-AML. However, a few limitations must be acknowledged. It is possible that, in addition to the 40 genes probed, other relevant co-occurring genetic mutations, epigenetic aberrations, or copy number variants may contribute to leukemogenesis, therapy resistance, and relapse. Additional analyses are needed to detect such events. Further, the prognostic data presented here must be interpreted conservatively, as relatively small numbers of patients were available for analysis, an inevitable consequence of subdividing a relatively small cohort. Furthermore, missing from this analysis is minimal residual disease (MRD) data, known to be predictive of outcome in CBF-AML.10 Therefore, before we can reach definitive, therapy-altering conclusions about the prognostic relevance of the presence and pattern of mutations in CBF-AML, integrated analyses of clinical, genetic, and MRD data in independent large cohorts are needed.
Conflict-of-interest disclosure: The author declares no competing financial interests.